Abstract
Manually labeled landmark sets are often required as inputs for landmark-based image registration. Identifying an optimal subset of landmarks from a training dataset may be useful in reducing the labor intensive task of manual labeling. In this paper, we present a new problem and a method to solve it: given a set of N landmarks, find the k(< N) best landmarks such that aligning these k landmarks that produce the best overall alignment of all N landmarks. The resulting procedure allows us to select a reduced number of landmarks to be labeled as a part of the registration procedure. We apply this methodology to the problem of registering cerebral cortical surfaces extracted from MRI data. We use manually traced sulcal curves as landmarks in performing inter-subject registration of these surfaces. To minimize the error metric, we analyze the correlation structure of the sulcal errors in the landmark points by modeling them as a multivariate Gaussian process. Selection of the optimal subset of sulcal curves is performed by computing the error variance for the subset of unconstrained landmarks conditioned on the constrained set. We show that the registration error predicted by our method closely matches the actual registration error. The method determines optimal curve subsets of any given size with minimal registration error.
1. Introduction
Registration of images consists of computing a transformation between two acquisitions, or equivalently, determining the point-to-point correspondence between the images [1, 2]. Registration algorithms are usually based either on aligning features extracted from the image or on the optimization of a similarity measure of the image intensities [3]. Landmarks are typically points or curves that are either labeled manually or extracted automatically from the image data [2, 4, 5, 6, 7]. Manual demarcation of landmarks can be a labor-intensive task, and therefore the set of landmarks to be labeled should be optimized. The locations of landmarks in the images imposes a correlation in them which can be used to reduce the size of the landmark set. In this paper, we explore the problem: given a set of N candidate landmark points, what is the best subset of k landmarks, from possible choices of such subsets? We discuss this problem as applied to the problem of sulcal set optimization for registration of cerebral cortices of human brains.
The cerebral cortex, or simply the cortex of the brain plays a key role in memory, attention, perceptual awareness, thought and language. The human cerebral cortex is 2 – 4 mm thick and thus for many applications it can be modeled as a surface rather than a volume. A sulcus is a depression or fissure in the cortex and is closely related to the function of the brain. Cortical surface registration has important applications in cross-sectional and longitudinal neuroanatomical studies, for example mapping and analyzing progression of disorders such as Alzheimer's disease [8] and studying growth patterns in developing human brains [9, 10]. There are two main categories of methods that align the cortex from a subject to an atlas: manual landmark based methods [11] and automatic methods based on alignment of geometric features [12, 13] or surface indices [14]. The main advantage of automatic methods is that there is no manual input required for performing the alignment. However they may be less reliable in the sense that they do not incorporate higher level knowledge of sulcal anatomy. Certain sulcal and gyral patterns are hard to identify automatically based on local geometric features, therefore expert delineation of such landmarks can be useful for accurate registration. The objective of landmark based manual registration methods is to minimize the misalignment error in the sulcal curves. Moreover, they allow the users to mark additional structures in certain areas of interest in order to achieve more control for accurate alignment in those regions. A disadvantage of the manual approaches is that the individual tracers need to be trained, and even then inter-rater variability introduces some uncertainty into the procedure. Additionally, for large scale studies, manual procedures may be infeasible unless we minimize the number of sulcal curves required in the manual tracing protocol. It is this latter issue that we address here.
We begin with a large set of sulcal curves that have been identified by the neuroanatomist on our team as candidate landmarks for cortical registration. Our objective is to select an optimal subset from this set such that, for a given number of curves, the sulcal registration error is minimized when computed over all sulci. One straightforward approach is to actually perform registration of the sulcal curves for a set of training images using all possible subsets and then measure the error in the remaining unconstrained sulcal curves. The difficulty with this approach is that there are a huge number of combinations possible. In our case we have 27 candidate curves. Supposing we want to define a protocol that uses 10 curves, the number of combinations to be tested is ≈ 8.4 million. If the error is to be estimated by performing pairwise registrations of 20 brains, i.e. registrations, then the total number of registrations required is ≈ 1.6 billion. This is a prohibitively large number.
