Summary
Opposing roles of Aurora kinases and protein phosphatase 1 (PP1) during mitosis have long been suggested. Here we demonstrate that Aurora kinases A and B phosphorylate a single residue on the kinetochore motor CENP-E. PP1 binds CENP-E via a motif overlapping this phosphorylation site and binding is disrupted by Aurora phosphorylation. Phosphorylation of CENP-E by the Auroras is enriched at spindle poles, disrupting binding of PP1 and reducing CENP-E’s affinity for individual microtubules. This phosphorylation is required for CENP-E-mediated towing of initially polar chromosomes toward the cell center. Kinetochores on such chromosomes cannot make subsequent stable attachment to spindle microtubules when dephosphorylation of CENP-E or rebinding of PP1 to CENP-E is blocked. Thus, an Aurora/PP1 phosphorylation switch modulates CENP-E motor activity as an essential feature of chromosome congression from poles and localized PP1 delivery by CENP-E to the outer kinetochore is necessary for stable microtubule capture by those chromosomes.
Introduction
Accurate chromosome segregation during mitosis requires the bipolar attachment of duplicated chromosomes to spindle microtubules emanating from opposite poles. Each time a cell divides, a specialized proteinaceous structure called the kinetochore assembles on the surface of each centromere, and it is the kinetochore that binds to spindle microtubules and directs chromosome motion during mitosis (Cleveland et al., 2003). Microtubule capture by the kinetochore is a stochastic process. Initial kinetochore attachment is often mediated via an interaction with the lateral surface of a microtubule, and kinetochores attached in this manner undergo rapid, dynein-mediated poleward motion (Rieder and Alexander, 1990). Although some chromosomes achieve biorientation without being transported to the spindle pole, dynein-mediated transport is an important mechanism to collect chromosomes to a common microtubule-dense region, where kinetochores have a greater chance of promoting efficient chromosome alignment.
Congression of polar-localized chromosomes to a metaphase position is powered by a processive, plus end-directed kinetochore motor CENP-E (Kinesin-7) (Kapoor et al., 2006; Kim et al., 2008). In various cell types and organisms, removal or inhibition of CENP-E leads to a failure in complete metaphase chromosome alignment, with a few unattached chromosomes found close to the spindle poles (Putkey et al., 2002; Wood et al., 1997; Yao et al., 2000; Yucel et al., 2000). Even the kinetochores that do become bioriented and fully aligned in the absence of CENP-E stably bind only half as many microtubules (Putkey et al., 2002). Our finding that CENP-E possesses a highly flexible and very long coiled-coil (Kim et al., 2008) raises the possibility that, while it can work advantageously for initial capture, CENP-E may also contribute, in part, to the inappropriate attachments of kinetochores. Indeed, the process of capturing spindle microtubules by kinetochores is prone to errors. Undesirable attachment frequently occurs in early prometaphase, with a single kinetochore capturing microtubules from both spindle poles (merotelic attachment), or both sister kinetochores attached to the same pole (syntelic attachment) (Cimini and Degrassi, 2005). These improper kinetochore attachments, if not resolved, can lead to chromosome missegregation and aneuploidy (Holland and Cleveland, 2009).
Correction of aberrant kinetochore attachment requires a conserved Ser/Thr kinase Aurora/Ipl1 (Lampson et al., 2004; Tanaka et al., 2002). While budding yeast has a single Aurora kinase Ipl1, metazoans express at least two Aurora kinases, Aurora A and B. Like Ipl1, Aurora B is a component of the chromosome passenger complex (together with INCENP, Survivin, and Borealin/Dasra) and is targeted to the inner centromere from prophase to metaphase (Ruchaud et al., 2007). Aurora B is thought to aid chromosome biorientation by destabilizing the kinetochore-microtubule interaction of improperly attached chromosomes (Cimini et al., 2006). Several proteins directly involved in microtubule capture at the kinetochore, including Dam1 in budding yeast and the core kinetochore microtubule binding components in metazoans (Ndc80 and KNL1), are known Aurora B substrates (Cheeseman et al., 2002; Cheeseman et al., 2006; Gestaut et al., 2008), and phosphorylation by Aurora B has been shown to decrease the affinity of these proteins for microtubules (Cheeseman et al., 2006; Gestaut et al., 2008).
Despite the high sequence similarity with Aurora B, Aurora A plays distinct roles during mitosis. Localized to the centrosomes during interphase and at the spindle poles during mitosis, Aurora A has been implicated in promoting mitotic entry and is required for centrosome maturation and separation (Marumoto et al., 2005). Inhibition of Aurora A has also been reported to cause chromosome congression defects (Hoar et al., 2007; Kunitoku et al., 2003; Marumoto et al., 2003); however, how Aurora A acts to promote chromosome alignment is unknown.
Genetic evidence in yeast (Francisco et al., 1994) and in vertebrates (Emanuele et al., 2008; Liu et al., 2010) suggest that the Aurora kinase activity is opposed by the ubiquitous Ser/Thr phosphatase, protein phosphatase 1 (PP1/Glc7). In vertebrates, PP1 isoforms α and γ can be detected at outer kinetochores (Trinkle-Mulcahy et al., 2006; Trinkle-Mulcahy et al., 2003), and PP1 has been shown to stabilize kinetochore-microtubule attachment by counteracting Aurora B kinase activity (Liu et al., 2010; Sassoon et al., 1999). Recently, the non-essential yeast protein Fin1 and conserved kinetochore protein KNL1 have been identified to target some PP1 to yeast and vertebrate kinetochores, respectively (Akiyoshi et al., 2009; Liu et al., 2010). However, whether the kinetochore possesses multiple docking modules for PP1 is not known.
Phosphorylation of the C-terminal tail of CENP-E by Cdk1, MAPK, or Mps1 has been previously proposed either to regulate CENP-E motor activity prior to its binding to kinetochores (Espeut et al., 2008) or inhibit a microtubule binding site in the tail that may link anti-parallel, midzone microtubules in anaphase (Liao et al., 1994; Zecevic et al., 1998). Additionally, among a proteomic screen of 260 mitotic phosphoproteins, CENP-E was identified to be multiply phosphorylated during mitosis (Nousiainen et al., 2006). However, the significance of the phosphorylations of CENP-E has not been established. Using purified components, selective inhibitors and a phospho-specific antibody, here we demonstrate that Aurora kinases, both A and B, phosphorylate a single conserved residue close to the CENP-E motor domain. We also identify a docking motif for PP1 that overlaps the site of phosphorylation and demonstrate that PP1 binding to CENP-E is disrupted by Aurora-mediated phosphorylation. Our findings establish an Aurora/PP1 phosphorylation switch that is required not only for congression of polar chromosomes through modulation of the intrinsic motor properties of CENP-E, but also for subsequent stable biorientation of those chromosomes by CENP-E’s delivery of PP1 to the outer kinetochore.
