Abstract
The myosin superfamily consists of more than 35 classes (each consisting of multiple isoforms) that have diverse cellular activities. The reaction pathway of the actin-activated myosin ATPase appears to be conserved for all myosin isoforms, but the rate and equilibrium constants that define the ATPase pathway vary significantly across the myosin superfamily, resulting in kinetic differences that that allow myosins to carry out diverse mechanical functions. Therefore, it is important to determine the lifetimes and relative populations of the key biochemical intermediates to obtain an understanding of a particular myosin’s cellular function. This chapter provides procedures for determining the overall and individual rate and equilibrium constants of the actomyosin ATPase cycle, including actomyosin binding and dissociation, ATP binding, ATP hydrolysis, phosphate release, and ADP release and binding. Many of the methods described in the chapter are applicable to the characterization of other ATPase enzymes.
1. INTRODUCTION
Myosins are motor proteins that use ATP hydrolysis to generate force and power motility along actin filaments. The myosin superfamily consists of more than 35 classes, each consisting of multiple isoforms, which have diverse cellular activities (Berg et al., 2001; Foth et al., 2006).
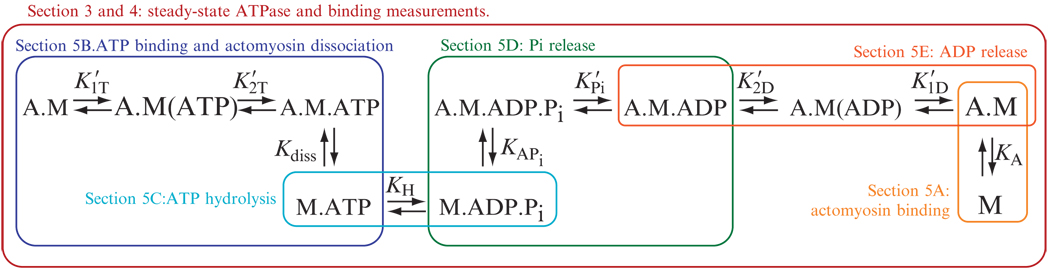
The reaction pathway for the actomyosin ATPase cycle appears to be conserved for all myosin isoforms (De La Cruz and Ostap, 2004); that is, the kinetic intermediates, and the order in which these intermediates are populated, are the same (Fig. 6.1). In the absence of ATP, myosin binds tightly to actin. ATP binding induces a conformational change in myosin that weakens its actin affinity and causes myosin to detach from actin. ATP is hydrolyzed to ADP and inorganic phosphate (Pi), and the hydrolysis products remain bound to myosin. Myosin rebinds to actin and the force generating power-stroke accompanies subsequent phosphate release. ADP is released, and the cycle repeats upon ATP binding (Bagshaw et al., 1974; Geeves and Holmes, 1999; Johnson and Taylor, 1978; Lymn and Taylor, 1971; Rosenfeld and Taylor, 1987).
Figure 6.1. The minimum actomyosin ATPase cycle reaction scheme.
The individual reaction steps and corresponding sections in which they are described are boxed and colored. For clarity, we have omitted some of the biochemical intermediates that are not significantly populated during steady-state ATP cycling in the presence of actin. The rate and equilibrium constants are defined as the reaction proceeds from left to right.
The rate and equilibrium constants that define the ATPase pathway vary significantly across the myosin superfamily, resulting in kinetic differences that allow myosins to carry out diverse mechanical functions (De La Cruz and Ostap, 2004; De La Cruz et al., 1999, 2001; El Mezgueldi et al., 2002; Kovacs et al., 2003). Therefore, it is important to determine the lifetimes and relative populations of the key biochemical intermediates to obtain an understanding of a particular myosin’s cellular role. The rates of the conversion between intermediates must be determined by transient kinetic analysis, and steady-state properties must be determined in terms of rate and equilibrium constants. This chapter provides procedures for determining the key rate and equilibrium constants of the actin-activated myosin ATPase outlined in Fig. 6.1. Single-molecule and actin-filament gliding assays provide valuable information regarding force generation and work output (i.e., motility) driven by myosin motors and can reveal how external loads affect specific ATPase cycle transitions (Laakso et al., 2008; Oguchi et al., 2008; Uemura et al., 2004; Veigel et al., 2003). However, we limit our discussion in this chapter to the experimental determination of the rate and equilibrium constants governing ATP utilization in solution, in the absence of external load. Although the assays described in the chapter are optimized for studying actomyosin interactions, many of the methods described are applicable to the characterization of other ATPase enzymes.
2. REAGENTS AND EQUIPMENT USED FOR ALL ASSAYS
A. Solution conditions and temperature
We typically perform experiments in KMg50 buffer (50 mM KCl, 2 mM MgCl2, 1mM EGTA, 2 mM dithiothreitol, and 10 mM imidazole, pH 7.0) at 25 °C so that we can compare the behavior of different myosin types. Similar experimental conditions have been used to measure numerous non-muscle myosins including myosin-Is (El Mezgueldi et al., 2002; Lewis et al., 2006; Ostap and Pollard, 1996), myosin-III (Dose et al., 2007), chicken myosin V (De La Cruz et al., 1999), porcine myosin VI (De La Cruz et al., 2001; Robblee et al., 2004, 2005), and human myosin VIIb (Henn and De La Cruz, 2005), but the relatively high ionic strength makes experiments with some myosins difficult.
B. Actin
Actin is purified from rabbit skeletal muscle (Spudich and Watt, 1971), pyrene-labeled with pyrenyl iodoacetamide as needed (Kouyama and Mihashi, 1981), gel-filtered over Sephacryl S-300HR equilibrated in G-buffer (5 mM Tris (pH 8.0), 0.2 mM ATP, 0.5 mM DTT, 1 mM NaN3, and 0.1 mM CaCl2) and stored at 4 °C. The bound Ca2+ should be exchanged by adding 0.2 mM EGTA and 50 µM MgCl2 (excess over [total actin]) immediately prior to polymerization by dialysis against KMg50 buffer. The dialysis step polymerizes actin and also removes free ATP, which could interfere with some of the assays. It is therefore important to ensure proper dialysis so that the final polymerized actin sample has essentially no free ATP. Phalloidin (1.1 molar equivalents) should be added to stabilize polymerized actin filaments. This stock should be stored at 4 °C and used within 2 days.
C. Myosin
Kinetic assays typically use myosin fragments prepared by proteolysis or recombinant myosins that contain only the motor and regulatory domains. Myosins that form filaments or higher-order oligomers may be problematic due to protein aggregation and lack of solubility, which complicates data analysis. Some myosins have very high-nucleotide affinities, resulting in the copurification of ATP or ADP. Apyrase (grade VII, <0.1 units mL−1) can be added to stock solutions to convert contaminating ADP and ATP to AMP. When using apyrase, ensure that low concentrations are used so as not to interfere with the kinetic measurements.
D. Nucleotides
We prepare stock solutions of nucleotides from dry powder of the free acid form, adjust the pH to 7.0 with KOH and store as 20–100 mM solutions at −20 °C or −80 °C. Nucleotide concentrations are determined by absorbance using molar extinction coefficients (ε) of 15,400 M−1 cm−1 at 259 nm for unlabelled (ATP and ADP), and 23,300 M−1 cm−1 at 255 nm for mant-labeled nucleotide (mantATP and mantADP, both mixed and single 2′-deoxy or 3′-deoxy isomers). We add a molar equivalent of MgCl2 to nucleotide solutions immediately before use. When characterizing myosins with high ADP affinities, it is critical to purify ATP from contaminating ADP (1%–2%) by HPLC (De La Cruz et al., 2000a). Similarly, it is critical to purify ATPγS from contaminating ADP generated by spontaneous hydrolysis immediately before use (Yengo et al., 2002).
E. Fluorescence spectrophotometer
Equilibrium-binding measurements that monitor changes in fluorescence or light scattering require an instrument with fluorescence detection capabilities. Numerous instruments are commercially available, both for measuring samples in optical cuvettes or multi-welled plates. Instruments equipped with excitation and emission monochromators are best because they permit evaluation of changes in wavelength as well as intensities, but instruments equipped with optical filters are also adequate provided changes in intensity are significant. These and all instruments used for kinetic and equilibrium analysis should be temperature controlled, as many of the ATPase cycle rate and equilibrium-binding constants have significant enthalpic components and are therefore sensitive to temperature.
F. Stopped-flow apparatus
Real-time acquisition of reaction time courses with millisecond time resolution is needed to measure the myosin ATPase cycle rate constants, as many of the experimentally observed and elementary rate constants are typically on the order of several hundred per second or faster. A variety of rapid mixers with absorbance and fluorescence detection are commercially available. Those driven by compressed air or stepper motors offer the most efficient perturbation and rapid mixing time. We have used instruments from KinTek, Applied Photophysics, and Hi-Tech, as well as in-house assembled instruments with satisfactory and reproducible results. Actin-activated Pi release from myosin is done following two mixing events (first with ATP then with actin; discussed subsequently) and requires an instrument capable of performing experiments in a sequential mixing configuration.
Manually driven rapid mixers that can be used with absorbance or fluorescence spectrophotometers are also available (De La Cruz and Pollard, 1994), but they are subject to longer mixing and dead times and are therefore only adequate for measuring slow reactions (<100 s−1). In addition, manually driven mixers require more material than the conventional stopped-flow instruments and are therefore not practical for characterization of myosin motors that can be purified in small quantities.
G. Quenched-flow apparatus
Several rapid mixing chemical-quench-flow instruments are commercially available. As with the stopped-flow, millisecond time resolution is needed to measure rapid rates and rate constants. We use the KinTek Model RQF-3 instrument.
3. STEADY-STATE ATPase ACTIVITY OF MYOSIN
3.1. High salt ATPase activity of myosin
The ATPase activities of some myosins are activated by the presence of high-salt and divalent cation chelators in the absence of actin (Chalovich and Eisenberg, 1982). These assays measure nonphysiological ATPase activities but are useful for determining the number of active myosins in a preparation and for measuring relative myosin concentrations in binding assays (e.g., section 4). The following procedure detects the steady-state hydrolysis of radioactive ATP and has the advantage of being insensitive to phosphate-contaminated myosin samples (Lynch et al., 1991). Nonradioactive methods for measuring phosphate are also available (Pollard, 1982), including a commercially available detection methods (Lanzetta et al., 1979; Webb, 1992).
Stock Solutions
2× K+/EDTA assay solution (1.0 M KCl, 30 mM Tris, pH 7.5, 10 mM EDTA) or 2× NH4+/EDTA assay solution (0.8 M NH4Cl, 50 mM Tris, pH 7.5, 70 mM EDTA).
Isobutanol-benzene at 1:1 mixture
Silicotungstic-sulfuric acid. Mix 2 parts of 10 N sulfuric acid with 5 parts of 6% aqueous silicotungstic acid.
Working solutions (freshly made)
Stop mixture. For each reaction tube, mix 1 mL of the isobutanolbenzene mixture with 0.25 mL of the silicotungstic-sulfuric acid mixture. This is most easily done using repipetter bottles. Aqueous and organic phases will form in each tube, with the organic phase on top.
Ammonium molybdate (10% solution). Dissolve 1 g of ammonium molybdate in 10 mL of water.
Prepare the ATPase assay solution by adding 1 mM [γ-32P]ATP (1 Ci mol−1) and diluting to 1×, keeping in mind the volume of myosin to be added (see subsequent sections).