Instead of performing actual brain registrations with multiple subsets of constrained sulci, we perform only pairwise unconstrained registrations using the elastic energy minimization procedure described in [11]. The resulting maps produce reasonable correspondences so that we can model the measured sulcal registration errors using a multivariate Gaussian distribution. Using conditional probabilities, we then analytically predict the registration error that would result if we constrained a subset of the curves to match using hand-labeled sulci. These errors can be rapidly computed using conditional covariances, and as we show below, lead to reasonably accurate estimates of the true errors that result when constraining the curves. For a fixed number of constrained curves, we estimate the errors for all possible subsets of that size and select the one with the smallest predicted error. We investigate the prediction accuracy of our model by doing actual registrations using the optimal sulcal constraint set. No searching strategy is required in our method, because for a particular subset of curves, our method gives an analytical expression for the registration error. This expression can be easily calculated for all possible subsets eliminating the need for a search.
2. Surface Registration Method
A class of surface registration based methods use an intermediate parameter space, either a square [8, 11] or a sphere [15], on which the sulcal landmarks or shape based features are aligned. In this paper, we use the flat-mapping surface registration method described in [11], however the same framework is readily adaptable to other surface registration methods [16, 17]. The details of the flat mapping based method are outlined in [11, 18].
Flat mapping based registration methods, combined with the highly variable cortical anatomy, introduce inherent correlation patterns in the locations of unconstrained sulci. In our maps to the unit square we map the corpus callosum to the edges of the unit square causing uneven deformations in different cortical regions, with the temporal lobe mapping to a much smaller area than the medial cortex for instance. Furthermore, minimizing elastic energy deforms the cortical anatomy in a smooth fashion so that one would expect to see significant correlations between sulcal errors in neighboring sulci. Our method takes advantage of this correlation structure to create a multivariate Gaussian model of sulcal registration errors when no curves are constrained. We then predict these errors when some sulci are constrained using conditional probabilities.
2.1. Sulcal Curves, Flat Mapping and Alignment
We used the BrainSuite software [19] to interactively trace the N = 27 candidate sulci depicted in Fig. 1 on both hemispheres of each of the 12 brains used in this study. The surface based registration method we used here performs simultaneous parameterization and registration of the manually traced sulcal landmarks. In order to generate such a flat map with prealigned sulcal curves, we model the cortical surface as an elastic sheet and solve the associated linear elastic equilibrium equation using the finite element method (FEM). Let ϕ = [ϕ1, ϕ2] be the two coordinates assigned to every point on a given cortical surface such that the coordinates ϕ satisfy the linear elastic equilibrium equation with Dirichlet boundary conditions on the boundary of each cortical hemisphere, represented by the corpus callosum. We constrain the corpus callosum to lie on the boundary of the unit square mapped as a uniform speed curve. The elastic strain energy E(ϕ) is given by:
Figure 1.