Results
A conserved phosphorylation site near the neck domain of CENP-E is phosphorylated by Aurora A and B in vitro
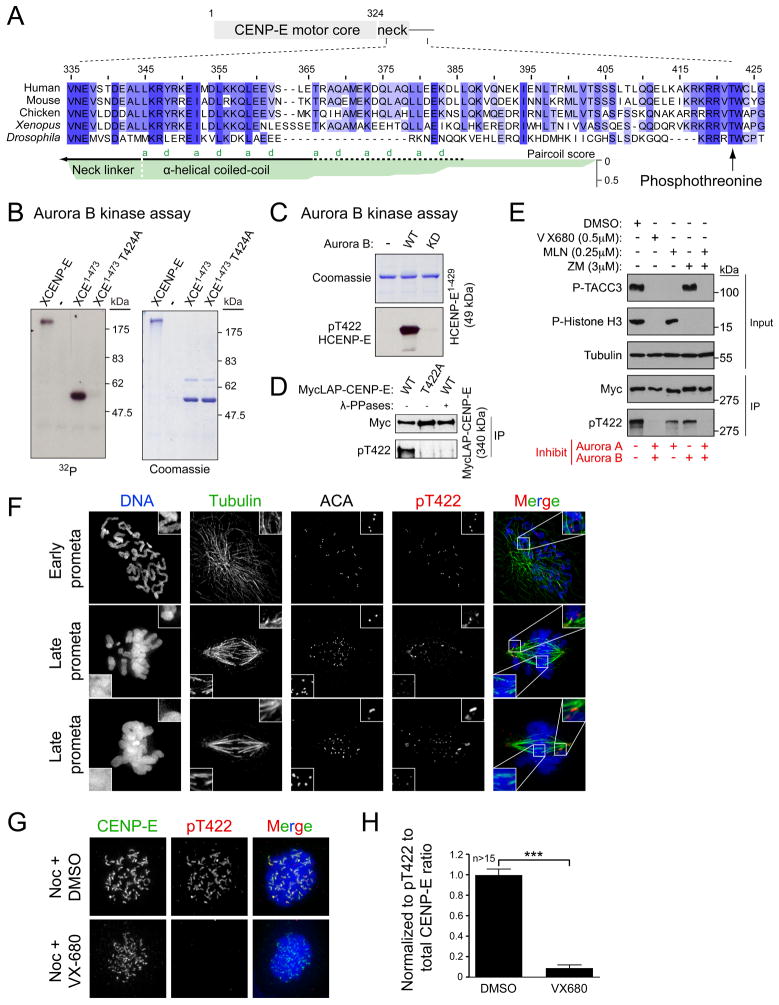
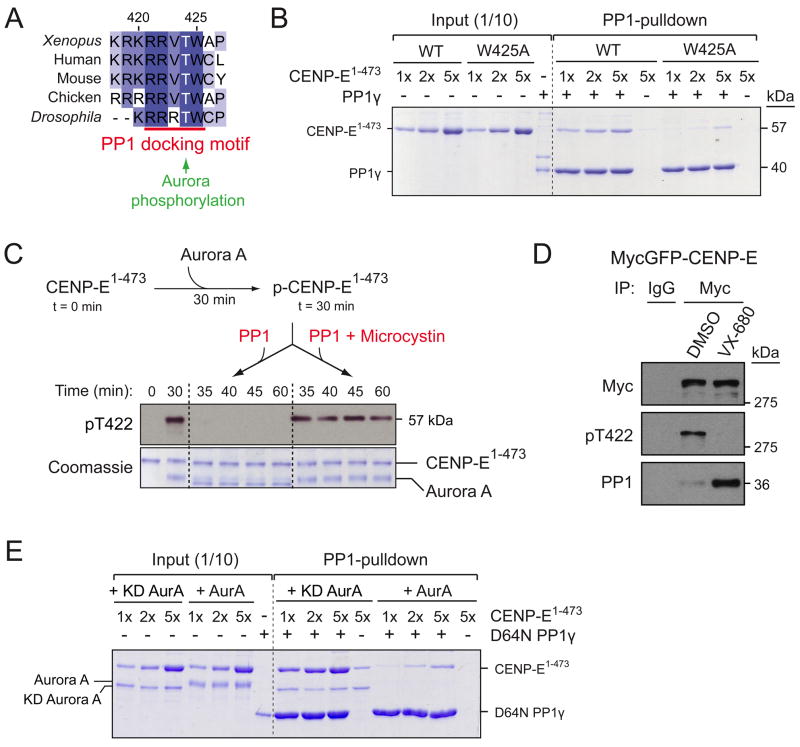
In searching for the origin of the one-dimensional diffusion found in CENP-E motility (Kim et al., 2008), we identified a highly conserved stretch of basic residues downstream of the CENP-E coiled-coil neck (Figure 1A). Consisting of 4 or more consecutive arginines or lysines, this basic stretch and the following threonine (422 in human and 424 in Xenopus laevis) are conserved in almost all the eukaryotes that possess a clear CENP-E homologue. Interestingly, the conserved threonine resides in a consensus motif for phosphorylation by Aurora kinase (Cheeseman et al., 2002) and has been previously mapped as a phosphorylation site in a mass spectrometry-based proteomic screen of mitotic spindles (Nousiainen et al., 2006).
Figure 1.
