Abstract
In this paper we present a new approach for the non-rigid registration of multi-modality images. Our approach is based on an information theoretic measure called the cumulative residual entropy (CRE), which is a measure of entropy defined using cumulative distributions. Cross-CRE between two images to be registered is defined and maximized over the space of smooth and unknown non-rigid transformations. For efficient and robust computation of the non-rigid deformations, a tri-cubic B-spline based representation of the deformation function is used. The key strengths of combining CCRE with the tri-cubic B-spline representation in addressing the non-rigid registration problem are that, not only do we achieve the robustness due to the nature of the CCRE measure, we also achieve computational efficiency in estimating the non-rigid registration. The salient features of our algorithm are: (i) it accommodates images to be registered of varying contrast+brightness, (ii) faster convergence speed compared to other information theory-based measures used for non-rigid registration in literature, (iii) analytic computation of the gradient of CCRE with respect to the non-rigid registration parameters to achieve efficient and accurate registration, (iv) it is well suited for situations where the source and the target images have field of views with large non-overlapping regions. We demonstrate these strengths via experiments on synthesized and real image data.
Keywords: information theory, Shannon entropy, multi-modal non-rigid registration, B-splines
1. Introduction
Image registration is a ubiquitous problem in medical imaging and many other applications of image analysis including but not limited to geo-spatial imaging, satellite imaging, movie editing, archeology etc. In medical imaging, non-rigid registration is particularly common in longitudinal studies such as in child development, ageing studies and also in comparisons between controls and pathologies to assess progress or remission of disease. There is an abundance of non-rigid registration algorithms in literature, the most popular approaches come in two varieties, those that assume brightness constancy in their cost function being optimized and others that use information theory based cost functions that don’t require the aforementioned restrictive assumption. The former are applicable only to same modality data sets while the latter can be applied to multi-modal data sets. There are many applications wherein use of multi-modality data sets is desired e.g., image-guided neurosurgery where an MR is used to locate the tumor and a registered high resolution CT is used for guidance. Another application is in cognitive studies where, MRI and fMRI registration is sought.
In this paper, we develop a multi-modal non-rigid registration technique which is based on a recently introduced information theoretic matching criterion (Wang et al., 2003) called cross cumulative residual entropy (CCRE) to measure the similarity between two images. In Wang et al. (2003), Wang et al. presented a new entropy measure defined on cumulative distributions rather than probability densities which is the usual norm. The mathematical properties of this new measure were developed in a follow up article in Rao et al. (2004). This new measure dubbed cumulative residual entropy (CRE) unlike the well known Shannon entropy was shown to be consistently valid across discrete and continuous domains i.e., the discrete version converges to the continuous one in the limit. Since its definition is based on cumulative distribution functions (CDFs) rather than probability densities and the former are more regular, this measure is more robust in the presence of noise. This property was borne out in the results depicted in Wang et al. (2003) for rigid and affine registration under a variety of noise levels.
In this paper, we use CCRE for achieving non-rigid registration between uni-modal and multi-modal data sets. We derive the analytic gradient of this match measure in order to achieve efficient and accurate non-rigid registration. The CCRE is then minimized over a class of smooth non-rigid transformations expressed in a B-spline basis. The key strengths of our proposed nonrigid registration scheme are: (1) it can accommodate images to be registered of varying contrast+brightness, and it is also robust in the presence of noise; (2) It can be empirically shown to converge faster in comparison to other registration methods that use information theory based cost functions; (3) The cost function and its derivative share common terms and this leads to computational savings being accrued in the numerical optimization process; (4) It is well suited for situations where the source and the target images have field of views with large non-overlapping regions (which is quite common in practice).
The rest of this paper is organized as follows. The remainder of Section 1 contains a brief review of the literature, focusing on the non-rigid registration methods. Section 2 contains a description of our model and all the associated details. Experimental results on synthetic and real image data sets are presented in Section 3. Finally, we draw conclusions in Section 4.
1.1. Previous Work
Non-rigid image registration methods in literature to date may be classified into feature-based and “direct” methods. Most feature-based methods are limited to determining the registration at the feature locations and require an interpolation at other locations. If however, the transformation/registration between the images is a global transformation e.g., rigid, affine etc. then, there is no need for an interpolation step. In the non-rigid case however, interpolation is required. Also, the accuracy of the registration is dependent on the accuracy of the feature detector.
Several feature-based methods involve detecting surfaces landmarks (Chui et al., 2003; Paragios et al., 2003; Audette et al., 2003; Leow et al., 2004), edges, ridges etc. Most of these assume a known correspondence with the exception of the work in Chui et al. (2003), Jian and Vemuri (2005), Wang (2006) and Guo and Rangarajan (2004). Work reported in Irani and Anandan (1998), it uses the energy (squared magnitude) in the directional derivative image as a representation scheme for matching achieved using the SSD cost function. Recently, Liu et al. (2002) reported the use of local frequency in a robust statistical framework using the integral squared error a.k.a., L2E. The primary advantage of L2E over other robust estimators in literature is that there are no tuning parameters in it. The idea of using local phase was also exploited by Mellor and Brady (2004), who used mutual information (MI) to match local-phase representation of images and estimated the non-rigid registration between them. However, robustness to significant non-overlap in the field of view (FOV) of the scanners was not addressed. For more on feature-based methods, we refer the reader to the recent survey by Zitová and Flusser (2003).
In the context of “direct” methods, the primary matching techniques for intra-modality registration involve the use of normalized cross-correlation, modified SSD, and (normalized) mutual information (MI). Ruiz-Alzola et al. (2000) presented a unified framework for non-rigid registration of scalar, vector and tensor data based on template matching. For scalar images, the cost function is the extension of modified SSD using a different definition of inner products. However this model can only be used on images from the same modality as it assumes similar intensity values between images. In Marroquin et al. (2002), Vemuri et al. (2000), a level-set based image registration algorithm was introduced that was designed to non-rigidly register two 3D volumes from the same modality of imaging. This algorithm was computationally efficient and was used to achieve atlas-based segmentation. Direct methods based on the optical-flow estimation form a large class for solving the non-rigid registration problem. Hellier et al. (2001) proposed a registration method based on a dense robust 3-D estimation of the optical flow with a piecewise parametric description of the deformation field. Their algorithm is unsuitable for multi-modal image registration due to the brightness constancy assumption. Variants of optical flow-based registration that accommodate for varying illumination maybe used for inter-modality registration and we refer the reader to Szeliski and Coughlan (1997) and Lai and Fang (1999) for such methods. Guimond et al. (2001) reported a multi-modal brain warping technique that uses Thirion’s Demons algorithm (Thirion, 1998) with an adaptive intensity correction. The technique however was not tested for robustness with respect to significant non-overlap in the FOVs. More recently, Cuzol et al. (2005) introduced a new non-rigid image registration technique which basically involves a Helmholtz decomposition of the flow field which is then embedded into the brightness constancy model of optical flow. The Helmholtz decomposition allows one to compute large displacements when the data contains such displacements. This technique is an innovation on accommodating for large displacements and not one that allows for inter-modality non-rigid registration. For more on intra-modality methods, we refer the reader to the comprehensive surveys (Toga and Thompson, 2001; Zitová and Flusser, 2003).