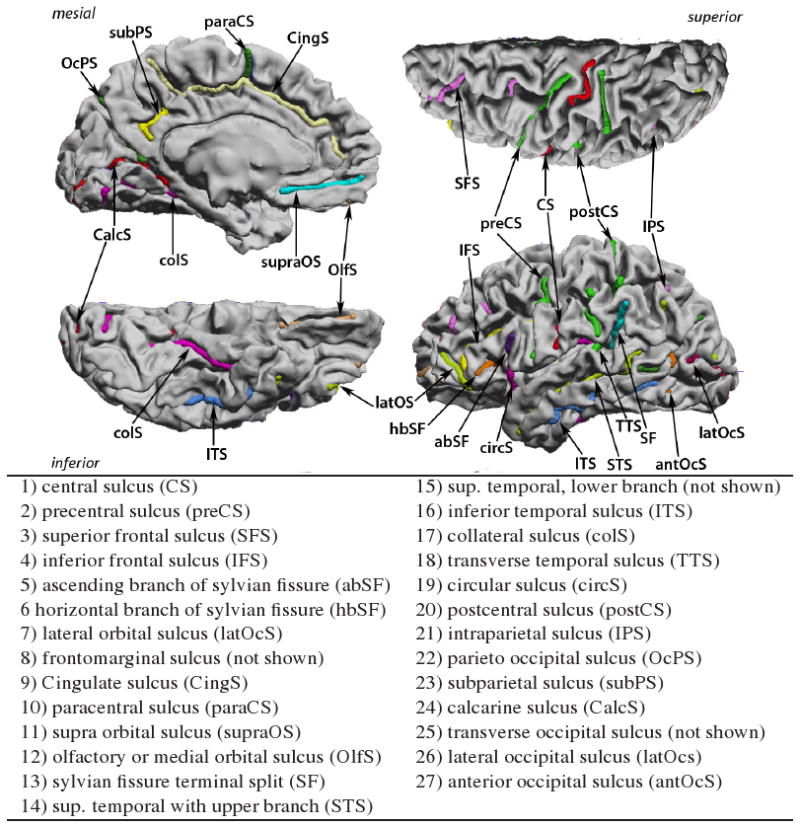
The complete set of candidate sulcal curves from which we select an optimal subset for constrained cortical registration
| (1) |
where Dϕ is the covariant derivative of the coordinate vector field ϕ. Let ϕB1 and ϕB2 denote the 2D coordinates to be assigned to corresponding hemispheres of two brains denoted by B1 and B2 respectively. We then define the Lagrangian cost function C(ϕB1, ϕB2) as
| (2) |
where ϕB1(xk) and ϕB2(yk) denote the coordinates assigned to the sulcal landmarks xk ∈ B1, yk ∈ B2 and σ2 is a Lagrange multiplier. The landmarks correspond to uniformly sampled points along each sulcal curve. We refer the reader to [11, 18] for implementation details. We use σ2 = 0 for unconstrained registration, which is used for estimation of the covariance of the sulcal registration errors, as described later. We then analytically predict the errors when some sulci are constrained, without doing actual constrained registrations, using conditional densities as described below. To test the accuracy of our predictions, we perform registrations with σ2 = 10 and C the set of constrained sulci, a subset of the 27 available curves. This method results in a bijective map of each brain hemisphere to unit squares such that the constrained sulci match in the flat space (Fig. 2a). The point correspondence from one cortical surface hemisphere to the other is then obtained using the coordinate system of the common mapping to the unit square.
Figure 2.
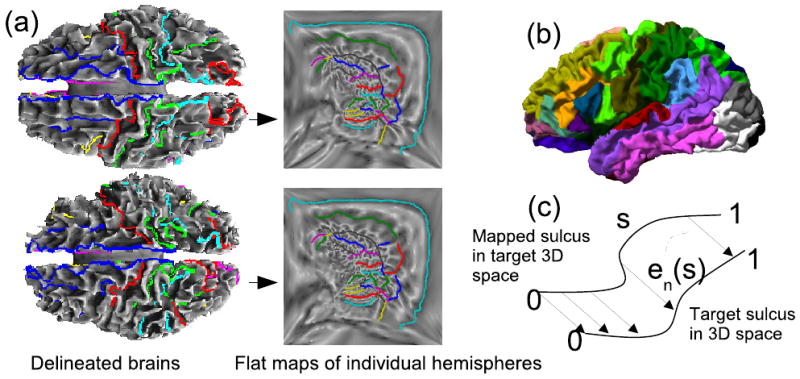
(a) Registration of two cortical surfaces based on the flat mapping method; (b) Parcellation of the cortex into regions surrounding the traced sulci; (c) Registration error for two corresponding sulci.