A conserved threonine close to the motor domain of CENP-E is phosphorylated by Aurora A and B on the kinetochores of unaligned chromosomes. (A) Alignment of CENP-E protein sequences using ClustalW algorithm. α-helical coiled-coil in the CENP-E neck were predicted for human CENP-E using Paircoil (Berger et al., 1995). Heptad repeat positions (a and d) in the coiled-coil are indicated. (B) In vitro kinase assays using Aurora B/INCENP to phosphorylate Xenopus full-length CENP-E or CENP-E1-473 with or without T424A mutation. Coomassie staining of purified proteins and autoradiogram showing incorporation of γ-32P ATP. (C) Coomassie staining and immunoblot for pT422 using human CENP-E1-429 incubated with wild-type (WT) or kinase dead (KD) Aurora B. (D) CENP-E immunoprecipitates from nocodazole-treated DLD-1 cells expressing either WT or T422A MycGFP-CENP-E were blotted with the pT422 antibody. Half the WT immunoprecipitate was treated with λ-phosphatase. (E) Nocodazole-arrested DLD-1 cells expressing MycGFP-CENP-E were treated with Aurora kinase inhibitors and MG132 and blotted for P-TACC3 (Aurora A substrate), P-Histone H3 (Aurora B substrate), and tubulin (loading control). CENP-E immunoprecipitates using a Myc antibody were blotted with the pT422 antibody. (F) PtK2 cells were stained for DNA (Blue), Tubulin (Green), ACA, and pT422 (Red). (G) Nocodazole-arrested HeLa cells were treated with VX-680 and MG132 and stained for CENP-E (Green), pT422 (Red) and DNA (Blue). (H) pT422 fluorescence intensity was normalized to the total CENP-E fluorescence. Plots show the mean of >15 cells per condition from two independent experiments. ***, P < 0.0001 by t test. Error bars represent SEM. See also Figure S1 and S2.
To test whether CENP-E T422 (424 in Xenopus laevis) is phosphorylated by Aurora kinases, we performed in vitro kinase assays using purified Aurora kinases and portions of Xenopus CENP-E as a substrate. Xenopus Aurora B, together with its activator INCENP, phosphorylated both full-length and a motor fragment of CENP-E (CENP-E1-473). However, Aurora B failed to phosphorylate CENP-E1-473 in which threonine 424 had been converted to alanine (Figure 1B). Xenopus CENP-E T424 was also readily phosphorylated by Aurora A (Figure S1A, B), confirming that the conserved threonine located close to the CENP-E motor domain is phosphorylated by both Aurora A and B in vitro. The stoichiometry of CENP-E1-473 phosphorylation by Aurora A saturated at two moles of PO4 per mole of CENP-E1-473, most likely with an additional phosphorylation site located C-terminal to T424, as a shorter CENP-E1-415 fragment was not phosphorylated by either Aurora kinase (Figure S1B).
Aurora A and B contribute to phosphorylation of CENP-E T422 in cells
To examine the phosphorylation of CENP-E T422 in vivo, a rabbit polyclonal antibody was generated against a phosphopeptide of human CENP-E surrounding T422 (Figure S1C). The affinity purified anti-pT422 antibody recognized recombinant human CENP-E1-429 only in the presence of active kinase (Figure 1C) and recognition of phosphorylated Xenopus CENP-E1-428 by the anti-pT422 antibody was abolished by the mutation T424A (Figure S1D). The anti-pT422 antibody also recognized wild-type (WT) CENP-E immunoprecipitated from nocodazole-arrested human (DLD-1) cells, but not CENP-E containing a T422A mutation or WT CENP-E that had been incubated with λ-phosphatase (Figure 1D). Together, these results demonstrate that the anti-pT422 antibody specifically recognizes CENP-E phosphorylated at T422.
To establish whether Aurora A or B phosphorylates CENP-E T422 in cells, we took advantage of the anti-pT422 antibody and a series of small molecule inhibitors that specifically inhibit either one or both of the Aurora kinases. As expected, treatment with the dual Aurora kinase inhibitor VX-680 (Harrington et al., 2004) abolished phosphorylation of the Aurora A substrate Transforming acidic coiled-coil 3 (p-TACC3) and the Aurora B substrate histone H3 (p-Histone H3) (Figure 1E). VX-680 treatment abolished phosphorylation of CENP-E at T422, whereas treatments with an Aurora A specific inhibitor (MLN8054) or an Aurora B specific inhibitor (ZM447439) (Ditchfield et al., 2003; Manfredi et al., 2007) resulted in only a partial reduction in T422 phosphorylation, indicating that inhibition of either Aurora kinase alone is not sufficient to eliminate the phosphorylation of CENP-E T422. However, when cells were treated with MLN8054 and ZM447439 together to inhibit both Aurora A and B, phosphorylation of T422 was completely inhibited (Figure 1E). Thus, we conclude that both Aurora A and B contribute to the phosphorylation of CENP-E at T422 in vivo.
Phosphorylation of CENP-E T422 is enriched on kinetochores close to the spindle poles
In unperturbed PtK2 cells, pT422 staining was uniformly detectable at individual kinetochores in early prometaphase, which colocalized with the centromere components recognized by autoantisera containing centromere antibodies (ACA) (Figure 1F). The kinetochore-localized pT422 signal was reduced on chromosomes congressed to the equator of the cells, but remained enriched at the kinetochores of unaligned chromosomes that are close to the spindle poles (Figure 1F). In nocodazole-treated HeLa cells, the pT422 antibody recognized a large crescent around kinetochore pairs, which colocalized with CENP-E and the outer kinetochore protein Bub1 (Figure S2A). Kinetochore-localized pT422 disappeared following depletion of CENP-E by siRNA (Figure S2B), confirming the specificity of the pT422 staining at kinetochores. Inhibition of Aurora kinases with VX-680 sharply reduced kinetochore-localized pT422 signal (Figure 1G). When normalized to the total level of CENP-E at the kinetochore (which is also reduced in VX-680 treated cells (Ditchfield et al., 2003)), a >90% reduction in T422 phosphorylation was seen following VX-680 treatment (Figure 1H), demonstrating that kinetochore-localized CENP-E is a substrate for Aurora kinases in vivo.