A popular framework for “direct” methods is based on the information theoretic measures (D’Agostino et al., 2004), among them, mutual information (MI) pioneered by Viola and Wells (1995) and Collignon et al. (1995) and modified in Studholme et al. (1996) has been effective in the application of image registration. Reported registration experiments in these works are quite impressive for the case of rigid motion. The problem of being able to handle non-rigid deformations in the MI framework is a very active area of research and some recent papers reporting results on this problem are Mellor and Brady (2004), Mattes et al. (2003), Rueckert et al. (2003), Hermosillo et al. (2002), Rueckert et al. (1999), Leventon and Grimson (1998), Gaens et al. (1998), Loeckx et al. (2004), Rohde et al. (2003), and Duay et al. (2004). In Mattes et al. (2003), Mattes et al. and in Rueckert et al. (2003), Rueckert et al. presented mutual information based schemes for matching multi-modal image pairs using B-Splines to represent the deformation field on a regular grid. Guetter et al. (2005) recently incorporated a learned joint intensity distribution into the mutual information formulation, in which the registration is achieved by simultaneously minimizing the KL divergence between the observed and learned intensity distributions and maximizing the mutual information between the reference and alignment images. Recently, D’Agostino et al. (2006), D’Agostino et al. presented an information theoretic approach wherein tissue class probabilities of each image being registered are used to match over the space of transformations using a divergence measure between the ideal case (where tissue class labels between images at corresponding voxels are similar) and actual joint class distributions of both images. This work expects a segmentation of either one of the images being registered. Computational efficiency and accuracy (in the event of significant non-overlaps) are issues of concern in most if not all the MI-based non-rigid registration methods.
Some registration methods under the direct approach are inspired by models from mechanics, either from elasticity (Davatzikos, 1997; Gee et al., 1993), or fluid mechanics (Bro-Nielsen and Gramkow, 1996; Christensen et al., 1996). Fluid mechanics-based models accommodate for large deformations, but are largely computationally expensive. Christensen (Geng et al., 2005) recently developed an interesting version of these methods, where the direct deformation field and the inverse deformation field are jointly estimated to guarantee the symmetry of the deformation with respect to permutation of input images. A more general and mathematically rigorous treatment of the non-rigid registration which subsumes the fluid-flow methods was presented in Trouve (1998). All these methods however are primarily applicable to intra-modality and not inter-modality registration.
In order to overcome the problems encountered in both feature-based and intensity-based methods, a “hybrid” approach was developed by Hellier and Barillot (2003), wherein they combined feature-based and intensity-based methods to register images in the context of inter-subject brain registration. An optical flow based intensity energy equation was incorporated with a local sparse constraint, which is, landmark-based correspondences located on the brain’s cortical sulci. The main limitation of this method is that there are several tuning parameters in the energy function, and the optical flow based energy function limits this algorithm to be applicable only to intra-modality registration tasks. Recently, Azar et al. (2006) presented an interactive hybrid nonrigid registration framework in which the intensity-based deformation field and feature-based deformation field are updated iteratively until convergence. The resulting transformation combines both intensity-based and feature-based deformation fields. This method also has many tuning parameters that need to be appropriately set for successful operation, which makes it rather unattractive for practical use.
2. The Registration Technique: Theory & Algorithm
In this section, we present the theoretical aspects of our registration model and the motivation for the use of a new information theoretic measure to drive the registration. We begin by introducing the energy function for the non-rigid registration and this is followed by a description of the non-rigid transformation model. We then present the derivative of the analytic gradient of the energy function with respect to the non-rigid transformation parameters. Finally we summarize our nonrigid registration algorithm at the end.
2.1. The Registration Model
An automatic registration method requires the choice of an image discrepancy criterion that measures the similarity of the test image to the reference image. The measure we choose is defined based on a new information theoretic measure called Cumulative Residual Entropy (CRE) which was introduced in Rao et al. (2004) and is reproduced here for convenience. Let x be a random variable in ℝ, and F(λ): = P(|x| > λ) is the cumulative residual distribution, which is also called survival function in the Reliability Engineering literature. The cumulative residual entropy (CRE) of x, is defined as:
| (1) |
Where ℝ+ = (x ∈ ℝ; x ≥ 0). The key idea in the definition is to use the cumulative distribution in place of the density function in Shannon’s definition of entropy. The distribution function is more regular because it is defined in an integral form unlike the density function, which is defined as the derivative of the distribution. The definition also preserves the well established principle that the logarithm of the probability of an event should represent the information content in the event. CRE can be related to the well-known concept of mean residual life function in Reliability Engineering which is defined as:
| (2) |
The mF(t) is of fundamental importance in Reliability Engineering and is often used to measure departure from exponentiation. CRE can be shown to be the expectation of mF(t) (Asadi and Zohrevand, 2006), i.e.
| (3) |
Based on CRE, cross-CRE (CCRE) between two random variables was defined, and applied to solve the image alignment problem, which is defined as: Given a pair of images I1(x) and I2(x′), where (x′)t = T(x)t and T is the matrix corresponding to the unknown parameterized transformation to be determined, define a match metric M(I1(x), I2(x′)) and maximize/minimize M over all T. The class of transformations can be rigid, affine, projective or non-rigid transformations. Several matching criteria have been proposed in the past, some of which were reviewed earlier. Amongst them, mutual information is very popular and is defined as follows for the continuous random variable case,
| (4) |
where h(X) is the differential entropy of the random variable X and is given by , where p(x) is the probability density function and can be estimated from the image data using any of the parametric and nonparametric methods. The reason for defining MI in terms of differential entropy as opposed to Shannon entropy is to facilitate the optimization of MI with respect to the registration parameters using any of the gradient based optimization methods. Note that MI defined using the Shannon’s entropy in discrete form will not converge to continuous case defined here due to the fact that Shannon’s entropy does not converge to the differential entropy (see Thomas and Cover, 1991).