2.2. Registration Error
For every pair of registered hemispheres, we map the traced curves of one brain to the other, which is arbitrarily defined as a target. The registration used is either unconstrained for error prediction, or constrained for validation. We parameterize each sulcal curve n over s ∈ [0, 1] and then compute the point-to-point average error en = means{en(s)}, as illustrated in Fig. 2, where en(s) is the registration error in 3D coordinates for location s on the nth curve. For symmetry, we repeat the procedure by interchanging subject and target brains.
The alignment error in a sulcus causes a registration error in the surrounding cortical area. Therefore, isolated sulci will have more impact on registration, since their misregistration will affect large cortical regions. To compensate for this effect, we parcellate the cortex into N = 27 regions by assigning each cortical point to the nearest sulcal curve (Fig 2b). The parcellation was performed for all M = 24 available brain hemispheres. We then defined a weight function for the nth sulcus to be , where is the surface area of the nth parcellated region in the ith brain, and Ai is the total surface area of the ith hemisphere.
Finally, the surface registration error metric was defined as
| (3) |
where
,
and
represents the x, y, and z components of en, and 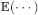 is the expectation operator. Alternatively, the errors could be calculated with respect to surface geometry rather than in 3D. While calculating errors based on geodesic distances is an interesting possibility, they are very difficult to model statistically because geodesics do not form a vector space. When the geodesic errors are small, they could be approximated by errors in 3D, as is done in this paper. Below, we substitute
in order to simplify subsequent analysis. The objective of the surface registration procedure is to minimize this registration error ER.
is the expectation operator. Alternatively, the errors could be calculated with respect to surface geometry rather than in 3D. While calculating errors based on geodesic distances is an interesting possibility, they are very difficult to model statistically because geodesics do not form a vector space. When the geodesic errors are small, they could be approximated by errors in 3D, as is done in this paper. Below, we substitute
in order to simplify subsequent analysis. The objective of the surface registration procedure is to minimize this registration error ER.
3. Probabilistic Model of the Sulcal Errors
We model the sulcal errors as jointly Gaussian random variables, since these errors are drawn from a large population of brain pairs. We describe computations for the x component of the error; similar computations are performed for y and z. The distribution model of for j ∈ {1, …, N} is:
| (4) |
where Σx denotes the covariance matrix of Ex. Therefore, the registration error can be expressed as:
| (5) |
We now want to predict the registration error when some of the sulci are explicitly constrained to register. We partition the curves into two sets: sulci F which are free and sulci N which are constrained so that {1…N} = F ∪ C. We assume that the registration algorithm is well behaved in a sense that it does not create unnatural deformations on the unconstrained sulci when a subset of them are constrained. In other words, if we constrain some sulci to register, the distribution of the remaining ones would be the same as if the constrained ones matched simply by chance, conditioned on the constrained sulci having zero error. Therefore, we model the registration errors in unconstrained sulci as the conditional distribution of the original joint Gaussian density. The probability density of a jointly Gaussian vector, conditioned on some of its elements being zero, is also jointly Gaussian. Therefore, the registration error after matching the sulci from C can be obtained using the conditional expectation:
| (6) |
where is the conditional covariance matrix of the error terms corresponding to free sulci. By rearranging sulci so that free sulci precede the constrained ones, we can partition the covariance matrix as:
| (7) |
where and are the error covariances for free sulci and constrained sulci respectively, and and are the cross-covariances.
The conditional covariance is given by:
| (8) |
which is the Schur complement of in Σx [20]. The estimated registration error after constraining a subset of sulci is then:
| (9) |
This formula allows us to estimate the x component of the registration error for a particular combination of constrained sulci and free sulci. The total registration error is evaluated by adding the x, y, and z components.
| (10) |
We use this formula to estimate the total registration errors for all combinations of sulcal subsets, where Nc is the number of constrained sulci, and choose the subset that minimizes this error.