Aurora-mediated phosphorylation of CENP-E T422 reduces its affinity for microtubules
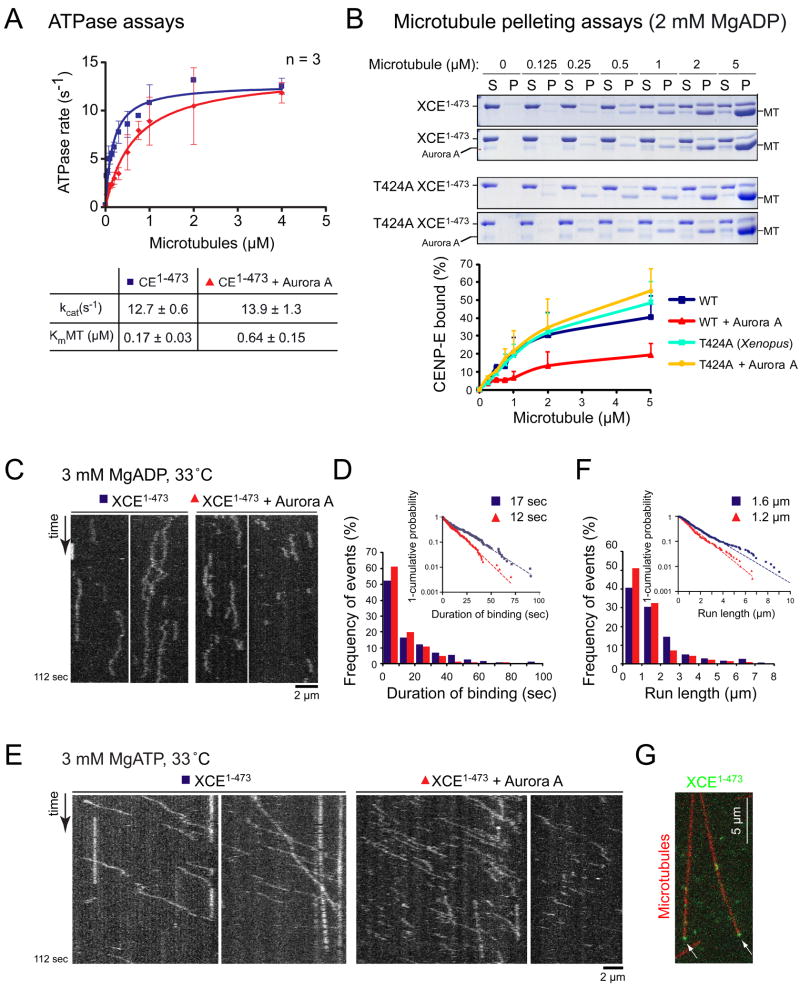
To determine if phosphorylation of T422 affects the motor properties of CENP-E, we phosphorylated T424 of Xenopus CENP-E motor (CENP-E1-473) and measured CENP-E’s microtubule-stimulated ATPase activity in the presence of an increasing concentration of microtubules (Figure 2A). The maximal ATP turnover rate (kcat) was not affected by Aurora A phosphorylation (13 ± 0.6 s−1 for CENP-E1-473, n=3; 14 ± 1.3 s−1 for CENP-E1-473 plus Aurora A, n=3) (Figure 2A). However, the concentration of microtubules required to reach the half maximal ATPase rate (KmMT) was increased by >3 fold following phosphorylation (0.17 ± 0.03 μM for CENP-E1-473, n=3; 0.64 ± 0.15 μM CENP-E1-473 plus Aurora A, n=3) (Figure 2A).
Figure 2.
Phosphorylation of T424 (Xenopus) by Aurora kinase reduces CENP-E’s affinity for microtubule and reduces its run length in vitro. (A) ATPase rates of Xenopus CENP-E1-473 and phosphorylated CENP-E1-473 measured with increasing concentrations of microtubules. Plots show the mean of three independent experiments and error bars represent SEM. kcat and KmMT values are represented ± SEM. (B) Equilibrium binding of WT or T424A CENP-E1-473 (incubated with or without Aurora A) to microtubules in the ADP state. S, supernatant; P, pellet. Percent of CENP-E1-473 bound is shown below (n=3; Error bars represent SEM). (C) Kymographs showing diffusive motion of CENP-E1-473-RFP pre-incubated with or without Aurora A in 3 mM MgADP at 33 °C. (D) Duration of binding (t) was distributed exponentially, and the mean binding time (tmean) was determined by fitting the data into a cumulative distribution function (exp [-t/tmean]). Inset shows 1-cumulative probability of CENP-E1-473-RFP binding time plotted on a log scale. The tmean is 17 ± 0.13 sec for CENP-E1-473-RFP, n = 231 and 12 ± 0.07 sec for CENP-E1-473-RFP plus Aurora A, n = 240. Values represented ± SE of the curve fit. Probability < 0.0001 that the duration of binding for CENP-E1-473-RFP plus Aurora A was distributed the same as the duration of binding for CENP-E1-473-RFP (by Kolmogorov-Smirnov Test). (E) Kymographs showing processive motion of CENP-E1-473-RFP in the presence of 3 mM MgATP at 33 °C. See Movie S1. (F) Run length of CENP-E1-473-RFP was determined by fitting the data into a cumulative distribution function. Inset shows 1-cumulative probability of CENP-E1-473-RFP run length plotted on a log scale. Mean run length is 1.6 ± 0.02 μm for CENP-E1-473-RFP, n=337 and 1.2 ± 0.02 μm for CENP-E1-473-RFP plus Aurora A, n=294. See Movie S1. Probability < 0.05 that the run lengths for CENP-E1-473-RFP plus Aurora A were distributed the same as the CENP-E1-473-RFP run lengths (by Kolmogorov-Smirnov Test). Values represent ± SE of the curve fit. (G) Accumulated CENP-E (green) at the end of the microtubules (red). See also Figure S3.
KmMT reflects CENP-E’s affinity for microtubules. In the absence of microtubules, kinesins are tightly bound to ADP in solution, and the rate of ADP release is extremely low (Hackney, 1988). However, binding of ADP-bound kinesin to microtubules greatly accelerates the rate of ADP release, and the kinesin proceeds to complete its enzymatic cycle. Since phosphorylation of CENP-E increased KmMT without significantly affecting kcat and the gliding speed (data not shown), it was likely that the phosphorylation of T424 reduces CENP-E’s microtubule affinity primarily in its ADP bound state without affecting the rate-limiting step in CENP-E enzymatic cycle (Woehlke et al., 1997). To test this hypothesis, the extent of Xenopus CENP-E1-473 binding to microtubules was determined with or without prior phosphorylation by Aurora kinase (Figure 2B). Phosphorylation of WT CENP-E1-473 by Aurora A reduced the amount of CENP-E that co-sedimented with microtubules by ~50% with a corresponding 50% increase in apparent Kd(MT) (2 μM for CENP-E1-473; 3 μM for CENP-E1-473 plus Aurora A). By contrast, Aurora A did not affect microtubule binding of T424A CENP-E1-473 (apparent Kd(MT) of 3.5 μM T424A CENP-E1-473; 3.4 μM for T424A CENP-E1-473 plus Aurora A), confirming that phosphorylation at T424 reduces the affinity of CENP-E for microtubules in the ADP state.