Method
ATPase reaction: Equilibrate the ATPase reaction mixture from which time points will be removed to the experimental temperature. Initiate the reaction by adding myosin.
Time points: Every minute for 5 min, remove 100 µL of the ATPase reaction, quench by adding to a predispensed stop-mixture tube, vortex vigorously for 10 s, add 100 µL of the ammonium molybdate solution, and vortex for 10 s.
Separate phases: Separate the organic and aqueous phases by centrifuging briefly for 1 min at ≈1000×g. This step minimizes background counts from unhydrolyzed ATP.
Determine phosphate concentration: Transfer 500 µL of the organic phase (the top phase that contains free phosphate) to a scintillation vial with the appropriate scintillation fluid. Determine the specific radioactive activity of the [γ-32P]-ATP by adding aliquots of the ATPase reaction solution directly to scintillation vials.
Determine the ATPase rate: The slope of a plot of the phosphate concentration as a function of time yields the ATPase rate of the reaction mixture in units of Pi liberated per unit time, typically seconds. Dividing the value of the ATPase rate by the concentration of myosin, yields the concentration-normalized ATPase rate in units of ATP hydrolyzed s−1 myosin−1.
3.2. Actin-activated Mg2+-ATPase activity of myosin
Determination of the actin-activated steady-state ATPase activity of myosin is the important first step in understanding the kinetic properties of the motor, and simply requires measuring the products of ATP hydrolysis (ADP or inorganic phosphate) as a function of time as explained for the high-salt ATPase activity of myosin described previously. When performed under the appropriate conditions, the assay reports the maximum rate at which myosin hydrolyses ATP in the absence of actin (vo), the maximum ATPase rate of myosin in the presence of saturating actin (kcat), and the actin concentration-dependence of the myosin ATPase activity (KATPase).
We prefer the NADH-coupled assay to measure the steady-state ATPase activity of myosin motors over other familiar detection methods including the colorimetric assay and radiolabeled ATP assay (discussed previously) because of the real-time detection, sensitivity, and regeneration of ATP from liberated ADP (see De La Cruz et al., 2000a). The assay relies on monitoring the change in absorbance or fluorescence that is coupled to the oxidation of NADH through a series of coupled enzymatic reactions. Pyruvate kinase converts phospho(enol )pyruvate (PEP) and the ADP generated from the steady-state ATP hydrolysis of myosin or actomyosin to ATP and pyruvate via a phosphoryl transfer reaction, which is subsequently converted to lactate by lactate dehydrogenase (LDH) in a reaction that is coupled to the oxidation of NADH to NAD+. NADH absorbs 340 nm of light, but NAD+ does not; NADH is also fluorescent. Therefore, the NADH concentration can be readily monitored by absorbance or fluorescence. The overall reaction stoichiometry is such that one NADH molecule is consumed per ADP, permitting the concentration of ADP liberated, and therefore ATP hydrolyzed by myosin or actomyosin, to be readily determined from the loss of NADH.
Stock solutions and instrumentation
Absorbance (or fluorescence) spectrophotometer equipped with time-based data acquisition
Lactate dehydrogenase (LDH; 4000 U mL−1 in KMg50 containing 50% glycerol)
Pyruvate kinase (PK; 10,000 U mL−1 KMg50 containing 50% glycerol)
Phospho(enol)pyruvate (PEP; 100 mM, pH adjusted to 7.0)
NADH (we use lyophilized 1-mg aliquots and prepare 5× cocktail solution in the vial)
5 × cocktail solution: KMg50 buffer supplemented with 1 mM NADH, 100 U mL−1 LDH, 500 U mL−1 PK, and 2.5 mM PEP.
Note, LDH, PK, and PEP can be stored at −20 °C. NADH should be stored in the dark.
Method
The method described is one for manual mixing using an absorbance (or fluorescence) spectrophotometer in time-based acquisition mode and optical microcuvette but can be easily adapted for other volumes or automated mixing using a stopped-flow apparatus.
In a cuvette, mix 20 µL of 5× cocktail solution with myosin (at a final concentration of 20–200 nM) with 60 µL of KMg50 buffer.
Add 20 µL of 10 mM ATP in KMg50 to the cuvette and mix by pipetting. We prefer to aliquot large volumes because it makes it easier to mix by pipetting.
Start recording the time course of absorbance (or fluorescence) change at 340 nm. Continue for 100–200 s. The time course should be linear with a negative slope and starting value corresponding to 200 µM NADH and the optical path length. If the initial absorbance is low, it is possible that contaminating ADP in one of the solutions consumed the NADH before the start of the experiment.
Repeat steps 1–4 in the presence of a range of [actin]. Actin should be added in place of KMg50 buffer as stated in step 2. Be certain that the [myosin] is identical in all samples and that the [actin] be the only variable among samples. Although the ATPase rate of actin alone is typically negligible, it can be significant at high concentrations and should be accounted for when determining the true ATPase rate of myosin.
Use the extinction coefficient of NADH (ε340 = 6220 M−1 cm−1) to convert absorbance at 340 nm to [ADP]. When using fluorescence detection, a standard curve with known ADP concentrations must be obtained.
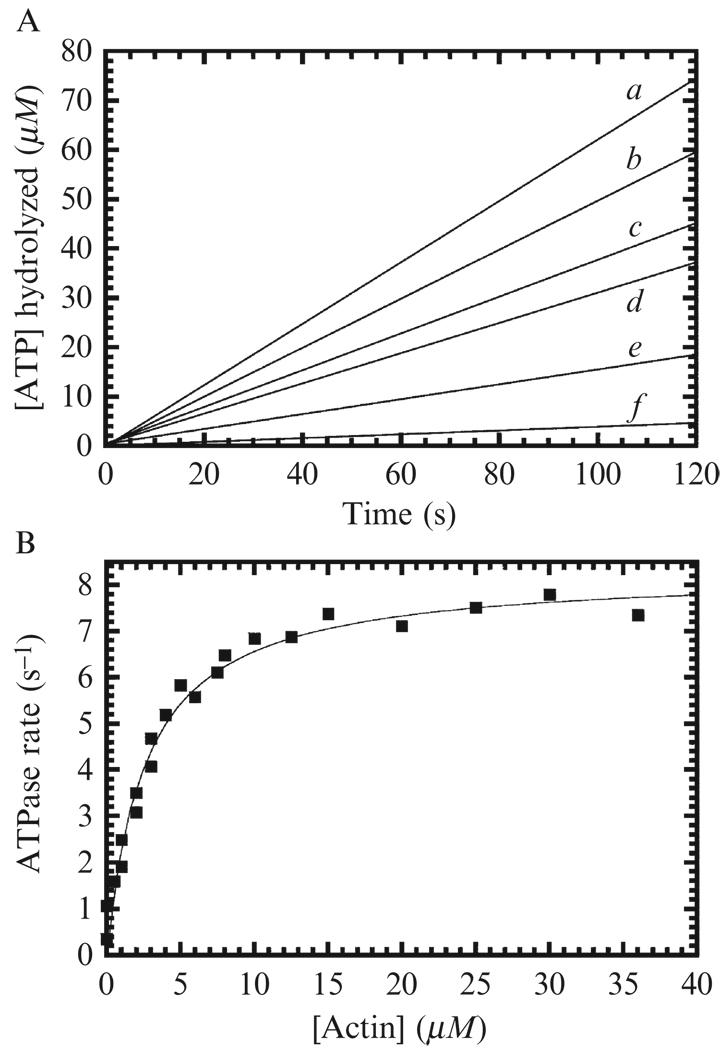
Generate a plot of the time course of ADP production ([ADP] versus time) and fit to a linear function (Fig. 6.2A). The slope yields the steady-state ATPase rate in units of [ADP] per unit time, typically expressed in seconds ([ADP] s−1). Divide this observed rate by the [myosin]. This normalized rate has units of ADP myosin−1s−1, but is often expressed in units of s−1, as one ATP is hydrolyzed per catalytic cycle of myosin.
-
Generate a plot of the steady-state rate of ADP production (in units of [ADP] [myosin]−1 s−1) versus the [actin]. A hallmark of myosin motors is that they are activated by binding to actin so the ATPase rate will increase as a function of the [actin]. The data should follow a rectangular hyperbola (Fig. 6.2B) and fitted to the Briggs-Haldane steady-state equation:
(6.1) The intercept is the ATPase rate of myosin alone in the absence of actin (v0), but it is usually indistinguishable from the origin and an unreliable measurement, and is best measured by a single turnover experiment (see De La Cruz et al., 1999). The kcat (determined from the best fit, not the data) is the maximum actin-activated ATPase rate of myosin (i.e., catalytic turnover number), KATPase is the concentration of actin needed to reach half maximal activation of myosin ATPase activity (i.e., apparent KM for actin). Note that reliable determination of the active myosin concentration is critical for determining the kcat and any uncertainties in myosin concentration or catalytically inactive motors will introduce uncertainties in kcat. If the data appear linear, then samples at higher [actin] are needed. If this cannot be achieved due to the high viscosity of the samples, lowering the ionic strength could lower the KATPase. Cases in which data points are acquired with the myosin concentration not at concentrations at least 10 times greater than that of actin should be fitted to a quadratic form of Eq. (6.1) (Henn et al., 2008).
Controls and troubleshooting. The final [ATP] according to this method is 2 mM. It is important that this be sufficient to saturate myosin and yield the maximum rate possible. Whether this condition is fulfilled can be evaluated by following the procedure described earlier, but measuring the KM for ATP from the [ATP]-dependence of the ATPase rate of myosin in the presence of saturating [actin]. When measuring the KATPase, the [ATP] in the samples should be >10 times greater than the KM for ATP. Similarly, when measuring the KM for ATP, the [actin] should be >10 times greater than the KATPase.
Figure 6.2. Steady-state ATPase activity of myosin VI.
(A) Time course of ATP turnover by 100 nM myosin VI at 16 (a), 8 (b), 4 (c), 2 (d), 1 (e), and 0 µM (f) actin filaments using the NADH-coupled assay. (B) Actin filament concentration dependence of the steady-state turnover rate of myosin VI. The solid line through the data points is the best fit to Eq. (6.1), with kcat = 8.3 ± 0.2 s−1 and KATPase = 2.8 ± 0.3 µM. Data are from (De La Cruz et al., 2001).
It is essential to confirm that the overall coupled assay reaction is more rapid than the ATPase rate of myosin or actomyosin. This is done by simply adding a small volume of concentrated ADP and monitoring the change in absorbance or fluorescence. The rate of absorbance change reflects the rate at which the coupled assay can convert free ADP in solution to the observed spectroscopic signal change. This rate must be more rapid than the ATPase rate measured for (acto)myosin in order for the experimentally measured ATPase rate to accurately reflect that of (acto)myosin.
Addition of ADP is also useful as a troubleshooting aid. If weak or no ATPase activity is detected, simply add ADP to the reaction mix. If there is a change in absorbance upon addition of ADP, then a lack of an observed ATPase activity is not due to a faulty reaction component in the assay mix or instrument configuration. Rather, the sample itself has little ATPase activity. Increasing the myosin concentration will accelerate the ATPase rate proportionally and could provide a reliable signal.