We now define the cross-CRE (CCRE) using CRE defined in Eq. (1).
| (5) |
We will use this quantity as a matching criterion in the image alignment problem. More specifically, let IT(x) be a test image we want to register to a reference image IR(x). The transformation g(x; μ) describes the deformation from VT to VR, where VT and VR are continuous domains on which IT and IR are defined, μ is the set of the transformation parameters to be determined. We pose the task of image registration as an optimization problem. To align the reference image IR(x) with the transformed test image IT(g(x; μ)), we seek the set of the transformation parameters μ that maximizes 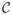 (IT, IR) over the space of smooth transformations i.e.,
(IT, IR) over the space of smooth transformations i.e.,
| (6) |
The computation of CCRE requires estimates of the marginal and joint probability distributions of the intensity values of the reference and test images. We denote p(l, k; μ) as the joint probability of (IT ◦ g(x; μ), IR). Let pT(l; μ) and pR(k) represent the marginal probability for the test image and reference images respectively, LT and LR are the discrete sets of intensities associated with the test image and reference image respectively. Then, we can rewrite the CCRE(IT ◦ g(x; μ), IR) as follows:
| (7) |
Let and . Using the fact that pT(l; μ) = Σk∈LR p(l, k; μ), we have P(i > λ μ) = Σk∈LRP(i > λ, k; μ). Equation (7) can be further simplified, which leads to,
| (8) |
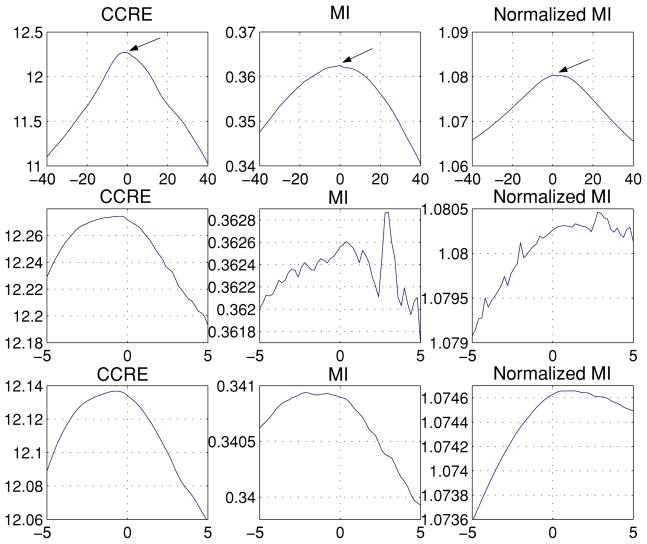
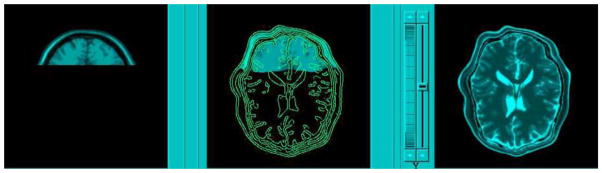
To illustrate the difference between CCRE and the now popular information theoretic cost functions such as MI and NMI, we choose to plot these functions against a parameter of the transformation, for illustrative purposes, say the rotations. The image pair we used here consists of MR and CT images that were originally aligned, and the MR and CT data intensities range from 0–255 with the mean 55.6 and 60.6 respectively. The cost functions are computed over the rotation angle that was applied to the CT image to misalign it with respect to the MR image. In each plot of the Fig. 1 the X-axis shows the 3D rotation angle about an arbitrarily chosen axis of rotation in 3D, while the Y-axis shows the values of CCRE, MI and NMI computed from the misaligned (by a rotation) image pairs. The second row shows a zoom-in view of the plots over a smaller region, so as to get a detailed view of the cost function. The following observations are made from this plot:
Figure 1.
CCRE, MI and NMI functions plotted for the misaligned MR and CT image pair where misalignment is generated by a 3D rotation of the CT image about an arbitrary axis in 3D. First row: over the range −40° to 40°. Second row: zoom in view (between −0.5° to 0.5°) of the region pointed to by the arrows in the first row; tri-linear interpolation was used in all the three cases in this row. Third row: All three cost functions implemented with partial volume interpolation (Collignon et al., 1995).
Similar to MI and NMI, the maximum of CCRE occurs at 0° of rotation, which confirms that our new information measure needs to be maximized in order to find optimum transformation between two misaligned images.
The CCRE shows much larger range of values than MI and NMI. This feature plays an important role in the numerical optimization since it leads to a more stable numerical implementation by avoiding cancelation, round off etc. that often plague arithmetic operations with smaller numerical values.
Upon closer inspection, we observe that CCRE is much smoother than MI and NMI in the registration of MR and CT data pair, this empirically validates that CCRE is more regular than MI and NMI. Theoretically, this justification stems from the fact that CCRE is based on CDFs which are more regular than density functions which are at the heart of the definitions of MI and NMI respectively.
2.2. Transformation Model for Non-Rigid Motion
We model the non-rigid deformation field between two 3D image pairs using a cubic B-splines basis in 3D. B-splines have a number of desirable properties for use in modeling the deformation field. (1) Splines provide inherent control of smoothness (degree of continuity). (2) B-splines are separable in multiple dimensions which provides computational efficiency. Another feature of B-splines that is useful in a non-rigid registration system is the “local control”. Changing the location of a single control point modifies only a local neighborhood of the control point.
The basic idea of the cubic B-spline deformation is to deform an object by manipulating an underlying mesh of control points γi. The deformation g is defined by a sparse regular control point grid. In 3D case, the deformation at any point x = [x, y, z]T in the test image can be interpolated with a linear combination of cubic B-spline convolution kernel.
| (9) |
where β(3)(x) = β(3)(x)β(3)(y)β(3)(z) and Δρ is spacing of the control grid. δj is the expansion B-spline coefficients computed from the sample values of the image. For the implementation details, we refer the reader to Forsey and Bartels (1988) and Mattes et al. (2003).
2.3. Optimization of CCRE
Calculation of the gradient of the energy function is necessary for its efficient and robust maximization. The gradient of CCRE is given as,
| (10) |
Each component of the gradient can be found by differentiating Eq. (7) with respect to a transformation parameters. We consider the two terms in Eq. (7) separately when computing the derivative. For the first term in Eq. (7), we have,
| (11) |
where , and
| (12) |
The derivative of the second term is given by,
| (13) |
where , and
| (14) |
Combining the derivatives of the two terms together, and using the fact that
| (15) |
we have the analytic gradient of CCRE,
| (16) |
note that in the derivation, we use the fact that P(i > λ; μ) = Σk∈LRP(i > λ, k; μ).
Comparing the expressions for CCRE and derivative of CCRE
| (17) |
we note that the two formulas in (17) are similar to each other and they share the common term . From a computational viewpoint, this is quite beneficial since the common term can not only save memory space, but also make the calculation of gradient more efficient. From the formulation, we can also see that calculation of CCRE and derivative of CCRE require us to find a method to estimate P(i > λ, k; μ) and . We will address the computation of these terms in the next subsection.
2.4. Computation of P(i > λ, k; μ) and
We will use the Parzen window technique to estimate the cumulative distribution function and its derivative. The calculation of P(i > λ, k; μ) requires estimate of the cumulative probability distributions of the intensity values of the reference and test images. Let β(0) be a zero-order spline Parzen window (centered unit pulse) and β(3) be a cubic spline Parzen window, the smoothed joint probability of (IR, IT ◦ g) is given by
| (18) |
where α is a normalization factor that ensures Σp(l, k) = 1, and IR(x) and IT(g(x; μ) are samples of the reference and interpolated test images respectively, which is normalized by the minimum intensity value, , and the intensity range of each bin, ΔbR, ΔbT.