4. Results
A total of 12 brains, or equivalently 24 hemispheres, were delineated by trained raters. Our tracings, consisting of 27 candidate sulci per hemisphere (Fig. 1), were verified and corrected whenever necessary by one of the authors, an expert neuroanatomist. We performed unconstrained mappings for all 24 hemispheres by directly minimizing Eq. 1 for each hemisphere separately, instead of doing pairwise registrations using Eq. 2 with σ = 0, since the optimization in 2 becomes separable in the unconstrained case σ = 0. Using the flat maps of the 24 hemispheres we computed pairwise registration errors , , and for all possible combinations of left and right hemispheres as described in Sec. 2.2. The resulting sample covariances are shown in Fig. 3.
Figure 3.
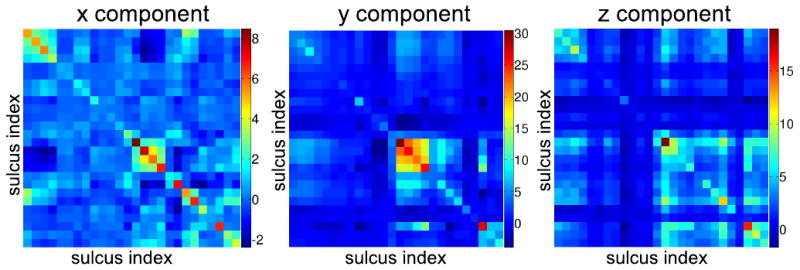
Sample covariance matrices for the x, y, and z components of the error represented as color coded images.
By applying Eq. 10 to all subsets of a given number of constrained curves, we identified the subset that minimizes the registration error. The optimal subsets of curves are given in Fig. 3 for numbers of constrained sulci from 1 to 27. We also calculated the sulcal registration errors for each of these optimal subsets by doing actual registrations. Comparing estimated and actual registration errors, also in Fig. 3, we see that the predicted values are close to those obtained when actually constraining the curves.
We performed a Lilliefors test which rejected the null hypothesis of normality for the errors , , and for many sulci. This was not surprising because the finite size of brains, and therefore the registration error, is bounded. Consequently, a small deviation from normality was expected. However, the distributions were unimodal and the predicted errors of our model are in accordance with the actual ones, which indicates that our assumptions are in the sense of achieving the desired minimum error behavior.
To further test our method, we manually selected the set of 6 sulci with indices (3,12,24,1,9,21) to be constrained, which seemed a reasonable a priori selection based on sulcal extent and spatial distribution around the cortex. The algorithm predicted an error of 67.72mm2 and the actual error was 64.19mm2. The optimal set (3,14,16,17,21,24) found by our method had predicted an error of 36.77mm2 and the actual error was 34.46mm2, which is significantly better than our manually selected subset. We anticipate that in general the curves selected by our method should be superior to those selected on an intuitive basis, since various confounding effects due to elastic flat mapping as well as correlations in errors are accounted for in the algorithm.
5. Discussion
We have described a general procedure for selecting subsets of sulcal landmarks for use in constrained cortical registration. The procedure can be used to reduce the time required for manual labeling of sulci in group studies of cortical anatomy and function.
Notice that the central sulcus is not selected for protocols with a small number of curves (less than 6). This is probably because the sulci that are most stable and consistent among brains, such as the central sulcus, may tend to align well even without explicitly aligning them. Therefore they may not improve the registration error significantly to justify their inclusion in the tracing protocol. Furthermore, short sulci neighboring other candidate curves are not preferred by the algorithm. For example, the paracentral sulcus, which is close to the cingulate sulcus, and the subparietal sulcus, which is close to the cingulate and the parieto-occipital sulcus are not preferred by the method. The algorithm tends to prefer long and variable sulci which are spatially distributed over different regions of the brain. This is consistent with intuition since the curves selected this way will help in overall brain registration.