4. STEADY-STATE MEASUREMENT OF ACTOMYOSIN BINDING AFFINITIES
The steady-state binding of actin and myosin is defined as:
| (6.2) |
where, Kd is the dissociation equilibrium constant, and < > signifies a distribution of chemical states. The Kd value depends on the conformational state of the myosin (as determined by the bound nucleotide, scheme 6.1), so the overall, observed Kd during steady-state ATPase cycling depends on the affinities and distribution of the myosin intermediate states. The affinity of myosin for actin is highly dependent on the solution ionic-strength, and in many cases, binding experiments are performed at ionic strengths lower than the physiological condition to allow for experimentally measurable Kd’s. Actin sedimentation assays pioneered by Chalovich and Eisenberg (1982) and pyrene-actin fluorescence quenching assay first described by Kouyama and Mihashi (1981) and developed for myosin by (Geeves, 1989; Criddle et al., 1985) are the most commonly used techniques to measure steady-state actomyosin affinity.
4.1. Sedimentation assays
Sedimentation assays quantitate the concentration of myosin that pellets with actin after high-speed centrifugation. This method is most useful for determining the effective binding constants of a population of myosin states, for example during steady-state ATP hydrolysis.
Method
Experimental mix. Mix myosin at a constant concentration that is at least 50-fold lower than the Kd for the interaction and actin directly in a 200-µL TLA-100 centrifuge tube (Beckman). Actin concentrations are titrated to a final concentration that is ≈5-fold greater than the Kd. Appropriate control samples include (a) myosin in the absence of actin, (b) actin in the absence of myosin, and (c) actin and myosin in the absence of nucleotide.
Add nucleotide. Nucleotides that are hydrolyzed by myosin (e.g., ATP and ATP-γ-S) should be added to the centrifuge tubes immediately before centrifugation. The actomyosin steady-state ATPase rate should be considered to ensure (a) the nucleotide substrate is not depleted during the experiment and (b) products of the ATP hydrolysis reaction do not affect the population of the steady-state intermediates. When assaying nucleotide analogues, one should ensure that the analogues are pure and do not contain contaminants that bind with a tighter affinity than the analog of interest. If the fraction of bound myosin is going to be determined via SDS-PAGE (see subsequent sections), a small amount of each sample should be saved before centrifugation (precentrifuged control).
Centrifuge. Centrifuge samples at 250,000×g for 20 min at the appropriate temperature. Carefully remove supernatants from the tubes as they are taken out of the rotor. Keep track of the expected location of the protein pellets, as they may be difficult to see. Transfer supernatants to 1.5-mL microfuge tubes on ice without disturbing or contacting the pellet. Save the centrifuge tubes containing the pellet.
- Detection of fraction of actin-bound myosin. The most common methods for determining the fraction of myosin bound to actin are (a) measurement of the ATPase activity of myosin in the supernatant and (b) resolution and quantitation via SDS-PAGE.
- ATPase activity. Measurement of the NH4/EDTA ATPase activity (section 3.1) provides a sensitive method for determining the concentration of myosin that remains in the supernatant after centrifugation and does not detect inactive myosins that may contaminate the protein preparation. The fraction of myosin bound (fb) at each condition is determined by:
(6.3) - SDS-PAGE. The protein pellets are resuspended in SDS-PAGE sample buffer to the original sample volume, heated to 100 °C for 2 min and resolved by SDS-PAGE. Pelleted samples are resolved side by side with the precentrifuged controls (see No. 3, earlier). The fb is determined by dividing the quantity of myosin in the pellet by the quantity of myosin in the precentrifuged control. Myosin concentrations are determined by scanning stained gels, so protein standards of known concentration must be run on every gel to ensure linearity of detection. Coomassie blue is the most commonly used gel stain, and it is easily quantitated using a standard gel-documentation system or flatbed scanner. However, we found Sypro protein dyes to be more sensitive to low protein concentrations and more precisely determined when scanned with a fluorescence scanner (e.g., Typhoon Imager; Lin et al., 2005; Manceva et al., 2007).
- Determination of the dissociation equilibrium-binding constant (Kd). Binding experiments are performed when the myosin concentration is kept at a constant concentration ≈10-fold lower than the Kd with the maximum actin concentration ≥5-fold the Kd. The actin affinities for different myosin isoforms vary >100-fold, so the protein concentrations depend on the isoform being investigated. Multiple experiments may be required to determine the appropriate concentration ranges. The Kd is determined by fitting the fraction bound (fb) to the hyperbolic relationship:
The Kd’s are determined by non-linear least-squares fitting using any of the widely available commercial or Web-based fitting programs. Linear transformations of the data are problematic in that they make error analysis difficult and, given the accessibility of computers and non–linear regression software, they are not recommended.(6.4)
4.2. Pyrene fluorescence measurements
The fluorescence of pyrene-labeled actin is linearly quenched by the binding of one myosin in the “strong binding” AM or AM.ADP state to one polymerized actin subunit (scheme 6.1). Pyrene-actin fluorescence is not quenched by binding of myosins in the “weak binding” AM.ATP or AM. ADP.Pi states (scheme 6.1). Thus, this method is best suited for (a) measuring that affinity of the strong AM or AM.ADP states, and (b) measuring the fraction of myosins in these states strong binding during steady-state ATPase cycling (De La Cruz et al., 2000b; Henn and De La Cruz, 2005). It is not suitable for measuring the total fractions of myosin bound during steady-state ATPase cycling.
Method
- Titration and detection of binding. Add pyrene-actin at the desired concentration to a fluorescence cuvette. Most fluorometers are very sensitive and detect linear changes of pyrene-actin fluorescence at concentrations <100 nM in a 100-µL cuvette. The excitation peak of pyrene-actin is 365 nm. Single-wavelength readings at ≈410 nm are sufficient for performing titrations, but scanning the emission wavelength for each sample between 375 nm and 450 nm is preferable, as it allows one to detect wavelength shifts and anomalous scattering due to air bubbles or protein aggregation. Experiments are performed by adding concentrated myosin solutions directly to the pyrene-actin-containing fluorescence cuvette or by preparing separate samples for each titration point. One must ensure that the actin concentration is not diluted as a result of myosin additions. Dilution of the sample is avoided by including pyrene-actin with the titrated myosin. The fractional saturation of pyrene-actin filaments (fb) is calculated by:
where Fo is the fluorescence signal in the absence of myosin, and F∞ is the signal at infinite myosin concentration.(6.5)
Determination of dissociation constant: Binding experiments are performed when the actin concentration is kept at a constant concentration of ≈10-fold lower than the Kd with the maximum myosin concentration of ≥5-fold the Kd. The actin concentration is kept constant to avoid changes in Fo and F∞ during the titration. The Kd is determined by fitting a plot of fb versus myosin concentration to the hyperbolic relationship Eq. (6.4).
If the AM and AM.ADP bind myosin with high affinity (Kd < 0.1 µM), the pyrene-actin concentration (as defined previously) may be below the detection limit of the fluorometer. If this is the case, the pyrene-actin in the cuvette is increased to a concentration ≈2-fold lower than the Kd. The binding curve is no longer hyperbolically related to the myosin concentration under these conditions, but rather is fit using the following quadratic Eq. (6.6):
| (6.6) |
where [Ao] and [Mo] are the total actin and myosin concentrations, respectively. It is important to ensure that the experiments are being performed at concentrations in which the curve is sensitive to the Kd.
5. TRANSIENT KINETIC ANALYSIS OF THE INDIVIDUAL ATPase CYCLE TRANSITIONS
Steady-state characterization of the actin-activated myosin ATPase is an important step to understanding motor activity. Quantitative determination of the kcat and (apparent) KM for actin and ATP describe reliably the overall cycling behavior of myosin and can reveal valuable insight to the physiological role of various myosin isoforms. However, it is immediately obvious that the simplified two- to three-step reaction mechanism generally assumed for Michaelis-Menten or Briggs-Haldane steady-state kinetic analysis does not reliably account for the minimal ATPase cycle of (acto)myosin (scheme 6.1) and that steady-state characterization will not provide any information regarding the identity and distribution of transiently populated intermediates that play critical roles in contractility, force generation and motility, nor will it provide any information regarding the lifetimes of these intermediates. As a result, diversity in function and motor activities that arise from differential population of biochemical cycling intermediates will be transparent to steady-state characterization. For example, the steady-state ATPase activity of high duty ratio myosin are qualitatively similar to some low-duty-ratio myosins in that they are activated by micromolar actin concentrations (De La Cruz et al., 1999, 2001), yet they have dramatically different kinetic behaviors that confer unique motor activity and allow them to perform different functions. Limiting the kinetic analysis of these myosin motors to steady-state ATPase characterization would never have revealed their important differences.
While steady-state characterization provides valuable information regarding the overall behavior of myosin, transient kinetics focuses on measuring the individual ATPase cycle reactions in isolation. The objective of transient kinetic analysis is to directly observe and identify biochemical intermediates populated during ATP cycling, determine the lifetime and distribution of these intermediates, and define the preferred reaction pathway through the ATPase cycle. A final goal of a thorough transient kinetic analysis is to account for the observed steady-state behavior and constants in terms of the fundamental rate and equilibrium constants of the ATPase cycle pathway shown in scheme 6.1.
The transient kinetic approach is rather straightforward and requires only three things: (1) a signal, chemical (e.g., chemical cleavage of ATP) or optical (e.g., fluorescence, light scattering, absorbance, anisotropy), (2) rapid physical (temperature or pressure) or chemical (changing reactant concentrations) disruption of a system at equilibrium, and (3) observation of the time course of approach to the new equilibrium with appropriate time resolution. We describe in the following sections the signals to monitor for particular ATPase cycle reactions and focus our discussions on rapid mixing techniques in which perturbation arises from the rapid mixing of reactants in single (two solutions) or double (three solutions) mixing events and the time course to new equilibria are measured as a function of the reactant concentration.
When performing rapid mixing, stopped-flow measurements, it is important to adjust the experimental conditions so that the concentration of one reactant is unchanged during the reaction. Otherwise, time courses of even simple, one-step reactions will deviate from single exponential behavior. This condition is easily achieved by ensuring that the concentration of one reactant is mixed at ≥10 times the concentration of the other so that the concentration at equilibrium of the reactant in excess will change by ≤10%. This condition is referred to as pseudo–first order condition and should be maintained whenever possible. It should be clarified that the concentration of the reactant in excess is used to determine the rate constants.
Reaction time courses will follow single or a sum of exponentials and should be fitted to the following function describing a linear sum of exponentials:
| (6.7) |
where S(t) is the signal at time t, S∞ is the final signal intensity, Ai is the amplitude, ki is the observed rate constant (kobs) characterizing the ith relaxation process, and n is the total number of observed kinetic phases. The value of n can vary among experiments but will usually be either one (single exponential) or two (double exponential). In some cases there will be an additional, linear component in the data. In this case, Eq. (6.7) will be modified to include a contribution to this linear phase (discussed subsequently).
5.1. Myosin binding to and dissociation from actin
Determination of the rate constants of myosin binding to and dissociation from actin allows the calculation of the actomyosin rigor affinity (KA; Fig. 6.1). These rate constants also reveal information about myosin structural transitions that occur upon actin binding (e.g., De La Cruz et al., 1999; Taylor, 1991). Binding is most simply measured by monitoring the fluorescence of pyrene-actin upon mixing with myosin, and dissociation is measured by mixing pyrene-actin-myosin complexes with excess unlabeled actin. It is essential for proteins to be free of contaminating ADP or ATP when measuring binding of rigor myosin, so treatment of samples with apyrase is advisable (see previous sections). Inclusion of saturating ADP permits measuring myosin-ADP binding to actin.