Since , we have the following,
| (19) |
where Φ() is the cumulative residual function of cubic spline kernel defined as follows,
| (20) |
Note that , we can then take the derivative of Eq. (19) with respect to μ, and we get
| (21) |
where is the image gradient.
2.5. Algorithm Summary
The registration algorithm can be summarized as follows,
For the current deformation field, interpolate the test image by IT ◦ g(x; μ). Calculate P(i > λ, k; μ) and using Eqs. (19) and (21) respectively.
Compute P(i > λ; μ) as Σk∈LRP(i > λ, k; μ), which is used to calculate the common term in both CCRE and gradient of CCRE, i.e., .
Compute the energy function and its gradient using the formulas given in Eq. (17), we can then use the Quasi-Newton method to numerically solve the optimization problem.
Update the deformation field g(x; μ). Stop the registration process if the difference in consecutive iterates is less than ε = 0.01, a pre-chosen tolerance, otherwise go to Step 1.
3. Implementation Results
In this section, we present the results of applying our non-rigid registration algorithm to several data sets. The results are presented for synthetic as well as real data. The first set of experiment was done with synthetic non-rigid motion. We show the advantage of using the CCRE measure in comparison to other information theoretic registration methods. We show that our algorithm is not only more robust, but also converges faster than others. We begin by applying our algorithm to register image pairs for which the ground truth was available.
3.1. Synthetic Motion Experiments
In this section, we demonstrate the robustness property of CCRE and will make a case for its use over Mutual Information in the alignment problem. The case will be made via experiments depicting faster convergence speed and superior performance under noisy inputs in matching the image pairs misaligned by a synthesized non-rigid motion. Additionally we will depict a larger capture range over MI-based methods in the estimation of the motion parameters.
The data we use for this experiment are corresponding slices from an MR T1 and T2 image pair, which were obtained from the brainweb site at the Montreal Neurological Institute (http://www.bic.mni.mcgill.ca/brainweb, 1997). They are originally aligned with each other. The two images are defined on a 1mm isotropic voxel grid in the Talairach space, with dimension (256 × 256). We applied a known non-rigid transformation to the T2 image so as to misalign it with respect to the T1 image, and the goal is to recover this deformation by applying our registration method. The mutual information algorithm/implementation which we will compare with here was originally reported in Mattes et al. (2003) and Thévenaz and Unser (2000). This implementation makes explicit use of the gradient of MI with respect to the transformation parameters. The analytic formula for this gradient was presented in Mattes et al. (2003) and Thévenaz and Unser (2000), thus allowing for the efficient application of gradient-based optimization methods.
3.1.1. Convergence Speed
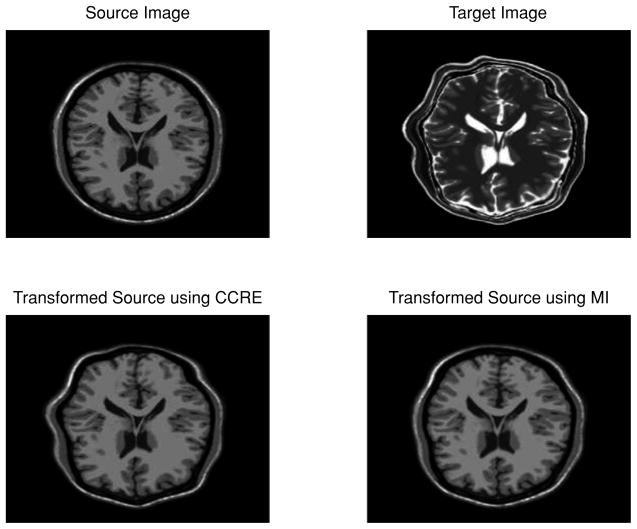
In order to compare the convergence speed of CCRE versus MI, we designed the experiment as follows: with the MR T1 and T2 image pair as our data, we chose the MR T1 image as the source, the target image was obtained by applying a known smooth non-rigid transformation that was procedurally generated. Notice the significant difference between the intensity profiles of the source and target images. For comparison purposes, we used the same gradient descent optimization scheme, and let the two registration methods execute for the same amount of time, and show the registration result visually and quantitatively.
The source and target image pair along with the results of estimated transformation using CCRE and MI applied to the source are shown in Fig. 2. As evident visually, we observe that the result generated by CCRE is more similar in shape to the target image than the one produced by MI.
Figure 2.
Upper left, MR T1 image as the source image; Upper right, deformed MR T2 image as the target image; Lower left and right, results of estimated transformations using CCRE and MI applied to the source respectively. Execution time for both the algorithms was set to 30 CPU seconds –time to convergence of our algorithm. Both algorithms used the same gradient descent technique.
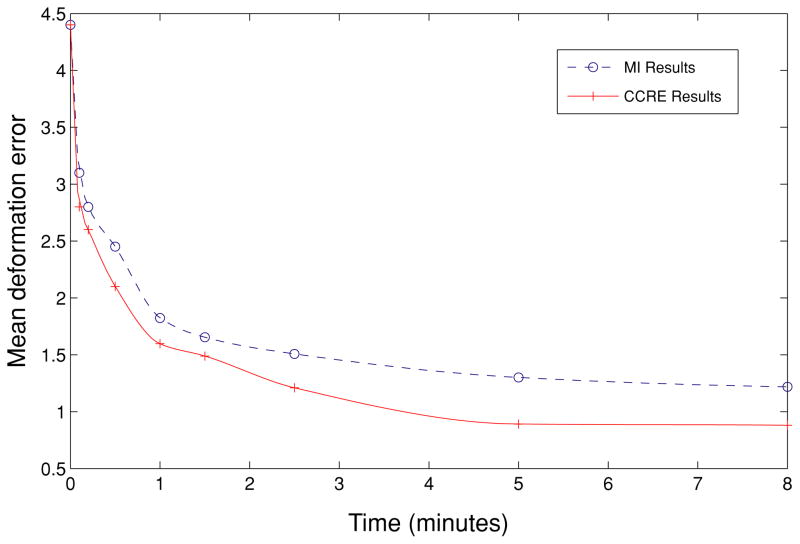
Quantitative assessment of accuracy of the registration is presented in Fig. 3, where we plotted the change of mean deformation error (MDE) obtained for the CCRE and the MI-based algorithms respectively. MDE is defined as , where g0(xi) and g(xi) are the ground truth and estimated displacements respectively at voxel xi. ||·|| denotes the Euclidean norm, and R is the volume of the region of interest. In both cases mean deformation errors are decreasing with time, but the solid curve is decreasing faster than the dotted curve. For example, it takes about 5 minutes for MI to reach an error level below 1.2 units, while CCRE only requires about half that time to achieve the same error level. This empirically validates the faster convergence speed of CCRE based algorithm over the MI-based algorithm.