Phosphorylation reduces CENP-E processivity
Total Internal Reflection Fluorescence (TIRF) microscopy was used to determine how Aurora phosphorylation affects properties of individual CENP-E molecules. Xenopus CENP-E1-473 was tagged with the monomeric, photostable red fluorescent protein TagRFP-T (CENP-E1-473-RFP) (Shaner et al., 2008). Oregon Green 488-labeled GMPCPP microtubules were tethered to a coverslip in a flow chamber (Figure S3A) and CENP-E1-473-RFP was added in the presence of apyrase to induce rigor binding. As expected, CENP-E1-473-RFP was stably bound in the absence of nucleotides, and fluorescence signals were photobleached in one or two steps 89% of the time (75 double steps and 68 single step; n=160), consistent with a dimeric state for the CENP-E1-473 motor (Figure S3B). When CENP-E1-473-RFP was introduced into the flow chamber in a buffer containing ADP, both phosphorylated and unphosphorylated CENP-E1-473-RFP remained loosely bound to microtubules without displaying directional motility (Figure 2C), supporting our previous observation that CENP-E motility contains a diffusive mode that does not require ATP hydrolysis (Kim et al., 2008). Following phosphorylation, the duration of CENP-E1-473-RFP binding to microtubules was shortened by ~30% in the presence of ADP (t = 17 ± 0.13 sec for CENP-E1-473-RFP, n = 231; t = 12 ± 0.07 sec for CENP-E1-473-RFP plus Aurora A, n = 240) (Figure 2D), consistent with the observation that phosphorylation of T424 reduces CENP-E’s affinity to microtubules in the ADP bound state.
Processivity of CENP-E in the presence of ATP was reduced after phosphorylation on T424, with run lengths of phosphorylated CENP-E1-473-RFP on individual microtubules 25% shorter than those of the unphosphorylated motor (1.6 ± 0.02 μm for CENP-E1-473-RFP, n=337; 1.2 ± 0.02 μm for CENP-E1-473-RFP plus Aurora A, n=294) (Figure 2E-F). Importantly, once reaching a microtubule end using its plus end-directed motility, individual CENP-E dimers did not immediately dissociate (Figure 2G), but remained bound there for 5.8 sec (± 0.8 SEM, n=26), a feature previously observed for several other processive kinesins (Case et al., 1997; Okada and Hirokawa, 1999; Varga et al., 2006).
Phosphorylation of CENP-E T422 is required for chromosome congression
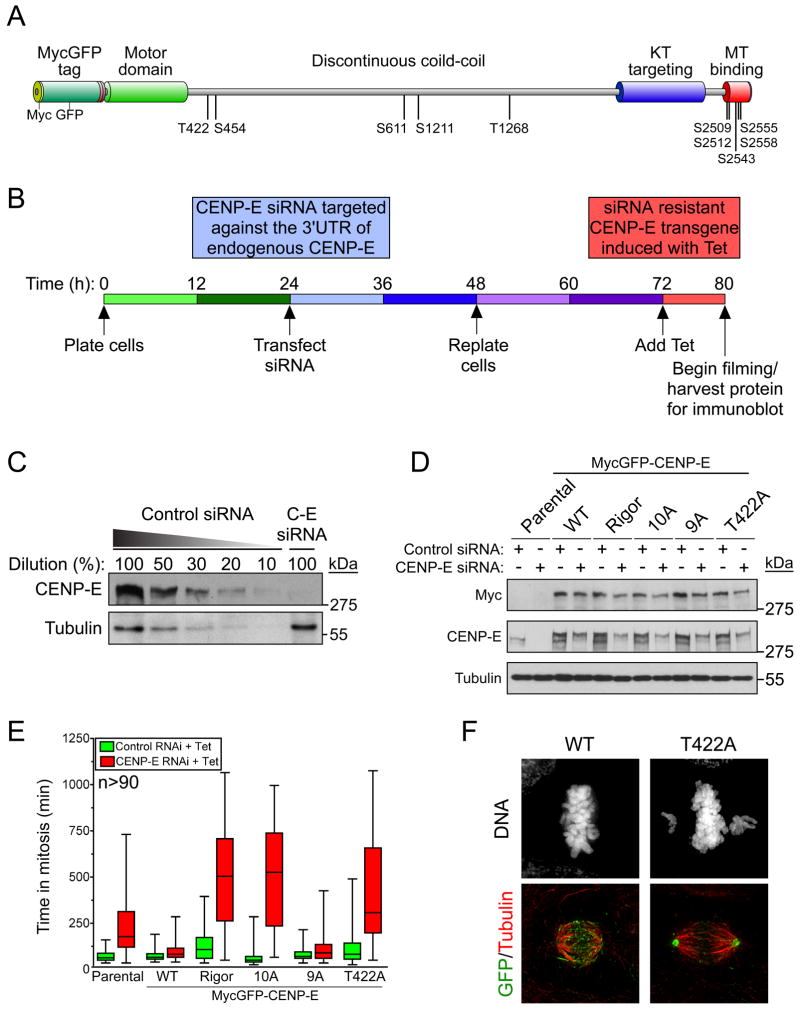
CENP-E is phosphorylated during mitosis on at least 10 sites (Figure 3A) (Nousiainen et al., 2006; Zecevic et al., 1998), albeit the significance of these phosphorylations has not been tested. To determine the consequence of preventing CENP-E phosphorylation in human cells, we developed a strategy to replace endogenous CENP-E with phosphorylation defective transgenes (Figure 3B). Full-length CENP-E fused at the N-terminus to a MycGFP epitope tag (Figure 3A) was integrated at a pre-defined genomic locus in DLD-1 cells using FRT/Flp-mediated recombination and expression was induced by addition of tetracycline (Figure S4A). Time-lapse microcopy revealed that the subcellular distribution of WT MycGFP-CENP-E closely mirrored that of endogenous CENP-E, localizing to kinetochores after nuclear envelope breakdown and relocating to the spindle midzone in anaphase and to the midbody during cytokinesis (Figure S5A).
Figure 3.