Myosin (and myosin-ADP) binding to actin can often be modeled as a simple one-step binding process:
| (Scheme 6.1) |
where A** is the high fluorescence state and A★ is the low fluorescence (quenched) state of pyrene actin. Although one-step binding is considered here, there is evidence for multistep (Henn and De La Cruz, 2005; Taylor, 1991) and additional actin-bound states for several myosins isoforms (Hannemann et al., 2005; McKillop and Geeves, 1993; Rosenfeld et al., 2000).
Method
Myosin binding to pyrene-actin. Myosin (syringe A) is mixed with phalloidin-stabilized pyrene-actin (syringe B) in a stopped-flow fluorometer. Fluorescence time courses are acquired at multiple pyrene-actin concentrations. The myosin concentration after mixing is typically ≈0.1 µM. To satisfy pseudo–first order conditions, the actin concentration should be ≈10-fold in excess of the myosin concentration. To retain adequate signal-to-noise in the fluorescence transients, the myosin concentration may have to be increased as the actin concentration is increased.
Dissociation of myosin from pyrene-actin. Myosin bound to pyreneactin is mixed with 50- to 100-fold higher concentrations of unlabeled actin in a stopped-flow fluorometer. The pyrene-actomyosin concentrations is typically ≈0.5 µM after mixing.
Data analysis. Fluorescence time courses of myosin binding to actin are usually best fit to single exponential functions, yielding actin concentration dependent kobs. A linear fit of a plot of kobs versus actin concentration yields an apparent second-order rate constant for binding in units of M−1s−1, which range from ≈1 × 106 M−1s−1 to ≈1 × 107 M−1s−1. Many myosins have a hyperbolic dependence of kobs on the actin concentration, indicating two step binding (see Taylor, 1991).
Fluorescence time courses of the dissociation of myosin from pyreneactin are usually best fit by a single exponential function. The dissociation rate constant varies significantly among myosins, ranging from ≈0.1 s−1 for muscle myosins to <0.01 s−1 for nonmuscle myosins.
5.2. ATP binding to actomyosin
ATP binding weakens the affinity of myosin for actin and dissociates the complex (Lymn and Taylor, 1971). ATP binding to all myosins (M) occurs in at least two steps with formation of a nonspecific collision complex, AM (ATP), that exists in rapid equilibrium (K1T′) with nucleotide-free actomyosin (AM), followed by an isomerization (K2T′) to a state (AMT) that dissociates rapidly from actin (kdiss):
| (Scheme 6.2) |
The three of the most efficient methods to measure ATP binding to actomyosin are (1) ATP-dependent dissociation of myosin from pyrene-labeled actin filaments, (2) monitoring the reduction in light scattering that occurs when myosin dissociates from (unlabeled) actin filaments, and (3) the fluorescence enhancement associated with binding of the nucleotide analogue mantATP (Hiratsuka, 1983). MantATP is valuable because one monitors the nucleotide and/or myosin (via energy transfer to mant from tryptophans at the nucleotide binding site) directly but can be used over a limited concentration range. Pyrene fluorescence provides the best signal-to-noise ratio and a broad ATP concentration working range (up to 20 mM). However, it has the disadvantage that it cannot distinguish between weakly bound, attached states (scheme 6.1) and those that are detached from actin. Light scattering is also very sensitive and has the advantages that it monitors actin attachment, regardless if myosin is strongly or weakly bound (scheme 6.1), and does not require fluorescent modification of actin or nucleotide. The combination of the three methods will identify if fluorescent modification of actin or nucleotides disrupts the intrinsic properties of the natural, unmodified substrates. We limit our discussion to the use of pyrene actin fluorescence or light scattering, as the detection methods are most sensitive and can be used to measure ADP binding (discussed subsequently).
Method
Determination of equilibrium/completely detached intensity: Before starting an experiment, the scattering or fluorescence intensity of actin alone. Rapidly mix (pyrene) actin (0.2–1 µM) with KMg50 buffer in a stopped-flow (λex = 365 nm, emission measured through a 400-nm long-pass filter for pyrene; light scattering can measured at 300–320 nm). The time course should be relatively flat. The signal intensity reflects that of actin alone after mixing and will be the final, equilibrium intensity of the experimental ATP-induced actomyosin dissociation transients, provided all of the bound myosin dissociates from actin upon addition of ATP. It should be noted that not all of the myosin need dissociate in the presence of ATP and an equilibrium signal that differs from actin alone can be indicative of physiological function. This behavior is a characteristic of high duty ratio motors that remain attached to actin (when present even at low micromolar concentrations) in the presence of ATP (De La Cruz et al., 1999, 2000b, 2001; Henn and De La Cruz, 2005). However, inactive myosin heads that bind but do not dissociate will also yield this behavior, so independent measurements of myosin activity should be acquired.
Determination of starting point: (Pyrene) actomyosin (0.2–1 µM at a 1:1 stoichiometry; syringe A) is rapidly mixed with buffer (syringe B). This establishes the low pyrene fluorescence or high light-scattering intensity from which kinetic transients should originate.
Experiment: Acquire time courses of fluorescence change after mixing with a range of ATP concentrations (0–several mM).
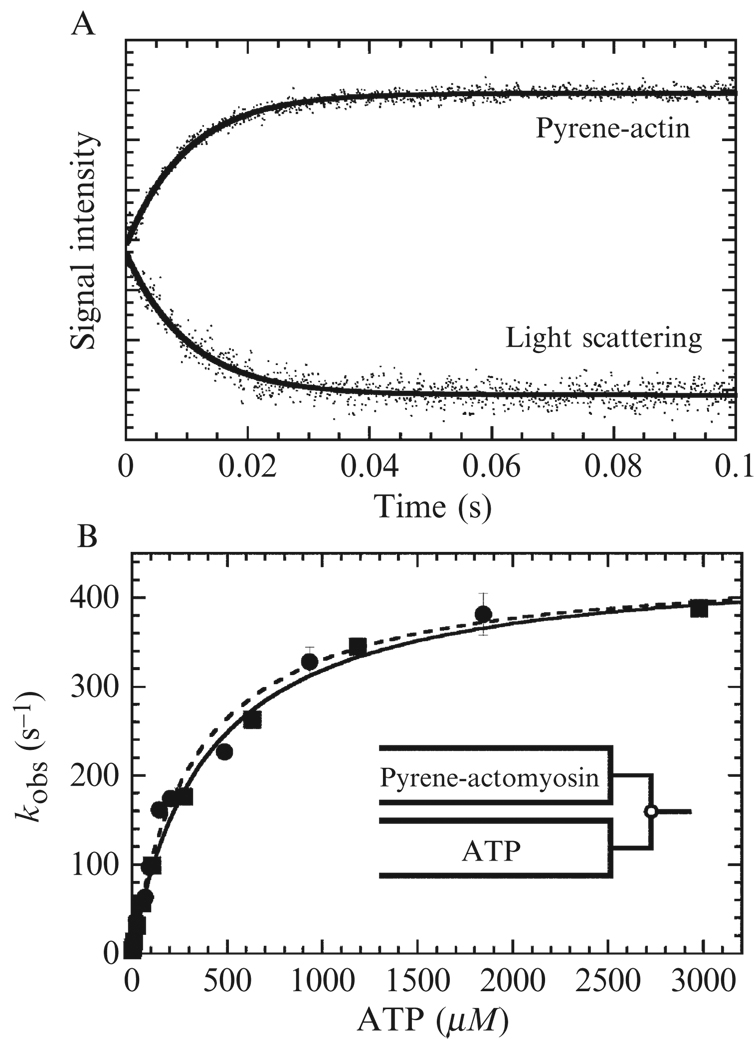
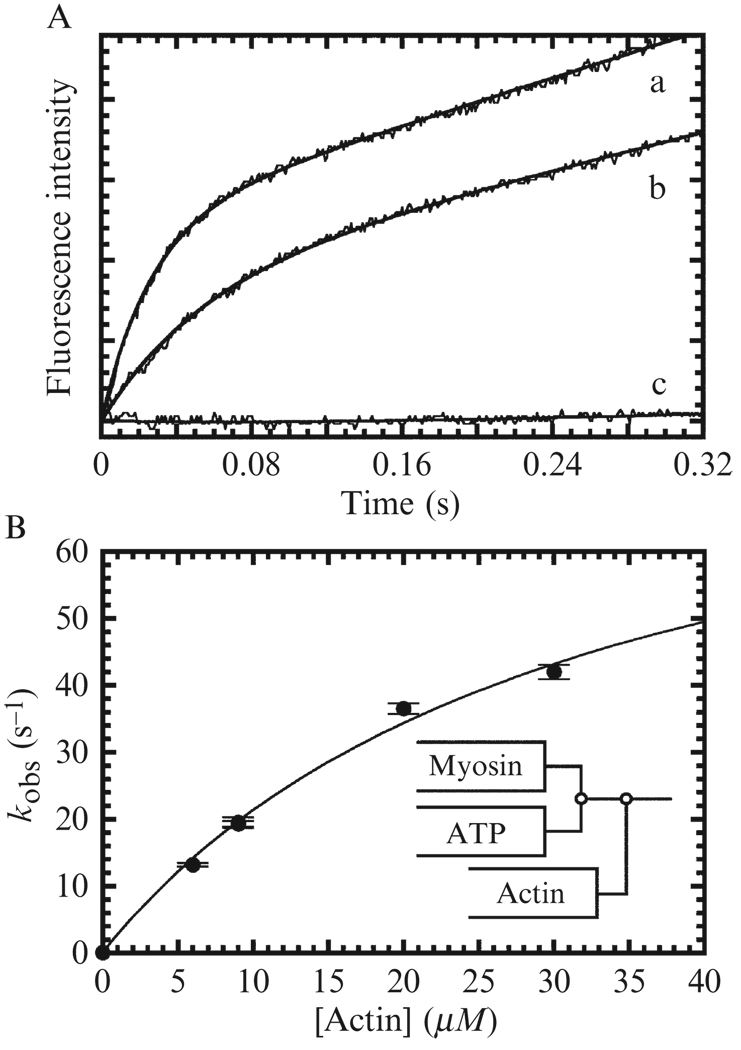
Data analysis and interpretation: Time courses of pyrene fluorescence enhancement (Fig. 6.3A), mantATP fluorescence, and reduction in light scattering (Fig. 6.3A) usually follow a single exponential with observed rate constants that depend hyperbolically on the ATP concentration (Fig. 6.3B) and should be fitted to the following equation:
| (6.8) |
Figure 6.3. ATP-induced dissociation of pyrene-actomyosin.
(A) Pyrene fluorescence and light-scattering transients obtained by mixing 0.25 µM actomyo1e with 100 µM ATP. Solids lines are single exponential fits (Eq. (6.7)) to determine kobs. (B) ATP concentration dependence of kobs determined from pyrene-fluorescence (●) and light scattering transients (■) is shown for the full range of ATP concentrations. Solid lines are fits to Eq. (6.8). Data are from (El Mezgueldi et al., 2002).
The best fit parameters yield K1T′ (as an association equilibrium constant in units of M−1) and the maximum isomerization rate constant achieved at saturating ATP (k+2T′).