Figure 3.
Plot demonstrating the Mean Deformation Error for CCRE and MI-based registration as a function of time. Solid curve shows the MDE for CCRE-based registration, while dotted curve illustrates the MDE for the MI-based registration.
3.1.2. Registration Accuracy
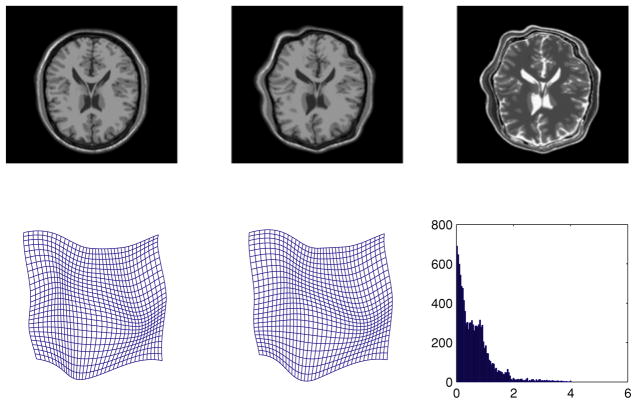
Using the same experimental setting as in the previous experiment, we present the registration error for our algorithm in the estimated non-rigid deformation field as an indicator of the accuracy of estimated deformations. Fig. 4 depicts the results obtained for this image pair. which is organized as follows, from left to right: the first row depicts the source image with the target image segmentation superposed to depict the amount of mis-alignment, the registered source image which is obtained using our algorithm superposed with the target segmentation, followed by the target image; second row depicts ground truth deformation field which we used to generate the target image from the MR T2 image, the estimated non-rigid deformation field followed by histogram of the estimated magnitude error. Note that the error distribution is mostly concentrated in the small error range indicating the accuracy of our method. As a measure of accuracy of our method, we also estimated the average, μ, and the standard deviation, σ, of the error in the estimated non-rigid deformation field. The error was estimated as the angle between the ground truth and estimated displacement vectors. The average and standard deviation are 1.5139 and 4.3211 (in degrees) respectively, which is quite accurate.
Figure 4.
Results from the synthetic data demonstrating the registration accuracy of our algorithm (see text for details).
Table 1 depicts statistics of the error in estimated non-rigid deformation when compared to the ground truth. For the mean ground truth deformation (magnitude of the displacement vector) in Column-1 of each row, 5 distinct deformation fields with this mean are generated and applied to the target image of the given source-target pair to synthesize 5 pairs of distinct data sets. These pairs (one at a time) are input to our algorithm and the mean (μ) of the mean deformation error (MDE) is computed over the five pairs and reported in Column-2 of the table. Column-3 depicts the standard deviation of the MDE for the five pairs of data in each row. As evident, the mean and the standard deviation of the error are reasonably small indicating the accuracy of our non-rigid registration algorithm. Note that this testing was done on a total of 20 image pairs (= 40) as there are 5 pairs of images per row.
Table 1.
Statistics of the error in estimated non-rigid deformation.
| μg | Mean μ of MDE | Standard deviation σ of MDE |
|---|---|---|
| 2.4 | 0.5822 | 0.0464 |
| 3.3 | 0.6344 | 0.0923 |
| 4.5 | 0.7629 | 0.0253 |
| 5.5 | 0.7812 | 0.0714 |
3.1.3. Noise Immunity
In the next experiment, we compare the robustness of the two methods, CCRE and MI, in the presence of noise. Still selecting the MR T1 image slice from the previous experiment as our source image, we generate the target image by applying a fixed smooth synthetic deformation field. We conduct this experiment by varying the amount of Gaussian noise added and then for each instance of the added noise, we register the two images using the two techniques. We expect both schemes are going to fail at some level of noise. (“failed” here means that the optimization algorithm primarily diverged.) By comparing the noise magnitude of the failure point, we can show the degree to which these methods are tolerant. The numerical schemes used in the implementation of these registration algorithms are based on the BFGS quasi-Newton algorithm (Nocedal and Wright, 2000).
The mean magnitude of the synthetic motion is 4.37 pixel, with the standard deviation at 1.8852. Table 2 show the registration results for the two schemes. From the table, we observe that the MI fails when the standard deviation of the noise is increased to 40, while CCRE is tolerant until 66, a significant difference when compared to the MI. This experiment conclusively depicts that CCRE has more noise immunity than MI when dealing with the non-rigid motion.
Table 2.
Comparison of the registration results between CCRE and other MI-based algorithms for a fixed synthetic deformation field.
| σ | CCRE |
MI |
||
|---|---|---|---|---|
| MDE | Standard deviation | MDE | Standard deviation | |
| 10 | 1.0816 | 0.9345 | 1.3884 | 1.4538 |
| 19 | 1.1381 | 1.1702 | 1.4871 | 1.5052 |
| 30 | 1.1975 | 1.3484 | 1.5204 | 1.5615 |
| 40 | 1.3373 | 1.6609 | FAIL | |
| 60 | 1.3791 | 1.9072 | ||
| 66 | FAIL | |||
3.1.4. Partial Overlap
Fig. 5 depicts an example of registration of the MR T1 and T2 data sets with large non-overlap. The left image of the figure depicts the MR T1 brain scan as the source image, and the right image shows the MR T2 data as the target. Note that the field of view (FOV) for the data sets are significantly non-overlapping. The non-overlap was simulated by discarding 66% of the MR T1 image (source image). The middle column depicts the transformed source image along with an edge map of the target (Deformed MR T2 image) superimposed on the transformed source. As is evident, the registration is visually quite accurate.
Figure 5.
Registration results of MR T1 and and T2 image slice with large non-overlap. (left) MR T1 source image before registration; (right) Deformed T2 target image; (middle) the transformed MR image superimposed with edge map from target image.
3.1.5. Convergence Range
To better demonstrate the convergence range of CCRE in comparison with Mutual Information based algorithms, we will apply them to estimate the 3D rigid motion parameters between image pairs that are known be misaligned by a 3D rigid motion. The data we use for this experiment is a pair of 3D MR T1 and T2 images from the brainweb (http://www.bic.mni.mcgill.ca/brainweb), and they are originally aligned with each other. The two volumes are defined on a 1mm isotropic voxel grid in Talairach space, with dimension (181 × 217 × 181). We fix the standard deviation (7) of noise added to the two images and vary the magnitude of the synthesized rigid motion until all of the methods fail. With this experiment, we can compare the convergence range of each registration algorithm. Notice that we used partial volume interpolation for all three methods in this implementation (Collignon et al., 1995). Six parameters are displayed in each cell of Table 3. The first three are rotation angles (in degrees), while the next three values show the translations (in mm). Both the rotation and translation parameters are in (x, y, z) order. From Table 3, we observe that the convergence range of MI and Normalized MI is estimated at (13°, 13°, 12°, 13, 13, 13) and (15°, 15°, 14°, 16, 16, 16) respectively, while our algorithm has a much larger capture range at (32°, 32°, 25°, 32, 32, 32). It is evident from this experiment that the capture range for reaching the optimum is significantly larger for CCRE when compared with MI and NMI in the presence of noise.