Finally, we note that the methodology presented here readily extends to other landmark based registration methods in which the goal is to select an optimal subset of given size. This can greatly reduce the manual landmark labeling time for large scale studies. It can possibly be applied to other areas of computer vision [21, 22, 23, 24] for aiding optimal landmark selection.
Figure 4.
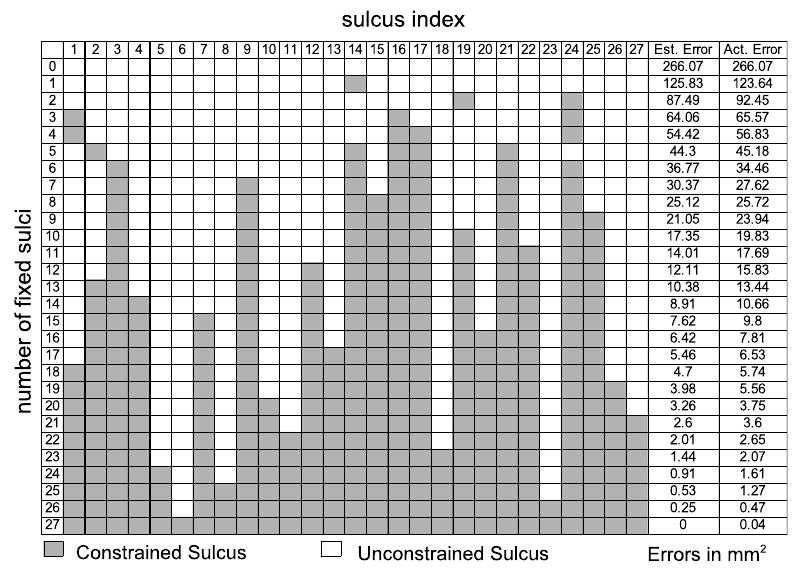
Each row gives the indices of the optimal subset of sulci that minimize the registration error against all other combinations with equal number of constrained curves. The two right columns show that the estimated (est.) error is close to the calculated (act.) error when actual registrations with the same constrained curves are performed.
Contributor Information
Anand Joshi, Email: ajoshi@sipi.usc.edu, Signal and Image Processing Institute, University of Southern California, Los Angeles 90089, USA.
Dimitrios Pantazis, Email: pantazis@sipi.usc.edu, Signal and Image Processing Institute, University of Southern California, Los Angeles 90089, USA.
Hanna Damasio, Email: hdamasio@college.usc.edu, Dornsife Neuroscience Imaging Center, University of Southern California, Los Angeles, CA 90089, USA.
David Shattuck, Email: shattuck@loni.ucla.edu, Laboratory of Neuro Imaging, University of California, Los Angeles, Los Angeles 90095, USA.
Quanzheng Li, Email: quanzhel@sipi.usc.edu, Signal and Image Processing Institute, University of Southern California, Los Angeles 90089, USA.
Richard Leahy, Email: leahy@sipi.usc.edu, Signal and Image Processing Institute, University of Southern California, Los Angeles 90089, USA.