The values of K1T′ and k+2T′ have significance. Myosins that bind ATP weakly and slowly (i.e., myosins I (Coluccio and Geeves, 1999; Lewis et al., 2006) and VI (De La Cruz et al., 2001; Robblee et al., 2005) do so because the observed K1T′ is very large and weak. Consequently, physiological ATP concentrations can be nonsaturating. Formation of a (nucleotide-free) actomyosin state that cannot bind nucleotide (closed versus open) contributes to the weak K1T′ of myosins I and VI. Actin-attached myosin VI dimmers communicate allosterically by shifting K1T′ to favor the open state, which accelerates ATP binding (Robblee et al., 2004). Intramolecular load can also affect the k+2T′ value of some myosins (Robblee et al., 2004).
There are some cases in which ATP binding to actomyosin does not follow a single exponential. Deviations from a single exponential including double exponential behavior (Coluccio and Geeves, 1999; Lewis et al., 2006) and lag phases (Robblee et al., 2005) can be indicative of an isomerization between a closed actomyosin state that cannot bind nucleotide and an open one that can. But one must be careful when interpreting data, as contaminating nucleotide could also yield deviations from a single exponential (De La Cruz et al., 2001).
5.3. ATP binding and hydrolysis by myosin
ATP hydrolysis (KH) is the biochemical transition closely linked to the structural change that results in the reverse power stroke, or repriming, step. The rate and equilibrium constants for this transition are most commonly determined in the absence of actin, because (a) ATP is normally hydrolyzed when myosins are detached from actin during normal cycling (scheme 6.1), and (b) the posthydrolysis M.ADP.Pi state has a dramatically longer lifetime in the absence of actin, due to slow phosphate release (k+Pi′), which simplifies experimental design and interpretation (Johnson and Taylor, 1978; Lymn and Taylor, 1970). The two most common methods for measuring the rate of ATP hydrolysis are detection of changes in the intrinsic tryptophan fluorescence of myosin using stopped flow, and the detection inorganic phosphate production using quenched flow.
5.3.1. Intrinsic tryptophan fluorescence
ATP binding to some myosins that contain a tryptophan at position 512 results in an enhancement in the intrinsic myosin fluorescence as follows:
| (Scheme 6.3) |
where the M* and M** are enhanced fluorescence states of myosin. The magnitude and origin of intrinsic tryptophan fluorescence enhancement is myosin-isoform dependent. For example, the fluorescence of skeletal and smooth-muscle myosin II increases upon population of the M.ATP state and increases further upon population of the M.ADP.Pi state. Vertebrate myosin I, Dictyostelium myosin II, and vertebrate myosin V have fluorescence enhancements that only correlate with the population of the M.ADP. Pi state, while the fluorescence of myosins VI is relatively insensitive to nucleotide binding (De La Cruz et al., 2001).
Method
Myosin (syringe A) is rapidly mixed with ATP (syringe B) in a stopped-flow fluorometer (λex = 280–295 nm, emission measured through a 320-nm long-pass filter). The required myosin concentration depends on the size of the signal change upon binding of nucleotide and on the sensitivity of the instrument. For most published investigations, 0.1–0.5 µM has been sufficient. Myosin must be free of contaminants, particularly contaminants that contain tryptophans that contribute to the fluorescence signal. The myosin preparation must also be homogeneous, otherwise the fluorescence transient may contain multiple components that are difficult to interpret.
Acquire time courses of fluorescence change at multiple ATP concentrations. Fluorescence time courses are acquired at concentrations from ≈1 µM to 1–2 mM ATP.
- Data analysis. Most myosins bind ATP rapidly and irreversibly, and release phosphate very slowly in the absence of actin. Therefore, interpretation of the ATP dependence of the fluorescence time courses is straightforward, in that the reaction can be considered a two step pathway, where an irreversible ATP-binding step is followed by a reversible first-order hydrolysis reaction. The two common cases for interpretation and analysis of the fluorescence signal are as follows.
-
Case 1: Individual high fluorescence stateWhen the ATP-induced change in the fluorescence originates from the population of the M.ADP.Pi state, the fluorescence time-course at each ATP concentration is best fit by a single exponential function Eq. (6.7). A plot of kobs versus the ATP concentration is hyperbolic, with the maximum value of kobs corresponding to the sum of the forward and reverse rates of ATP hydrolysis (k+H + k−H). A linear fit of the plot at low ATP concentrations yields the apparent second-order rate constant for ATP binding (K1T′k+2T′).
-
Case 2: Multiple high fluorescence states.Data analysis is more complicated when the fluorescence signal in the presence of ATP is the linear combination of multiple conformational states. The best characterized example is skeletal-muscle myosin II, where the M*.ATP state has a fluorescent enhancement intermediate between the M and M**.ADP.Pi state (Johnson and Taylor, 1978). As the ATP concentration is increased, the fluorescence time course will be best fit to the sum of two exponentials, where kobs1 reports the rate of population of the M*.ATP state and kobs2 reports the rate of population of the M**.ADP.Pi state. A linear fit of kobs1 versus the ATP concentration reveals the apparent second-order rate constant for ATP binding, and the maximum rate of kobs2 yields k+H + k−H. At high ATP concentrations (>500 µM), the time course of the population of the M*.ATP state might be too fast (>1000 s−1) to be recorded by most stopped-flow instruments, resulting in the resolution of only a signal that is best fit by a single exponential function, corresponding to k+H + k−H.
-
5.3.2. Quench flow
The rate of ATP hydrolysis and the equilibrium constant are determined by measuring the time dependence of phosphate production using a quench-flow apparatus. In this technique, myosin is mixed with ATP, aged for a specified time, and then quenched with acid, which denatures the myosin and stops the ATPase reaction. Because myosin is denatured, phosphate that was sequestered in the active site in the M.ADP.Pi state is also measured.
Quenched-flow experiments are more labor-intensive than stopped-flow, as a single time course requires phosphate determinations from multiple time points at relatively high myosin concentrations. However, the advantage is that it is a direct measurement of ATP hydrolysis.
Method
Myosin is mixed with [γ-32P]-ATP (1 Ci mol−1) in a quenched-flow instrument. When using the KinTek RQF-3 instrument, 35 µL of myosin is mixed with 35 µL of [γ-32P]-ATP, the reaction is aged for a specified time, then mixed with quenching solution (2 M HCl, 0.35 mM NaH2PO4). The myosin concentration is typically >1 µM after mixing, with higher concentrations being better. The ATP concentration should be high enough so that rate of ATP binding does not limit the rate of ATP hydrolysis. For example, if the rate of ATP binding is 5 × 106 M−1s−1, and the rate of ATP hydrolysis is 50 s−1, one would select an ATP concentration of at least 25 µM.
Acquire multiple time points. Enough points are acquired to resolve the time-course of ATP hydrolysis. For example, if ATP hydrolysis occurs at a rate of 50 s−1, points every ≈10 ms are acquired for 100–150 ms. Longer time courses are acquired to resolve the steady-state phase of ATP hydrolysis. Quenched time points are kept on ice, and the free phosphate concentration is determined as soon as possible to minimize acid hydrolysis of the ATP.
Determine phosphate concentration. There are multiple methods for determining the free phosphate concentration, including thin layer chromatography (Gilbert et al., 1995; Henn et al., 2008) and the molybdate method described earlier. We prefer the method developed by White (White and Rayment, 1993), in which equal volumes of the quenched reaction are mixed with a 10% activated charcoal slurry in quench solution and centrifuged at ≈12,000×g. The supernatant contains phosphate, and the charcoal fraction contains ATP and ADP. The supernatant and a volume of the total reaction mix are separately added to scintillation vials and counted. The radioactivity counted from each time point is normalized against the total counts in the total reaction mix to account for pipetting errors.
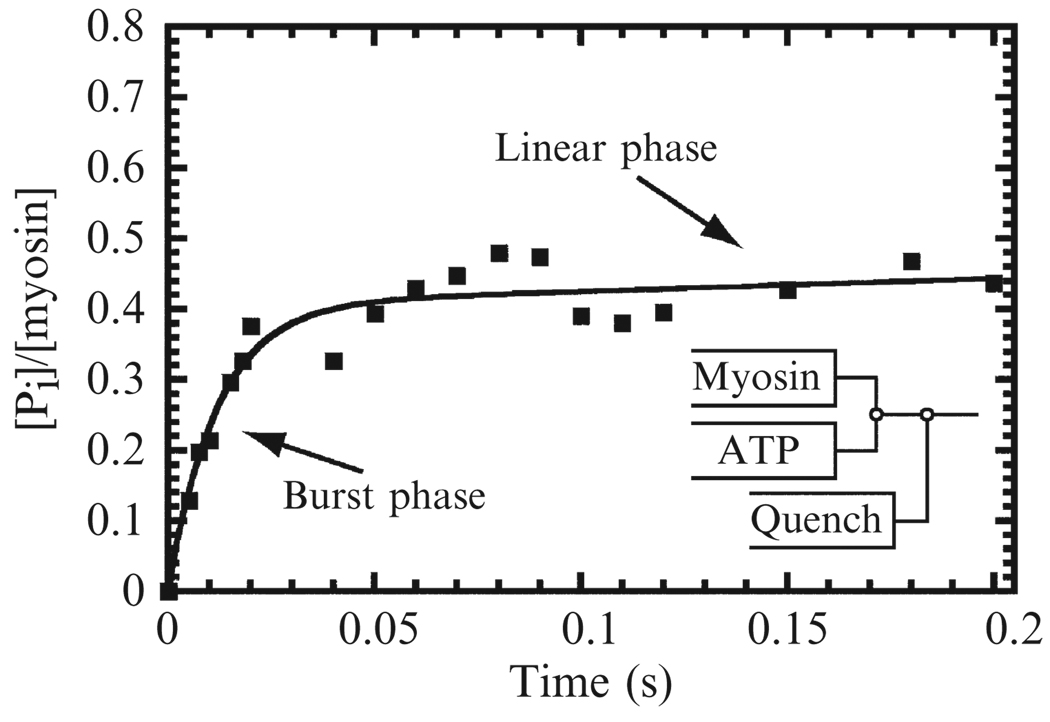
- Data analysis. Phosphate release from most myosins in the absence of actin (k+Pi′) is very slow, so time courses of phosphate concentration formed is composed of burst and linear phases (Fig. 6.4). The burst phase reports formation of the M.ADP.Pi state, and the linear phase reports the rate of steady-state ATP turnover. When the phosphate concentration is normalized by dividing by the myosin concentration, the time course is fit by:
where kobs = (k+H + k−H) and kss is the steady-state turnover rate. The burst amplitude is given by (KH/(1 + KH)). In most cases, kss is slow and can be ignored. Knowing KH and kobs, one can calculate the hydrolysis (k+H) and ATP resynthesis (k−H) rate constants:(6.9) (6.10)
Figure 6.4. Time course of ATP hydrolysis and ADP-Pi burst of myosin V.
Time course of ADP-Pi formation by a single-headed myosin V construct after mixing with 100 µM ATP. The solid line is the best fit of the data to Eq. (6.9) with an observed rate constant of 84 ± 13 s−1 and burst amplitude of 0.43 ± 0.03 Pi/myosin. Data are from (De La Cruz et al., 2000b).
5.4. Actin-activated Pi release
Multiple methods for measuring Pi are available, including the molybdate and charcoal extraction assays described earlier. These assays are not suited for real-time measurements and offer poor time resolution because they are typically done by manual mixing. In addition, the assays involve denaturation of the myosin and determination of total Pi formed, and can therefore not distinguish bound Pi from free Pi released to the solution. The MESG/phosphorylase assay (Webb, 1992) is advantageous in that it provides real-time acquisition of free Pi in solution that can be monitored by absorbance. However, it has a sensitivity of ≈1 µM and can only measure rates up to 90 s−1 when the purine nucleoside phosphorylase enzyme is present at very high concentrations (>50 µM). While this is adequate for many experimental systems, it is too slow for measuring actin-activated Pi release from many myosin isoforms, which can be ≥100 s−1 (De La Cruz et al., 1999; White et al., 1997).