Phosphorylation of CENP-E at T422 is required for chromosome alignment. (A) Diagram of MycLAP-CENP-E transgene with previously identified phosphorylation sites. Splice variations exist within the CENP-E coiled-coil domain and phosphorylation sites are numbered with respect to their position in the CENP-E clone used in this study. (B) Schematic of replacing endogenous CENP-E with a siRNA resistant transgene. (C) Immunoblot showing depletion of CENP-E by siRNA. (D) Immunoblot showing knockdown with control (GAPDH) or CENP-E siRNA in cells expressing various MycLAP-CENP-E transgenes. (E) Box and whisker plots showing the time spent in mitosis for cells expressing MycLAP-CENP-E following transfection of control (GAPDH, green) or CENP-E (red) siRNA. > 90 cells per condition are plotted from at least three independent experiments. (F) Immunofluorescence images of cells in which endogenous CENP-E has been replaced with WT or T422A MycGFP-CENP-E. GFP (green); Tubulin (red). See also Figure S4–6 and movie S2 and S3.
Transfection of siRNA targeting the 3′ untranslated region (UTR) of CENP-E mRNA depleted endogenous CENP-E by ~90% across the population, yielding it undetectable at the kinetochores of most mitotic cells (Figure 3C and data not shown). As expected, depletion of CENP-E extended the average duration of mitosis (to 221 min) compared to control transfected cells (71 min) (Figure 3D, E). Importantly, this delay was largely rescued by the expression of MycGFP-CENP-E (producing an average of 100 min in mitosis). Replacing endogenous CENP-E with a rigor mutant (T93N) (Figure 3D) strongly exacerbated the mitotic delay (to 506 min on average) with a few chromosomes chronically misaligned near the spindle poles (Figure 3E), confirming our previous finding that the motor activity of CENP-E is essential for metaphase chromosome alignment (Kim et al., 2008). Replacement of endogenous CENP-E with a variant (10A) with all 10 phosphorylation sites abolished (by substitution to alanines) produced a robust mitotic delay (505 min on average). On the other hand, abolishing phosphorylation of the 9 sites other than T422 (9A) (Figure 3A, D) had little effect on mitotic progression (114 min on average in mitosis) (Figure 3E). Surprisingly, preventing phosphorylation of T422 alone was sufficient to produce a substantial mitotic delay (425 min on average), demonstrating that of these 10 CENP-E phosphorylation sites, phosphorylation at T422 makes the largest contribution to timely mitotic progression.
Replacing endogenous CENP-E with the T422A mutant prevented complete metaphase chromosome alignment, with a few chromosomes remaining close to the spindle poles in > 85% of cells (Figure 3F, S4B–D, S5B), a phenotype highly reminiscent of that observed with diminished levels of CENP-E (Weaver et al., 2003). Phosphorylation of T422 was not required for the kinetochore recruitment of CENP-E (Figure S6A, B). To eliminate the possibility that mutation of T422 caused defects other than simply preventing phosphorylation, we created an additional CENP-E phospho-deficient mutant, in which two arginines in the Aurora consensus motif were converted to lysines (RR:KK) (Figure S6C). Mutation of RR:KK did not abolish the epitope of the pT422 antibody (data not shown). However, recombinant Xenopus CENP-E1-428 carrying the RR:KK mutation could not be efficiently phosphorylated by Aurora A and B in vitro (Figure S6D and data not shown) and the RR:KK mutant was not phosphorylated on T422 in human cells (Figure S6E). Indeed, replacing endogenous CENP-E with the RR:KK mutant caused a mitotic delay (Figure S6F) similar to that observed with the T422A mutant with a few chromosomes remaining close to the spindle poles, confirming that phosphorylation of CENP-E at T422 is required for chromosome congression.
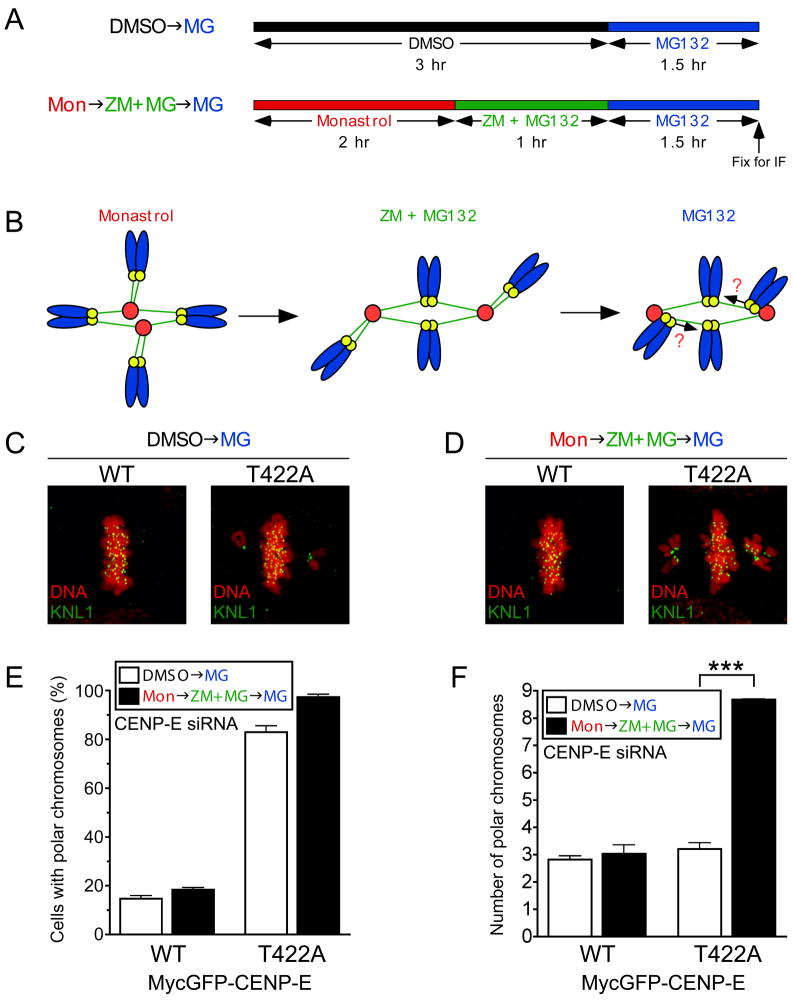
CENP-E phosphorylation by Aurora is required for congression of polar chromosomes
CENP-E has been implicated in powering chromosome congression by transporting mono-oriented chromosomes to the spindle equator along mature kinetochore fibers of already bioriented chromosomes (Kapoor et al., 2006). To test whether phosphorylation of T422 is required for this process, we adopted a method to enrich mono-oriented, polar chromosomes (Kapoor et al., 2006; Lampson et al., 2004) in cells in which endogenous CENP-E was replaced with the WT or T422A MycLAP-CENP-E (Figure 4A, B). Cells were first treated with monastrol to generate monopolar spindles with a high frequency of syntelically-attached chromosomes and released from monastrol in the presence of an Aurora kinase inhibitor (ZM) to allow bipolar spindles to form while preserving improper kinetochore attachments. Following the removal of ZM, congression of mal-oriented chromosomes was assessed (Figure 4A, B). As a control, cells were treated in parallel with DMSO to determine the extent of chromosome misalignment in an unperturbed mitosis. The enrichment of improper kinetochore attachments significantly increased the number of polar chromosomes in cells defective in T422 phosphorylation, but not in cells expressing WT CENP-E (Figure 4C–F). Live cell imaging demonstrated that, following reactivation of the Aurora kinases, improperly attached chromosomes were frequently moved to either spindle pole in cells expressing WT or T422A CENP-E (Figure S7A–C). However, these chromosomes remained closely associated with those poles in cells expressing T422A CENP-E (Figure S7C), establishing that phosphorylation of CENP-E on T422 by Aurora kinases is required for the congression of polar chromosomes.