Table 3.
Comparison of the convergence range of the rigid registration between CCRE and other MI-based schemes for a fixed noise standard deviation of 7.
| GroundTruth | CCRE | Traditional MI | Normalized MI | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7° | 7° | 7° | 6.970° | 6.912° | 7.014° | 6.973° | 7.003° | 7.057° | 6.955° | 7.186° | 6.954° |
| 5 | 5 | 5 | 4.952 | 5.001 | 5.073 | 5.003 | 4.942 | 5.069 | 5.015 | 4.931 | 5.097 |
| 13° | 13° | 12° | 12.961° | 13.033° | 12.055° | FAILED | 13.025° | 12.980° | 11.990° | ||
| 13 | 13 | 13 | 13.013 | 12.921 | 13.034 | 12.843 | 13.094 | 12.916 | |||
| 15° | 15° | 14° | 15.031° | 15.008° | 14.012° | FAILED | |||||
| 16 | 16 | 16 | 16.015 | 15.911 | 16.027 | ||||||
| 32° | 32° | 25° | 32.072° | 32.054° | 25.022° | ||||||
| 32 | 32 | 32 | 32.047 | 31.852 | 31.767 | ||||||
| 33° | 33° | 33° | FAILED | ||||||||
| 33 | 33 | 33 | |||||||||
3.2. Real Data Experiments
In this section, we present the performance of our method on a series of CT and MR data containing real non-rigid misalignments. For the purpose of comparison, we also apply the MI-based registration algorithm implemented as was presented in Mattes et al. (2003) to these same data sets. The CT image is of size (512, 512, 120) while the MR image size is (512, 512, 142), and the voxel dimensions are (0.46, 0.46, 1.5) mm and (0.68, 0.68, 1.05) for the CT and MR images respectively. The registration was performed on reduced volumes (210 × 210 × 120) with the control knots placed every 16 × 16 × 16 voxels. The algorithm was coded in the C++ and all experiments were performed on a 2.6 GHZ Pentium PC.
We used a set of eight volumes of CT data sets and the task was to register these eight volumes to the MR data chosen as the target image for all registrations, by using both CCRE and the MI algorithms. Note that all the CT and MR volume pairs were acquired from different subjects and thus would involve non-rigid registration in order to align them. The parameters used in both the algorithms were identical. For both algorithms, the iterative optimization of the cost functions was halted when improvements of at least 0.01 in the cost function could not be detected. The time required for registering all data sets for our algorithm as well as MI method are given in Table 4. This table shows that, on the average, our CCRE algorithm is about 2.5 times faster than the MI-based approach for this set of experiments. For brevity, we only show one registration result in Fig. 6. Here, one slice of the volume is shown in the first row with the source CT image on the left and reference image on the right. The middle image shows the transformed CT image slice superimposed with edge map from target image. In the second row, the source image superimposed with edge map from target image is shown on the left, while shown in the middle and right are the heads reconstructed from the transformed source using CCRE method and the target MR image respectively. From this figure, we can see that the source and target image depict considerable non-rigid changes in shape, nevertheless our method was able to register these two images quite accurately. To validate the conformity of the two reconstructed surfaces, we randomly sample 30 points from the surface of the transformed source using CCRE, and then estimate the distances of these points to the surface in the target MR volume. The average of these distances is about 0.47 mm, which indicates a very good agreement between two surfaces. The resemblance of the reconstructed shapes from transformed source with the target indicates that our CCRE algorithm succeeded in matching the source CT volume to the target MR image.
Table 4.
Comparison of total time taken to achieve registration by CCRE and MI algorithms.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| CCRE time (s) | 4827 | 3452 | 4345 | 4038 | 3910 | 4510 | 5470 | 3721 |
| MI time (s) | 9235 | 6344 | 10122 | 17812 | 12157 | 11782 | 13157 | 10057 |
Figure 6.
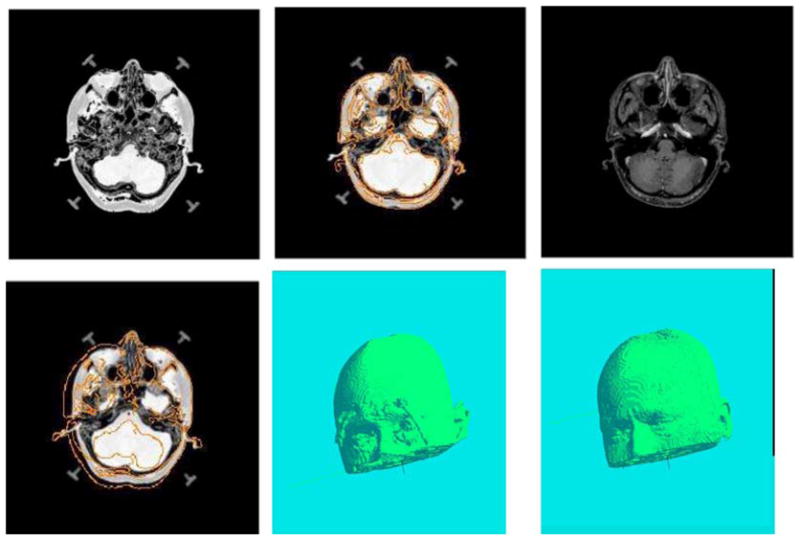
Registration results of different subjects using MR and CT brain data with real non-rigid motion. (see text for details).
The accuracy of the information theoretic based algorithm for non-rigid registration problems was assessed quantitatively by means of an region-based segmentation task (Chan and Vesse, 1999). ROIs (whole brain, eyes) were segmented automatically in these eight CT data sets used as the source image and binary masks were created. The deformation fields between the CT and MR volumes were computed and used to project the masks frsom each of the CT to the MR volume. Contours were manually drawn on a few slices chosen at random in MR volume (four slices/volume). Manual contours on MR and contours obtained automatically were then compared using an accepted similarity index defined as two times the number of pixels in the intersection of the contours divided by the sum of the number of pixels within each contour (Rohde et al., 2003). This index varies between zero (complete disagreement) and one (complete agreement) and is sensitive to both displacement and differences in size and shape. Table 5 lists mean values for the similarity index for each structure. It is customarily accepted that a value of the similarity index above 0.80 indicates a very good agreement between contours. Our results are well above this value. For comparison purposes, we also computed the same index for the MI method. We can conclude from the table that our CCRE can achieve better registration accuracy than the MI for the task of non-rigid registration of real multi-model images.
Table 5.