Actin-activated Pi release from myosin-ADP-Pi can be rapid (>100 s−1) and be measured only using the fluorescently labeled mutant of the Pi-binding protein (MDCC-labeled PiBiP; (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin)-labeled phosphate binding protein) developed by Martin Webb (Brune et al., 1994; White et al., 1997) with the stopped flow with the instrument in sequential mixing mode. PiBiP has the advantage over other detection methods in its sensitivity (10 nM) and ability to measure rapid rates and rate constants (>700 s−1) in real time, though it can be difficult if significant Pi contaminates the solutions and glassware (which it always does). Background Pi must be removed from all solutions, syringes and the instrument with Pi “mop” solution: 7-methylguanosine (0.2–0.5 mM) and purine nucleoside phosphorylase (0.1 units mL−1), for at least 1 h. We treat the instrument with mop solution overnight before performing an experiment.
To accurately measure transient Pi release from actomyosin, the ATP binding (K1k+2) and hydrolysis rate constant (k+H + k−H) must be known. The experiment is done by first mixing myosin with ATP under single or multiple turnover conditions, ageing for sufficient time to allow ATP binding and hydrolysis to occur (typically ms–s), and then rapidly mixing with a range of actin filament concentrations in the second mix. There is ≈5-fold enhancement in the fluorescence of PiBiP with Pi binding (λex = 430 nm, 455 nm long pass emission filter). PiBiP should be present at 5–10 µM and preferably included in the myosin, nucleotide, and actin solutions so that Pi binding to PiBiP is more rapid than Pi release from (acto)myosin. The rate and equilibrium constants of Pi binding to MDCC-PiBiP in KMg50 buffer and 25 °C are: k+ = 117 (± 8) µM−1 s−1,k− = 24 s−1, and Kd = 0.20 µM (Henn and De La Cruz, 2005).
Method
Configure the stopped-flow instrument into sequential (i.e., double mixing) mixing mode
Treat instrument with mop solution
First mix and aging time: Rapidly mix myosin (syringe A) with ATP (syringe B) and age for sufficient time that ATP binding and hydrolysis (but not Pi release from myosin) occur. The myosin concentration needed will depend on the enzymatic behavior of the myosin, particularly the equilibrium constant for ATP hydrolysis (KH). In our experience, an initial mix with ≈ 4 µM myosin is a good starting point if KH favors the post hydrolysis states. If the value of KH is such that a significant fraction of the myosin-bound nucleotide will remain as ATP, higher myosin concentrations will be needed. The ATP concentration can be less than (single turnover) or greater than (multiple turnover) that of myosin. Myosin motors that bind ADP rapidly and with high affinity can be forced to undergo a single turnover (De La Cruz et al., 2001; De La Cruz et al., 1999) even when [ATP] ≫ [myosin] by including excess (mM) ADP in the actin syringe (discussed subsequently).
Second mix: Rapidly mix the aged myosin-ATP/ADP.Pi with actin (syringe C) over a broad concentration range (0–tens of micromolar). If the myosin motor being characterized binds ADP strongly and rapidly, including mM ADP with the actin solution will ensure that ATP will not bind myosin after the initial round of product release and that a single turnover of Pi release will be measured.
Convert fluorescence intensity to Pi concentration by acquiring a standard curve.
Data analysis: Experimental time courses will follow single or double exponentials under single turnover conditions and single (or double) exponentials with a slope (steady-state) under multiple turnover conditions in the presence of actin. Time courses in the absence of actin will appear flat over the seconds timescale, as Pi release from myosin alone is very slow (≈0.02 s−1). We will limit our discussion of the analysis to time courses that follow either a single exponential or a single exponential with a steady-state slope (Fig. 6.5A) but refer the reader to notable exceptions (White et al., 1997).
Figure 6.5. Pi release from actomyosin VI.
(A) Time course of transient Pi release from a truncated myosin VI construct after mixing with 20 (curve a), 9 (curve b), or 0 µM (curve c) actin filaments. (B) Actin filament concentration dependence of the Pi release burst rate. Final concentrations at t = 0 were 1.5 µM myosin VI, 4.5 µM PiBiP, 100 µM ATP, and the indicated actin filament concentrations. Data are from (De La Cruz et al., 2001).
Plot the observed rate constant of the exponential phase versus the [actin]. The [actin] dependence should either be linear or hyperbolic (Fig. 6.5B). If linear, the slope yields the second-order association rate constant of myosin-ADP-Pi binding to actin (KAPik+Pi′; scheme 6.1) and the maximum rate of actin-activated Pi release (k+Pi′) is more rapid than the fastest experimentally observed rate constant. If hyperbolic, the maximum observed rate constant reflects the rate of Pi release from actomyosin (k+Pi′; Pi rebinding, k−Pi′, does not contribute since Pi release is irreversible in the presence of PiBP) and the [actin] needed to reach half of k+Pi′ reflects the dissociation constant of myosin-ADP-Pi binding to actin (KAPi). The slope under multiple turnover conditions reflects the steady-state ATPase rate activity at the actin and ATP concentrations present and should compare to that measured by other methods.
5.5. ADP release
The most reliable and informative way to measure ADP binding to actomyosin is by evaluating how it affects the pyrene fluorescence enhancement associated with ATP binding (described previously). The fluorescence of mantADP can also be monitored, as described for mantATP earlier, but it is less sensitive than the pyrene fluorescence assays we describe subsequently.
ADP binding to pyrene actomyosin is not associated with a fluorescence change, so competition approaches are used to obtain the kinetic and equilibrium binding parameters. There are two different ways to design an experiment, both of which involve keeping the [ATP] constant and varying the [ADP]. One approach is to measure how ATP binds to a preequilibrated mixture of actomyosin and ADP (i.e., mix actomyosin ± ADP with a solution of ATP). In this case, the [ADP] equilibrated with actomyosin is varied (Geeves, 1989). The second is to see how actomyosin responds to addition of a solution containing both ATP and ADP (i.e., mix actomyosin with a solution of ATP and ADP). The observed time courses will follow single or multiple exponentials in either case, depending on the ADP (and ATP) binding properties (Hannemann et al., 2005). Measuring exactly how the time courses vary with [ADP] permits determination of the binding mechanism and constants.
We note that performing the ADP/ATP competition experiments both ways can serve as a useful diagnostic tool. If ADP binds actomyosin in a rapid equilibrium, time courses of pyrene fluorescence enhancement will follow single exponentials with observed rate constants that become slower as [ADP] increases regardless of how the mixing is done. If ADP dissociation from actomyosin is slow and not a rapid equilibrium, time courses will follow multiple exponentials at [ADP] that are ≈Kd for ADP binding and single exponentials at high (i.e., saturating) [ADP], regardless of how the mixing is done. Subsequently we describe how time courses could behave for each experiment and how to analyze each possible case.
5.5.1. ATP binding to an equilibrated mixture of actomyosin and ADP
The rate of ADP release, and the affinity of ADP for actomyosin is most commonly determined by measuring the rate of ATP-induced dissociation of myosin from pyrene-actin in the presence of ADP as given by the following scheme:
| (Scheme 6.4) |
The nucleotide concentrations and the method of data analysis depends on the affinity of ADP for the actomyosin complex. Two approaches are described subsequently.
Case 1
This procedure is used for myosins that have relatively weak ADP affinities. Experimental conditions are set so the rates of binding and dissociation of ADP are rapid compared with the rate of ATP binding.
Method
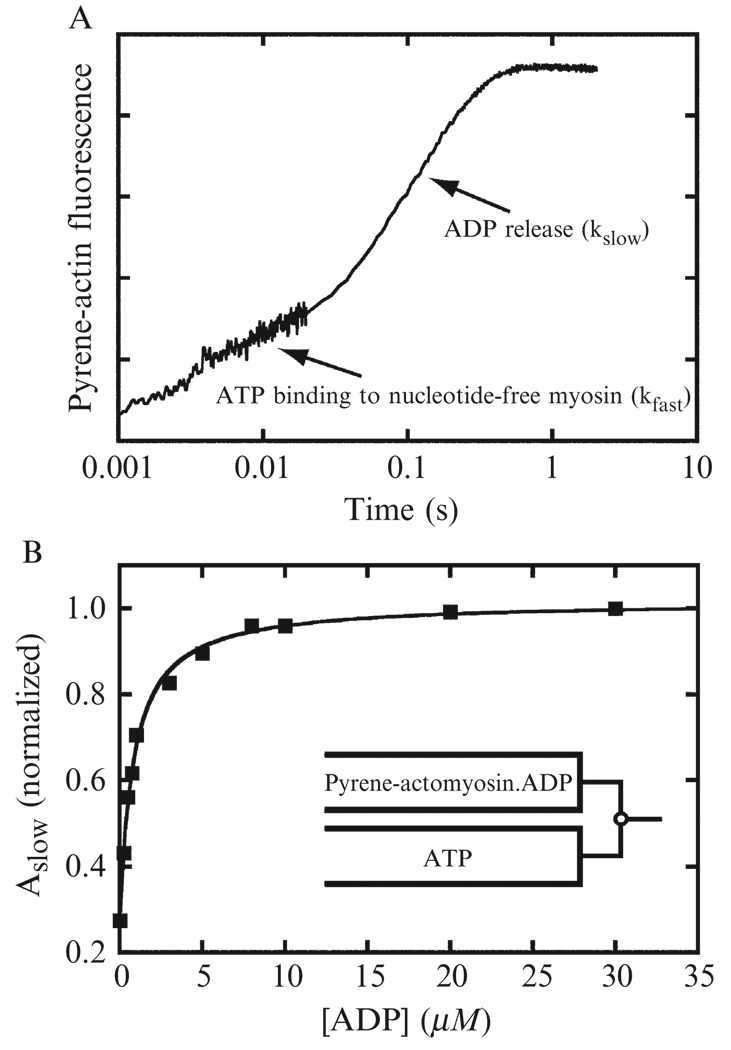
Determination of ADP affinity. Pyrene-actomyosin (≈1.0 µM) equilibrated with ADP is mixed with ATP in a stopped-flow fluorometer. Final ADP and ATP concentrations depend on the myosin properties, but 50 µM ATP and 0–500 µM ADP are suitable starting concentrations.
- Fluorescence intensities of transients increase due to the dissociation of myosin from pyrene-actin, and the transients should fit a single exponential function to obtain a rate (kobs). If the time course is best fit by two exponential rates, see Case 2. A plot of kobs versus ADP concentration should be hyperbolic (Fig. 6.6), where kobs is related to the ADP concentration by:
where k0 is the observed rate constant of ATP binding and actomyosin dissociation in the absence of ADP at a given ATP concentration (K1T′k+2T′ [ATP]; Fig. 6.1), and K2D′ K1D′ is the overall dissociation constant for ADP binding to actomyosin (Fig. 6.1).(6.11) Determination of the ADP dissociation rate constant (k+2D′). Pyreneactomyosin (≈1.0 µM) equilibrated with ADP is mixed with ATP in a stopped-flow fluorometer. The ADP concentration should be high enough that all myosin active-sties have a bound ADP. For example, if the Kd is 20 µM, the ADP concentration should be ≥100 µM. The ATP concentration should be high enough to displace the ATP. In this example, 1 mM ATP is sufficient.