References
- 1.Zitova B, Flusser J. Image registration methods: a survey. Image and Vision Computing. 2003 October;21(11):977–1000. [Google Scholar]
- 2.Brown LG. A survey of image registration techniques. ACM Computing Surveys. 1992;24(4):325–376. [Google Scholar]
- 3.Maintz AJB, Viergever MA. A survey of medical image registration. Medical Image Analysis. 1998 March;2(1):1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- 4.Rohr K, Sprengel R, Stiehl H, et al. Incorporation of landmark error ellipsoids for image registration based on approximating thin-plate splines. Proc Computer Assisted Radiology and Surgery (CAR'97); Berlin, Germany. June 1997; pp. 25–28. [Google Scholar]
- 5.Chui H, Rambo J, Duncan J, Schultz R, Rangarajan A. Registration of cortical anatomical structures via robust 3D point matching. Lecture Notes in Computer Science. 1999:168–181. [Google Scholar]
- 6.Besl P, McKay H. A method for registration of 3D shapes. IEEE Transactions on pattern analysis and machine intelligence. 1992;14(2):239–256. [Google Scholar]
- 7.Chui H, Rangarajan A. A new point matching algorithm for non-rigid registration. Computer Vision and Image Understanding. 2003;89(2-3):114–141. [Google Scholar]
- 8.Thompson PM, Mega MS, Vidal C, Rapoport J, Toga AW. Detecting disease-specific patterns of brain structure using cortical pattern matching and a population-based probabilistic brain atlas. IPMI2001 LNCS. 2001:488–501. doi: 10.1007/3-540-45729-1_52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gogtay N, et al. Dynamic mapping of human cortical development during childhood through early adulthood. PNAS. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffery KJ. Learning of landmark stability and instability by hippocampal place cells. Neuropharmacology. 1998;37(4-5):677–687. doi: 10.1016/s0028-3908(98)00053-7. [DOI] [PubMed] [Google Scholar]
- 11.Joshi AA, Shattuck DW, Thompson PM, Leahy RM. Surface-constrained volumetric brain registration using harmonic mappings. IEEE Trans Med Imaging. 2007;26(12):1657–1669. doi: 10.1109/tmi.2007.901432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Chiang MC, Thompson PM. Automated surface matching using mutual information applied to Riemann surface structures. MICCAI 2005, LNCS. 2005;3750:666–674. doi: 10.1007/11566489_82. [DOI] [PubMed] [Google Scholar]
- 13.Fischl B, Sereno MI, Tootell RBH, Dale AM. High-resolution inter-subject averaging and a coordinate system for the cortical surface. Human Brain Mapping. 1998;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosun D, Rettmann ME, Prince JL. Mapping techniques for aligning sulci across multiple brains. Medical Image Analysis. 2005;8(3):295–309. doi: 10.1016/j.media.2004.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurdal MK, Stephenson K, Bowers PL, Sumners DWL, Rottenberg A. Coordinate system for conformal cerebellar flat maps. NeuroImage. 2000;11:S467. [Google Scholar]
- 16.Thompson PM, Mega MS, Toga AW. Disease-specific probabilistic brain atlases. Procedings of IEEE International Conference on Computer Vision and Pattern Recognition. 2000:227–234. doi: 10.1109/MMBIA.2000.852382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ju L, Hurdal MK, Stern J, Rehm K, Schaper K, Rottenberg D. Quantitative evaluation of three cortical surface flattening methods. NeuroImage. 2005;28(4):869–880. doi: 10.1016/j.neuroimage.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 18.Joshi AA, Shattuck DW, Thompson PM, Leahy RM. A finite element method for elastic parameterization and alignment of cortical surfaces using sulcal constraints. Proc of ISBI. 2007 [Google Scholar]
- 19.Shattuck DW, Leahy RM. Brainsuite: An automated cortical surface identification tool. Medical Image Analysis. 2002;8(2):129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 20.Mardia K, Kent J, Bibby J. Multivariate Analysis. Academic Press; 1979. [Google Scholar]
- 21.Madsen CB, Andersen CS. Journal of Robotics and Autonomous Systems. 1998;23:277–292. [Google Scholar]
- 22.Olson C. Landmark selection for terrain matching. Proceedings of the IEEE International Conference on Robotics and Automation. 2000:1447–1452. [Google Scholar]
- 23.Greiner R, Isukapalli R. Learning to select useful landmarks. National Conference on Artificial Intelligence. 1994:1251–1256. [Google Scholar]
- 24.End G, Treuer H, Boesecke R. Optimization and evaluation of landmark-based image correlation. Phys Med Biol. 1992;37:261–271. doi: 10.1088/0031-9155/37/1/019. [DOI] [PubMed] [Google Scholar]