Comparison of the S-value of several brain structures for CCRE and MI.
| Volume | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|
| CCRE | Whole brain | 0.987 | 0.996 | 0.974 | 0.962 | 0.975 | 0.967 | 0.988 | 0.981 |
| Left eye | 0.925 | 0.935 | 0.925 | 0.907 | 0.875 | 0.890 | 0.834 | 0.871 | |
| Right eye | 0.840 | 0.940 | 0.891 | 0.872 | 0.851 | 0.829 | 0.910 | 0.921 | |
| MI | Whole brain | 0.986 | 0.981 | 0.976 | 0.96 | 0.950 | 0.961 | 0.942 | 0.952 |
| Left eye | 0.911 | 0.893 | 0.904 | 0.791 | 0.853 | 0.810 | 0.851 | 0.853 | |
| Right eye | 0.854 | 0.917 | 0.889 | 0.814 | 0.849 | 0.844 | 0.897 | 0.854 |
4. Conclusion
In this paper, we presented a novel way to non-rigidly register multi-modal data sets based on a recently introduced matching criterion called the cross cumulative residual entropy(CCRE) (Wang et al., 2003) to measure the similarity between two images. The matching criterion is defined based on a new information measure, namely the cumulative residual entropy (CRE), which is defined based on the probability distributions as opposed to probability densities. Since distributions are more regular than densities, there is inherent robustness in the definition of CRE. Furthermore, CCRE also inherits this robustness property from the CRE.
In this work, CCRE between two images to be registered is maximized over the space of smooth and unknown non-rigid transformations. For efficient and robust computation of the non-rigid deformations, a tensor product of tri-cubic B-spline based representation of the deformation function is used. The key strengths of combining CCRE with the tri-cubic B-spline representation in addressing the non-rigid registration problem are that, not only do we achieve the robustness due to the nature of the CCRE measure but we also achieve computational efficiency in estimating the non-rigid registration. The salient features of our algorithm are that: (i) it accommodates images to be registered of varying contrast+brightness, (ii) it has a faster convergence speed compared to other information theory-based matching measures used for non-rigid registration in literature, (iii) the use of analytic gradient of CCRE with respect to the non-rigid registration parameters achieves efficient and accurate registration, (iv) it is well suited for situations where the source and the target images have field of views with large non-overlapping regions.
Finally comparisons were made between CCRE and MI (Mattes et al., 2003; Forsey and Bartels, 1988) and all the experiments depicted significantly better performance of CCRE over the MI-based methods currently used in literature. Our future work will focus on reducing the computational load by using adaptive meshing schemes for computing the B-spline coefficients representing the non-rigid deformations. Validation of non-rigid registration on real data with the aid of segmentations and landmarks obtained manually from a group of trained anatomists is another one of the goals of our future work.
Acknowledgments
This research was supported in part by the NIH grant RO1 NS046812. Authors would like to thank Dr. Frank Bova of the UF Neurosurgery Dept. for the MR-CT data sets.
Contributor Information
FEI WANG, IBM Almaden Research Center, 650 Harry Road, San Jose, CA 95120.
BABA C. VEMURI, Department of CISE, University of Florida, Gainesville, FL 32611
References
- Asadi M, Zohrevand Y. On the dynamic cumulative residual entropy. Unpublished Manuscript 2006 [Google Scholar]
- Audette MA, Siddiqi K, Ferrie FP, Peters TM. An integrated range-sensing, segmentation and registration framework for the characterization of intra-surgical brain deformations in image-guided surgery. Comput Vis Image Underst. 2003;89(2–3):226–251. [Google Scholar]
- Azar A, Xu C, Pennec X, Ayache N. An interactive intensity- and feature-based non-rigid registration framework for 3D medical images. Proceedings of the Third IEEE International Symposium on Biomedical Imaging (ISBI 2006).2006. [Google Scholar]
- Bro-Nielsen M, Gramkow C. Fast fluid registration of medical images. VBC ’96: Proceedings of the 4th International Conference on Visualization in Biomedical Computing; London, UK: Springer-Verlag; 1996. pp. 267–276. [Google Scholar]
- Chan T, Vesse L. An active contour model without edges. Intl. Conf. on Scale-space Theories in Computer Vision; 1999. pp. 266–277. [Google Scholar]
- Christensen GE, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Transactions On Image Processing. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- Chui H, Win L, Schultz R, Duncan J, Rangarajan A. A unified non-rigid feature registration method for brain mapping. Medical Image Analysis. 2003;7(2):112–130. doi: 10.1016/s1361-8415(02)00102-0. [DOI] [PubMed] [Google Scholar]
- Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Automated multimodality image registration based on information theory. In: Bizais Y, Barillot C, Di Paola R, editors. Information Processing in Medical Imaging. 1995. [Google Scholar]
- Cuzol A, Hellier P, Memin E. A novel parametric method for non-rigid image registration. In: Christensen G, Sonka M, editors. ser LNCS, no 3565; Proc. Information Processing in Medical Imaging (IPMI’05); Colorado, USA: Glenwood Springes; 2005. pp. 456–467. [DOI] [PubMed] [Google Scholar]
- D’Agostino E, Maes F, Vandermeulen D, Suetens P. Non-rigid atlas-to-image registration by minimization of class-conditional image entropy. MICCAI. 2004;(1):745–753. doi: 10.1007/978-3-540-73273-0_27. [DOI] [PubMed] [Google Scholar]
- D’Agostino E, Maes F, Vandermeulen D, Suetens P. An information theoretic approach for non-rigid image registration using voxel class probabilities. Medical Image Analysis. 2006;10(3):413–431. doi: 10.1016/j.media.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput Vis Image Underst. 1997;66(2):207–222. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- Duay V, D’Haese P-F, Li R, Dawant BM. Non-rigid registration algorithm with spatially varying stiffness properties. ISBI. 2004:408–411. [Google Scholar]
- Forsey DR, Bartels RH. Hierarchical b-spline refinement. Computer Graphics. 1988;22(4):205–212. [Google Scholar]
- Gaens T, Maes F, Vandermeulen D, Suetens P. Non-rigid multimodal image registration using mutual information. Proc. Conference on Medical Image Computing and Compter—Assisted Intervention (MICCAI); 1998. pp. 1099–1106. [Google Scholar]
- Gee JC, Reivich M, Bajcsy R. Elastically deforming 3d atlas to match anatomical brain images. J Comput Assist Tomogr. 1993;17(2):225–236. doi: 10.1097/00004728-199303000-00011. [DOI] [PubMed] [Google Scholar]
- Geng X, Kumar D, Christensen GE. Information Processing in Medical Imaging. 2005. Transitive inverse-consistent manifold registration; pp. 468–479. [DOI] [PubMed] [Google Scholar]