Figure 6.6. ADP dissociation determined by ATP-induced dissociation of pyrene-actomyo1b.
(A) Pyrene fluorescence transient obtained by mixing 0.15 µM myo1b equilibrated with 2 µM ADP with 1 mM ATP. The time course is presented on a log scale to show the slow and fast exponential phases. (B) Normalized amplitude of the slow phase obtained by fitting pyrene transients to double exponential functions (Eq. (6.7)) as a function of ADP concentration. The solid line is a fit of the data to Eq. (6.12). Data are from (Lewis et al., 2006).
The fluorescence time course should fit a single exponential function, where the rate is equal to rate of ADP dissociation (Fig. 6.1). The rate of ADP association can now be calculated by dividing the dissociation rate by the Kd determined earlier.
Case 2
This procedure is used for myosins that have tight ADP affinities. Experimental conditions are set so the rate of ADP dissociation is slow compared with the rate of ATP binding.
Method
Pyrene-actomyosin (≈0.20 µM) equilibrated with ADP is mixed with ATP in a stopped-flow fluorometer, and the pyrene fluorescence is monitored as a function of time. Final ADP and ATP concentrations depend on the myosin properties, but 1.0 mM ATP and 0–50 µM ADP are suitable starting concentrations.
The fluorescence of the time courses increases due to ATP-induced dissociation of myosin from pyrene-actin. At low ADP concentrations, the data should be best fit by the sum of two exponential rate functions (Eq. (6.7)). The observed rate constant of the fast component (kfast) reports rapid binding of ATP to the fraction of nucleotide-free pyrene-actomyosin (Fig. 6.6A). The slow observed rate constant (kslow) reports ADP release. At high ADP concentrations, the pyrene-actomyosin should be saturated with ADP, and the transient is dominated by the slow component.
A plot of the relative amplitude of the slow component (Aslow) versus the ADP concentration is hyperbolic (Fig. 6.6B), and the overall ADP affinity (K2D′K1D′) is obtained by fitting the data to:
| (6.12) |
5.5.2. ATP and ADP binding to actomyosin
ADP binding can also be measured by kinetic competition in which a solution of ATP and ADP race to bind free actomyosin. In this case, nucleotide-free actomyosin is rapidly mixed with solutions of ATP supplemented with ADP. The ADP concentration is varied over a broad range. As described for the experiments earlier, time courses will follow single or double exponentials depending on the ADP-binding mechanism and constants. We discuss both possible cases and how to analyze the data to extract the actomyosin-ADP binding constants.
Case 1. Time courses follow single exponentials with kobs that gets slower with [ADP]
If time courses of fluorescence change after mixing pyrene actomyosin with a solution of ATP and ADP follow single exponentials at all [ADP] examined, ADP binds in a rapid equilibrium. As observed for mixing ATP to a pre-equilibrated actomyosin and ADP solution, the observed rate constants should become slower as [ADP] increases. Identical results are obtained by both mixing methods because ADP equilibrates rapidly with actomyosin during the mixing time in this experiment. That is, equilibrium between actomyosin and ADP is reached during the mixing time as it would if the sample were allowed to equilibrate before mixing with ATP. The actomyosin-ADP binding affinity can be determined as described earlier.
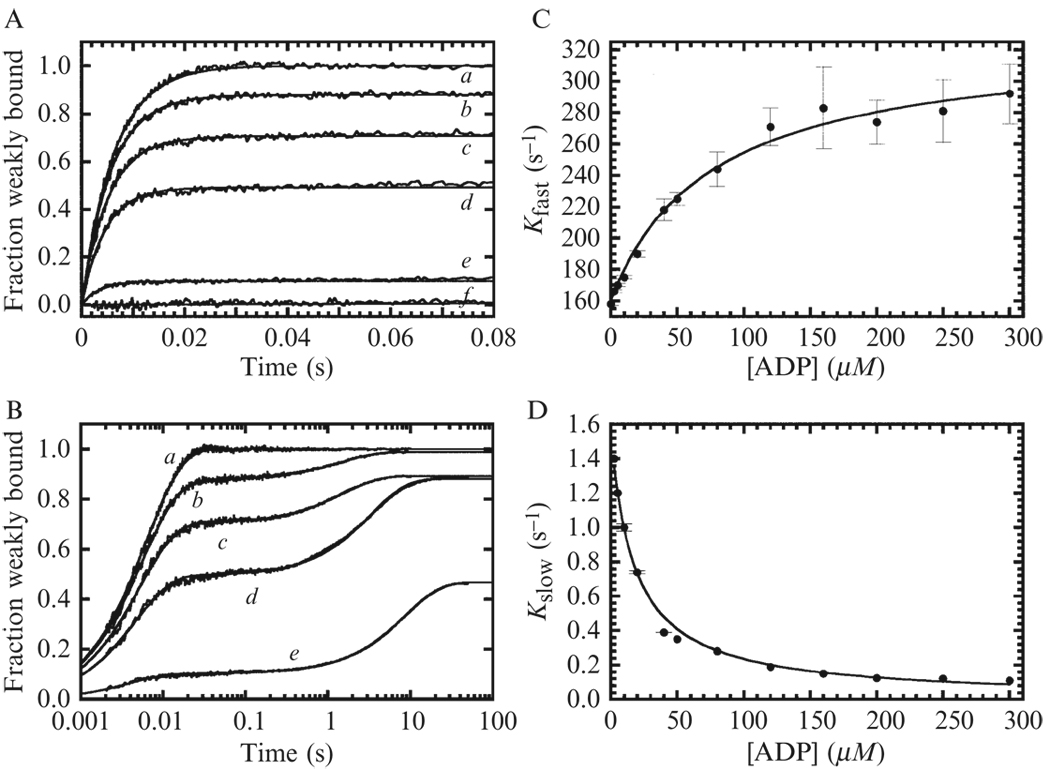
Case 2. Time courses follow double exponentials
When ADP release is slower than ATP binding and not in rapid equilibrium, time courses of pyrene fluorescence enhancement after mixing a solution of ADP and ATP actomyosin are biphasic and follow double exponentials (Figs. 6.7A–7B) with fast and slow phases that depend on the [ADP] when the [ATP] is held constant (Figs. 6.7C–7D). The method simply involves mixing actomyosin with a solution of ATP in which ADP is varied over a broad concentration range. The [ADP] dependence of the fast phase observed rate constant may depend hyperbolically on the [ADP] (Fig. 6.7C), indicating that ADP binding, like ATP binding occurs via a two-step binding process and that competitive ATP and ADP binding to actomyosin (AM) can be described by the following parallel reaction mechanism:
| (Scheme 6.5) |
where A** denotes a high (unquenched) pyrene fluorescence and the parentheses indicate collision complexes in rapid equilibrium with dissociated species. K1T′ denotes an association constant and K1D′ denotes a dissociation constant to reflect progression through the ATPase cycle (Fig. 6.1). Note that this nomenclature differs from the papers in which this method was described (Henn and De La Cruz, 2005; Olivares et al., 2006; Robblee et al., 2004, 2005).
Figure 6.7. ADP binding and dissociation from actomyosin VIIb measured by kinetic competition with ATP.
(A) Time courses of fluorescence enhancement after mixing 0.1 µM pyrene actomyosin VIIb with 100 µM ATP supplemented with either no Mg·ADP (curve a), 10 µM (curve b) 20 µM (curve c) 50 µM (curve d) or 300 µM (curve e) Mg·ADP (curve b, c, d, e). Curve f is actomyosin VIIb mixed with no nucleotide. Concentrations are final after mixing. Smooth lines through the data represent best fits to a double exponential. (B) Time courses shown on a logarithmic time scale. (C) [ADP]-dependence of the fast observed rate constants measured by kinetic competition. The solid line is the best fit to Eq. (6.14). (D) [ADP]-dependence of the slow phase observed rate constant. The solid line is the best fit to Eq. (6.15). Data are from (Henn and De La Cruz, 2005).
The observed rate constant of the fast phase (kfast) reflects the depletion of free actomyosin and therefore depends on the sum of the observed rate constants for ATP (kATP) and ADP (kADP) binding, which can be expressed as:
| (6.13) |
when nucleotide binding is irreversible which is fulfilled when nucleotide dissociation is slower than binding.
The [ADP]-dependence of the fast phase observed rate constants (kfast) should be fitted to a rectangular hyperbola in the form of the following expression:
| (6.14) |
with the ATP binding constants (K1T′ and k+2T′) constrained to the values obtained independently from ATP binding experiments (described previously). These constraints allow the ADP binding parameters (K1D′ and k−2D′) to be readily obtained.
The slow phase of the reaction arises from actomyosin-ADP formed from kinetic partitioning during the fast phase that subsequently dissociates bound ADP then binds ATP to populate the high fluorescence, weak binding states. The [ADP]-dependence of the slow phase observed rate constant (kslow) also follows a hyperbola (Fig. 6.7D), but with negative amplitude (i.e., it becomes slower as [ADP] increases). The slow observed rate constant is equal to the rate constant of ADP dissociation (k+2D′) times the probability that ATP will bind instead of ADP (kATP/(kATP + kADP)) and should be fitted to the following equation with the ATP-binding parameters (K1T′ and k+2T′) constrained, as when fitting the fast phase (described previously):
| (6.15) |
When kATP ≫ kADP (e.g., when [ADP] approaches zero and/or [ATP] ≫ [ADP]), ADP rebinding (kADP) is insignificant; ADP release is essentially irreversible; and kslow simplifies to k+2D′. The rate constant of ADP release from actomyosin (k+2D′) can therefore be readily obtained by extrapolating the best fit of kslow versus [ADP] to the limit of [ADP] = 0 (i.e., the intercept, Fig. 6.6D).
The overall ADP binding affinity is given by the product of both equilibrium constants (K1D′K2D′) and can be determined from the values of K1D′, k−2D′ (obtained from fast-phase analysis), and k+2D′ (obtained from slow-phase analysis). The final amplitudes reflect the equilibrium partitioning among strong and weak binding states as dictated by the nucleotide binding affinities and concentrations (Henn and De La Cruz, 2005; Robblee et al., 2004).
6. KINETIC SIMULATIONS
We have focused most of this chapter on designing and carrying out steady-state binding and transient kinetic experiments to directly measure the individual myosin ATPase cycle reactions and analyzing the concentration-dependence of the observed behavior by non–linear regression to extract the fundamental rate and equilibrium constants. There are often instances in which individual ATPase cycle reactions cannot be measured and/or experimental conditions required for the fitting equations to apply are not fulfilled (i.e., pseudo–first order conditions). In these cases, one must rely on kinetic simulations and global fitting to analyze the experimental data. Although analyzing experimental data with analytical solutions of the rate equations is ideal, deriving them can be complex and labor intensive (Henn et al., 2008; Johnson, 1986), particularly if analysis of the amplitudes is desired (Hannemann et al., 2005). Kinetic simulations can therefore be viewed as a practical and extremely valuable alternative. We would argue that kinetic simulations should be considered an essential part of any transient kinetic analysis, at a minimum to confirm that the derived model and associated binding constants reliably account for the experimentally observed data, including the amplitudes. We also regularly use kinetic simulations to help design experiments (e.g., what concentrations and timescales to collect data).