Figure 4.
CENP-E phosphorylation by Aurora is required for congression of polar chromosomes. (A) Drug treatment scheme to enrich mono-oriented chromosomes near spindle poles. (B) Diagram illustrating attachment status of chromosomes following treatment with drug regime in (A). (C, D) Immunofluorescence images of drug-treated cells in which endogenous CENP-E was replaced with either WT or T422A MycLAP-CENP-E. KNL-1 (Green); DNA (Red). (E) Percentage of cells from (C,D) with polar chromosomes. (F) Number of polar chromosomes in cells from (C,D) displaying chromosome misalignment. Bars represent the mean of four independent experiments. ***, P < 0.0001 by t test. Error bars represent SEM. See also Figure S7 and Movie S4.
Aurora-mediated phosphorylation of CENP-E T422 opposes direct binding of CENP-E to the catalytic subunit of PP1
Following CENP-E T422 is a highly conserved tryptophan, thereby producing a RRVTW sequence that conforms to the docking motif for protein phosphatase 1 (PP1) (Figure 5A) (Hendrickx et al., 2009). Indeed, our mass spectrometry analysis of tandem affinity purified CENP-E from mitotic human cells identified the catalytic subunit of PP1 to be associated with CENP-E (data not shown) and PP1 was also present in CENP-E immunoprecipitates from nocodazole-arrested DLD-1 cells (Figure 5D). The interaction between CENP-E and PP1 is direct, as recombinant CENP-E motor (CENP-E1-473) was recovered together with PP1γ in a pulldown experiment using Microcystin-beads (Figure 5B). Recovery of a stoichiometric (1:1) complex of CENP-E and PP1 required addition of >5 molar excess of CENP-E over PP1, indicating a weak affinity between CENP-E and PP1. Further, CENP-E with a W425A substitution had markedly reduced binding to PP1 (Figure 5B), demonstrating that the interaction between CENP-E and PP1 is mediated through the PP1 docking motif. To test whether phosphorylated T422 is a substrate for PP1, phosphorylated CENP-E1-473 was incubated with either PP1γ or PP1γ pre-inactivated with the inhibitor Microcystin (Figure 5C). Monitoring of CENP-E’s phosphorylation status with the pT422 antibody revealed that PP1γ rapidly dephosphorylated CENP-E T422 (Figure 5C).
Figure 5.
Aurora phosphorylation of CENP-E T422 opposes direct binding of CENP-E to PP1. (A) CENP-E T422 flanks the conserved PP1 docking motif (R/K-R/K-V-X-F/W). (B) Coomassie staining of the PP1-bound complexes purified with Microcystin-agarose. 1, 2, or 5 molar excess of WT or W425A Xenopus CENP-E1-473 was incubated with Xenopus PP1γ. (C) In vitro phosphatase assay using PP1γ (with or without Microcystin) and phosphorylated Xenopus CENP-E1-473 as substrate. Immunoblot shows phosphorylation of T422 and coomassie stain shows purified proteins. (D) Nocodazole-arrested DLD-1 cells expressing MycGFP-CENP-E were treated with VX-680 and MG132 before harvesting. CENP-E immunoprecipitates were blotted for pT422 and PP1. (E) Coomassie staining of the PP1-bound complexes purified with Microcystin-agarose. 1, 2 or 5 molar excess of WT CENP-E1-473 was pre-incubated with either WT or kinase-dead Aurora A before incubating with catalytically inactive (D64N) PP1γ.
Previous reports have shown that phosphorylation of serine or threonine flanking the PP1 docking motif impairs the binding to PP1 (Hubbard and Cohen, 1989). Given that CENP-E T422 is overlapped by a consensus motif for Aurora kinases and a conserved motif for PP1 binding, we tested whether Aurora phosphorylation at T422 disrupts PP1’s binding to CENP-E. Following in vivo inhibition of T422 phosphorylation with the pan Aurora inhibitor VX-680, the amount of PP1 associated with CENP-E was dramatically increased (Figure 5D). Moreover, phosphorylation of CENP-E1-473 by Aurora A resulted in a ~10-fold reduction in the binding of CENP-E to the catalytically inactive (D64N) PP1γ in vitro (Figure 5E), demonstrating that Aurora-mediated phosphorylation of CENP-E T422 opposes direct binding of CENP-E to PP1.