- Guetter C, Xu C, Sauer F, Hornegger J. Learning based non-rigid multi-modal image registration using kullback-leibler divergence. MICCAI. 2005;(2):255–262. doi: 10.1007/11566489_32. [DOI] [PubMed] [Google Scholar]
- Guimond A, Roche A, Ayache N, Menuier J. Three-dimensional multimodal brain warping using the demons algorithm and adaptive intensity corrections. IEEE Trans on Medical Imaging. 2001;20(1):58–69. doi: 10.1109/42.906425. [DOI] [PubMed] [Google Scholar]
- Guo SJH, Rangarajan A. A new joint clustering and diffeomorphism estimation algorithm for non-rigid shape matching. IEEE Computer Vision and Pattern Recognition. 2004:16–22. [Google Scholar]
- Hellier P, Barillot C, Mmin E, Prez P. Hierarchical estimation of a dense deformation field for 3d robust registration. IEEE Transaction on Medical Imaging. 2001;20(5):388–402. doi: 10.1109/42.925292. [DOI] [PubMed] [Google Scholar]
- Hellier P, Barillot C. Coupling dense and landmark-based approaches for non rigid registration. IEEE Trans Med Imaging. 2003;22(2):217–227. doi: 10.1109/TMI.2002.808365. [DOI] [PubMed] [Google Scholar]
- Hermosillo G, Chefd’hotel C, Faugeras O. Variational methods for multimodal image matching. Int J Comput Vision. 2002;50(3):329–343. [Google Scholar]
- Irani M, Anandan P. International Conference on Computer Vision. Bombay, India: 1998. Robust Multi-sensor Image Alignment; pp. 959–965. [Google Scholar]
- Jian B, Vemuri BC. A robust algorithm for point set registration using mixture of gaussians. International Conference on Computer Vision; 2005. pp. 1246–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SH, Fang M. Robust and efficient image alignment with spatially-varying illumination models. IEEE Conference on Computer Vision and Pattern Recognition; 1999. pp. 167–172. [Google Scholar]
- Leow A, Thompson PM, Protas H, Huang S-C. Brain warping with implicit representations. ISBI. 2004:603–606. [Google Scholar]
- Leventon ME, Grimson WEL. Multimodal volume registration using joint intensity distributions. Proc. Conference on Medical Image Computing and Compter–Assisted Intervention (MICCAI); Cambridge, MA. 1998. pp. 1057–1066. [Google Scholar]
- Liu J, Vemuri BC, Marroquin JL. Local frequency representations for robust multimodal image registration. IEEE Transactions on Medical Imaging. 2002;21(5):462–469. doi: 10.1109/TMI.2002.1009382. [DOI] [PubMed] [Google Scholar]
- Loeckx D, Maes F, Vandermeulen D, Suetens P. Nonrigid image registration using free-form deformations with a local rigidity constraint. MICCAI. 2004;(1):639–646. [Google Scholar]
- Marroquin L, Vemuri B, Botello S, Calderon F, Fernandez-Bouzas A. An accurate and efficient bayesian method for automatic segmentation of brain mri. IEEE Trans Med Imaging. 2002:934–945. doi: 10.1109/TMI.2002.803119. [DOI] [PubMed] [Google Scholar]
- Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. Pet-ct image registration in the chest using free-form deformations. IEEE Trans Med Imaging. 2003;22(1):120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- Mcconnell Brain Imaging Centre Brain Database. 1997 http://www.bic.mni.mcgill.ca/brainweb/
- Mellor M, Brady M. Non-rigid multimodal image registration using local phase. Medical Image Computing and Computer-Assisted Intervention—MICCAI; 2004; Saint-Malo, France. 2004. pp. 789–796. [Google Scholar]
- Nocedal J, Wright SJ. Numerical Optimization 2000 [Google Scholar]
- Paragios N, Rousson M, Ramesh V. Non-rigid registration using distance functions. Comput Vis Image Underst. 2003;89(2–3):142–165. [Google Scholar]
- Rao M, Chen Y, Vemuri BC, Wang F. Cumulative residual entropy, a new measure of information. IEEE Trans on Information Theory. 2004;50(6):1220–1228. [Google Scholar]
- Rohde GK, Aldroubi A, Dawant BM. The adaptive bases algorithm for intensity based nonrigid image registration. IEEE Trans Med Imaging. 2003;22(11):1470–1479. doi: 10.1109/TMI.2003.819299. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: Application to breast mr images. IEEE Trans on Medical Imaging. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Frangi AF, Schnabel JA. Automatic construction of 3d statistical deformation models of the brain using non-rigid registration. IEEE Trans Med Imaging. 2003;22(8):1014–1025. doi: 10.1109/TMI.2003.815865. [DOI] [PubMed] [Google Scholar]
- Ruiz-Alzola J, Westin C-F, Warfield SK, Nabavi A, Kikinis R. In: DiGioia AM, Delp S, editors. Nonrigid registration of 3d scalar vector and tensor medical data; Proceedings of MICCAI 2000, Third International Conference on Medical Image Computing and Computer-Assisted Intervention; Pittsburgh. 2000. pp. 541–550. [Google Scholar]
- Studholme C, Hill D, Hawkes DJ. Automated 3D registration of MR and CT images in the head. Medical Image Analysis. 1996;1(2):163–175. doi: 10.1016/s1361-8415(96)80011-9. [DOI] [PubMed] [Google Scholar]
- Szeliski R, Coughlan J. Spline-based image registration. Int J Comput Vision. 1997;22(3):199–218. [Google Scholar]
- Thévenaz P, Unser M. Optimization of mutual information for multiresolution image registration. IEEE Transactions on Image Processing. 2000;9(12):2083–2099. doi: 10.1109/83.887976. [DOI] [PubMed] [Google Scholar]
- Thirion JP. Image matching as a diffusion process: An analogy with maxwell’s demons. Medical Image Analysis. 1998;2(3):243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- Thomas JAT, Cover M. Elements of Information Theory. John Wiley and Sons; 1991. [Google Scholar]
- Toga AW, Thompson PM. The role of image registration in brain mapping. Image Vision Comput. 2001;19(1–2):3–24. doi: 10.1016/S0262-8856(00)00055-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouve A. Diffeomorphisms groups and pattern matching in image analysis. Int J Comput Vision. 1998;28(3):213–221. [Google Scholar]
- Vemuri BC, Ye J, Chen Y, Leonard CM. A level-set based approach to image registration. IEEE Workshop on Mathematical Methods in Biomedical Image Analysis. 2000:86–93. [Google Scholar]
- Viola PA, Wells WM. Alignment by maximization of mutual information. Fifth Intl. Conference on Computer Vision; Cambridge: MIT; 1995. [Google Scholar]
- Wang F, Vemuri BC, Rao M, Chen Y. A new & robust information theoretic measure and its application to image alignment. Information Processing in Medical Imaging. 2003:388–400. doi: 10.1007/978-3-540-45087-0_33. [DOI] [PubMed] [Google Scholar]
- Wang F, Vemuri BC, Rangarajan A, Eisenschenk IMSSJ. Simultaneous nonrigid registration of multiple point sets and atlas construction. European Conference on Computer Vision; 2006. pp. 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitová B, Flusser J. Image registration methods: A survey. Image Vision Comput. 2003;21(11):977–1000. [Google Scholar]