Because of space limitations, we will not discuss the use of kinetic simulations and global fitting in the kinetic analysis of myosin motors. Rather, we direct the reader to some key papers in which kinetic simulations have been used to characterize complex reaction pathways (Frieden, 1983; Moore and Lohman, 1994), authoritative reviews (Frieden, 1994), and tutorials (Wachsstock and Pollard, 1994), and we recommend various kinetic simulation programs that can be readily accessed for free through the Internet. We also remind readers that it is very unlikely that the constants used to fit a complex, multistep mechanism are a unique solution to the data, as only a small subset of the kinetic parameters may influence the observed signal of a given experiment. Therefore, constraining measured constants to the experimentally determined values will minimize the number of unknown fitting parameters and increase the likelihood of reliably identifying and characterizing unobservable chemical transitions.
Most kinetic simulation programs available are modern, user-friendly programs based on the original KINSIM program developed by Carl Frieden and colleagues (Dang and Frieden, 1997). These programs simulate reaction time courses of a molecular mechanism provided by the user by deriving and numerically solving the differential equations for the concentrations and flux of all chemical species identified in the mechanism. KINSIM has a companion program, FITSIM, which permits fitting mechanism parameters to real data. In general, we use KINSIM to identify plausible mechanisms that can account for the experimental data (and eliminate many that cannot) then use FITSIM to fit the data and extract the rate and equilibrium constants that best account for the data according to the defined reaction mechanism. The more recent kinetic simulation programs incorporate both simulation and fitting modules into a single program. We recommend KINSIM/FITSIM (www.biochem.wustl.edu/cflab/message.html), Tenua (http://www.geocities.com/tenua4java/), and Dynafit (http://www.biokin.com/dynafit/). We also recommend the KinTek Global Kinetic Explorer (http://www.kintek-corp.com/) and Berkeley-Madonna (http://www.berkeleymadonna.com/), but these must be purchased to access the complete software package with importing and saving options.
ACKNOWLEDGMENTS
We gratefully acknowledge support from the various funding agencies that support the research activities of our laboratories. E.M.D.L.C. thanks the National Institutes of Health for supporting myosin research activities under award GM071688, and the National Science Foundation (MCB-0546353), the American Heart Association Grant (0655849T), and the Hellman Family Foundation for supporting other research. E.M.D.L.C. is an American Heart Association Established Investigator (0940075N) and recipient of a National Science Foundation CAREER Award. E.M.O. is supported by grants from the National Institutes of Health (GM057247 and AR051174).
REFERENCES
- Bagshaw CR, Eccleston JF, Eckstein F, Goody RS, Gutfreund H, Trentham DR. The magnesium ion-dependent adenosine triphosphatase of myosin. Two-step processes of adenosine triphosphate association and adenosine diphosphate dissociation. Biochem. J. 1974;141:351–364. doi: 10.1042/bj1410351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol. Biol. Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- Chalovich JM, Eisenberg E. Inhibition of actomyosin ATPase activity by troponin-tropomyosin without blocking the binding of myosin to actin. J. Biol. Chem. 1982;257:2432–2437. [PMC free article] [PubMed] [Google Scholar]
- Coluccio LM, Geeves MA. Transient kinetic analysis of the 130-kDa myosin I (MYR-1 gene product) from rat liver. A myosin I designed for maintenance of tension? J. Biol. Chem. 1999;274:21575–21580. doi: 10.1074/jbc.274.31.21575. [DOI] [PubMed] [Google Scholar]
- Criddle AH, Geeves MA, Jeffries T. The use of actin labelled with N-(1-pyrenyl)iodoacetamide to study the interaction of actin with myosin subfragments and troponin/tropomyosin. Biochem. J. 1985;232:343–349. doi: 10.1042/bj2320343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Q, Frieden C. New PC versions of the kinetic-simulation and fitting programs, KINSIM and FITSIM. Trends Biochem. Sci. 1997;22:317. doi: 10.1016/s0968-0004(97)01062-1. [DOI] [PubMed] [Google Scholar]
- De La Cruz E, Pollard TD. Transient kinetic analysis of rhodamine phalloidin binding to actin filaments. Biochemistry. 1994;33:14387–14392. doi: 10.1021/bi00252a003. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr. Opin. Cell. Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM, Sweeney HL. Kinetic mechanism and regulation of myosin VI. J. Biol. Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Sweeney HL, Ostap EM. ADP inhibition of myosin V ATPase activity. Biophys. J. 2000a;79:1524–1529. doi: 10.1016/S0006-3495(00)76403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Actin and light chain isoform dependence of myosin V kinetics. Biochemistry. 2000b;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- Dose AC, Ananthanarayanan S, Moore JE, Burnside B, Yengo CM. Kinetic mechanism of human myosin IIIA. J. Biol. Chem. 2007;282:216–231. doi: 10.1074/jbc.M605964200. [DOI] [PubMed] [Google Scholar]
- El Mezgueldi M, Tang N, Rosenfeld SS, Ostap EM. The kinetic mechanism of Myo1e (human myosin-IC) J. Biol. Chem. 2002;277:21514–21521. doi: 10.1074/jbc.M200713200. [DOI] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C. Polymerization of actin: Mechanism of the Mg2+induced process at pH 8 and 20 degrees C. Proc. Natl. Acad. Sci. USA. 1983;80:6513–6517. doi: 10.1073/pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C. Analysis of kinetic data: Practical applications of computer simulation and fitting programs. Methods Enzymol. 1994;240:311–322. doi: 10.1016/s0076-6879(94)40053-9. [DOI] [PubMed] [Google Scholar]
- Geeves MA. Dynamic interaction between actin and myosin subfragment 1 in the presence of ADP. Biochemistry. 1989;28:5864–5871. doi: 10.1021/bi00440a024. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC. Structural mechanism of muscle contraction. Annu. Rev. Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Webb MR, Brune M, Johnson KA. Pathway of processive ATP hydrolysis by kinesin. Nature. 1995;373:671–676. doi: 10.1038/373671a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannemann DE, Cao W, Olivares AO, Robblee JP, De La Cruz EM. Magnesium, ADP, and actin binding linkage of myosin V: Evidence for multiple myosin V-ADP and actomyosin V-ADP states. Biochemistry. 2005;44:8826–8840. doi: 10.1021/bi0473509. [DOI] [PubMed] [Google Scholar]
- Henn A, Cao W, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J. Mol. Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, De La Cruz EM. Vertebrate myosin VIIb is a high duty ratio motor adapted for generating and maintaining tension. J. Biol. Chem. 2005;280:39665–39676. doi: 10.1074/jbc.M507667200. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T. New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim. Biophys. Acta. 1983;742:496–508. doi: 10.1016/0167-4838(83)90267-4. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Taylor EW. Intermediate states of subfragment 1 and actosubfragment 1 ATPase: Reevaluation of the mechanism. Biochemistry. 1978;17:3432–3442. doi: 10.1021/bi00610a002. [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamidelabelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Kovacs M, Wang F, Hu A, Zhang Y, Sellers JR. Functional divergence of human cytoplasmic myosin II: Kinetic characterization of the non-muscle IIA isoform. J. Biol. Chem. 2003;278:38132–38140. doi: 10.1074/jbc.M305453200. [DOI] [PubMed] [Google Scholar]
- Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Lewis JH, Lin T, Hokanson DE, Ostap EM. Temperature dependence of nucleotide association and kinetic characterization of myo1b. Biochemistry. 2006;45:11589–11597. doi: 10.1021/bi0611917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Tang N, Ostap EM. Biochemical and motile properties of Myo1b splice isoforms. J. Biol. Chem. 2005;280:41562–41567. doi: 10.1074/jbc.M508653200. [DOI] [PubMed] [Google Scholar]
- Lymn RW, Taylor EW. Transient state phosphate production in the hydrolysis of nucleoside triphosphates by myosin. Biochemistry. 1970;9 doi: 10.1021/bi00817a007. 2975-2583. [DOI] [PubMed] [Google Scholar]
- Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Brzeska H, Baines IC, Korn ED. Purification of myosin I and myosin I heavy chain kinase from Acanthamoeba castellanii. Methods Enzymol. 1991;196:12–23. doi: 10.1016/0076-6879(91)96004-b. [DOI] [PubMed] [Google Scholar]
- Manceva S, Lin T, Pham H, Lewis JH, Goldman YE, Ostap EM. Calcium regulation of calmodulin binding to and dissociation from the myo1c regulatory domain. Biochemistry. 2007;46:11718–11726. doi: 10.1021/bi700894h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: Evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Lohman TM. Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 2. Application of a kinetic competition approach. Biochemistry. 1994;33:14565–14578. doi: 10.1021/bi00252a024. [DOI] [PubMed] [Google Scholar]
- Oguchi Y, Mikhailenko SV, Ohki T, Olivares AO, De La Cruz EM, Ishiwata S. Load-dependent ADP binding to myosins V and VI: Implications for subunit coordination and function. Proc. Natl. Acad. Sci. USA. 2008;105:7714–7719. doi: 10.1073/pnas.0800564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares AO, Chang W, Mooseker MS, Hackney DD, De La Cruz EM. The tail domain of myosin Va modulates actin binding to one head. J. Biol. Chem. 2006;281:31326–31336. doi: 10.1074/jbc.M603898200. [DOI] [PubMed] [Google Scholar]
- Ostap EM, Pollard TD. Biochemical kinetic characterization of the Acanthamoeba myosin-I ATPase. J. Cell Biol. 1996;132:1053–1060. doi: 10.1083/jcb.132.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Assays for myosin. Methods Enzymol. 1982;85(Pt. B):123–130. doi: 10.1016/0076-6879(82)85015-5. [DOI] [PubMed] [Google Scholar]
- Robblee JP, Cao W, Henn A, Hannemann DE, De La Cruz EM. Thermodynamics of nucleotide binding to actomyosin V and VI: A positive heat capacity change accompanies strong ADP binding. Biochemistry. 2005;44:10238–10249. doi: 10.1021/bi050232g. [DOI] [PubMed] [Google Scholar]
- Robblee JP, Olivares AO, de la Cruz EM. Mechanism of nucleotide binding to actomyosin VI: Evidence for allosteric head-head communication. J. Biol. Chem. 2004;279:38608–38617. doi: 10.1074/jbc.M403504200. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Taylor EW. The mechanism of regulation of actomyosin subfragment 1 ATPase. J. Biol. Chem. 1987;262:9984–9993. [PubMed] [Google Scholar]
- Rosenfeld SS, Xing J, Whitaker M, Cheung HC, Brown F, Wells A, Milligan RA, Sweeney HL. Kinetic and spectroscopic evidence for three actomyosin:ADP states in smooth muscle. J. Biol. Chem. 2000;275:25418–25426. doi: 10.1074/jbc.M002685200. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Taylor EW. Kinetic studies on the association and dissociation of myosin subfragment 1 and actin. J. Biol. Chem. 1991;266:294–302. [PubMed] [Google Scholar]
- Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat. Struct. Mol. Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell. Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- Wachsstock DH, Pollard TD. Transient state kinetics tutorial using the kinetics simulation program, KINSIM. Biophys. J. 1994;67:1260–1273. doi: 10.1016/S0006-3495(94)80598-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HD, Belknap B, Webb MR. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 1997;36:11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- White HD, Rayment I. Kinetic characterization of reductively methylated myosin subfragment 1. Biochemistry. 1993;32:9859–9865. doi: 10.1021/bi00088a042. [DOI] [PubMed] [Google Scholar]
- Yengo CM, De la Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of myosin V. Biochemistry. 2002;41:8508–8517. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]