Dephosphorylation of CENP-E T422 by PP1 is required for stable biorientation of the chromosomes congressed by CENP-E
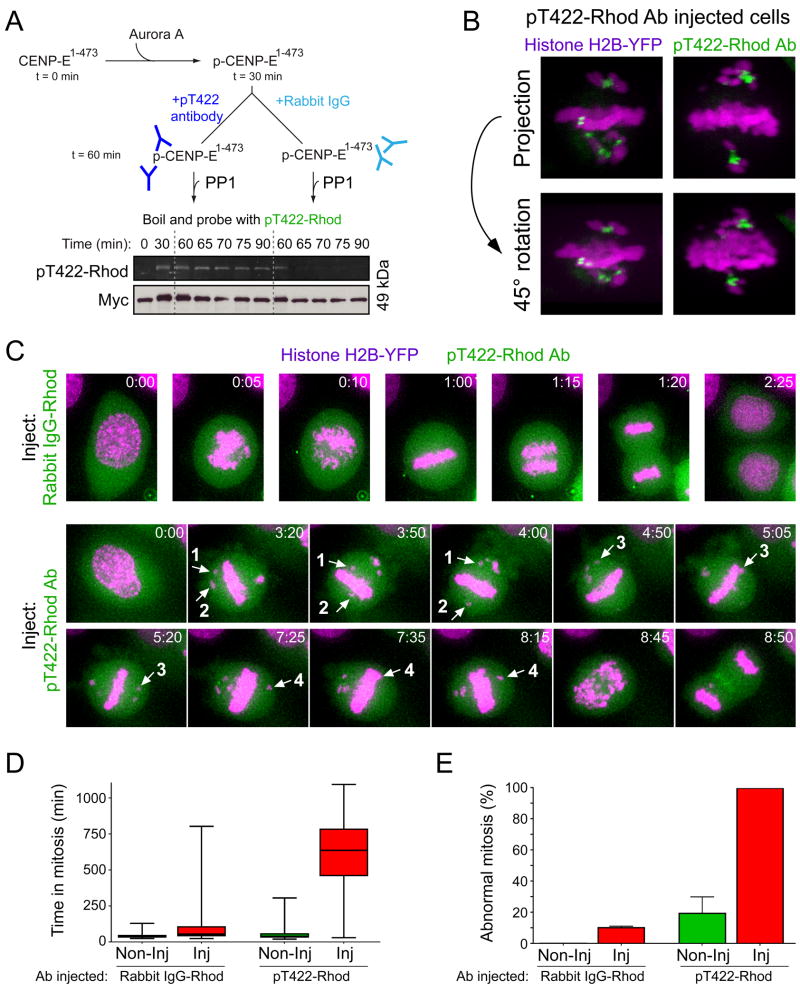
The pT422 antibody inhibited PP1-mediated dephosphorylation of Xenopus CENP-E1-473 at T424 (the position homologous to T422 in human CENP-E) in vitro (Figure 6A). Thus, to test the in vivo significance of the dephosphorylation of CENP-E T422 by PP1, we microinjected rhodamine-labeled pT422 antibodies into HeLa cells stably expressing histone H2B-YFP. Consistent with our immunofluorescence analysis (Figure 1F), the microinjected rhodamine-labeled pT422 antibody was virtually absent from aligned kinetochores, but accumulated to high levels at the kinetochores of chromosomes positioned close to the spindle poles (Figure 6B). Microinjection of the pT422 antibody substantially delayed the duration of mitosis compared to control injected cells (average 125 min for rhodamine-labeled rabbit IgG injected cells; 619 min for rhodamine-labeled pT422 injected cells) (Figure 6C–E). Interestingly, antibody-mediated preservation of phosphorylation on CENP-E T422 promoted dynamic chromosome movements distinct from the chromosome behaviors observed when T422 phosphorylation is abolished (Figure S5B). Polar chromosomes congressed to the equator of the cell, but most failed to make stable microtubule attachments and fell back out of the spindle equator or continued to move forward to the other pole (Figure 6C). Consistently, the microinjected pT422 antibody remained enriched on the kinetochores of chromosomes juxtaposed to the metaphase plate that did not form stable microtubule attachments (Figure 6B). Thus, despite CENP-E-mediated congression of chromosomes to the proximity of the spindle equator, stable kinetochore attachment (and subsequent bi-orientation) does not occur when dephosphorylation of CENP-E by PP1 is blocked.
Figure 6.
Dephosphorylation of CENP-E T422 by PP1 is required for the stable biorientation of chromosomes congressed by CENP-E. (A) Phosphorylated Xenopus CENP-E1-473 (1 μM) was pre-incubated with rabbit IgG or the pT422 antibody (3 μM) and subsequently mixed with PP1γ (0.2 μM). Phosphorylation of T422 was determined using rhodamine-labeled pT422 antibody and visualizing the rhodamine fluorophore. Anti-Myc immunoblot shows the CENP-E1-473 loading. (B) Reconstructed Z-sections of rhodamine-labeled pT422 antibody-injected live HeLa cells expressing Histone H2B-YFP. Histone H2B-YFP (purple); pT422-Rhod antibody (green). (C) Time-lapse images of antibody-microinjected (green) HeLa cells stably expressing Histone H2B-YFP (purple). Numbered arrows track the movement of four individual chromosomes which congress to the equator, but fail to stably biorient. Stills are taken from Movie S5 and S6 (D) Box and whisker plots showing the time spent in mitosis for microinjected cells. Uninjected cells in the same field of view were also examined. For each antibody, >120 microinjected cells were observed from two independent experiments. (E) Bar graphs showing the percentage of abnormal mitosis for the cells observed in (D).
Discussion
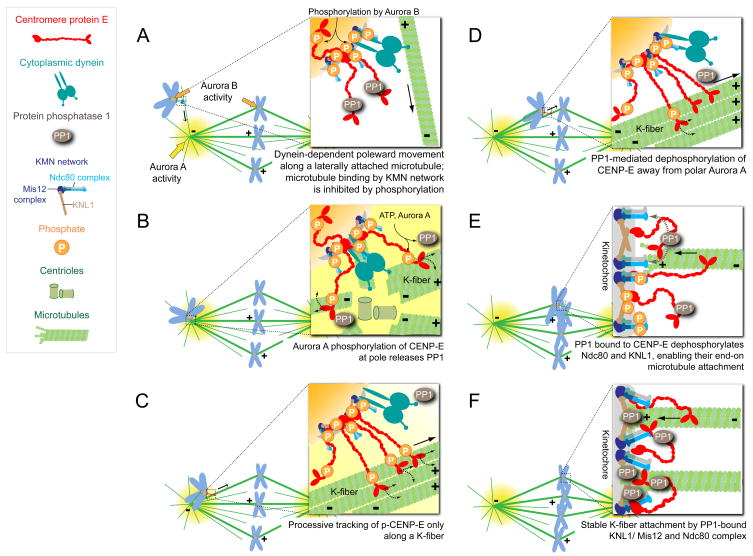
Here we demonstrate that phosphorylation by Aurora kinases of a single conserved residue (T422) close to the CENP-E motor domain is essential to promote the congression of polar chromosomes and dephosphorylation of this site is required for the stable biorientation of these kinetochores. Aurora-mediated phosphorylation of this site regulates the intrinsic motor properties of CENP-E and disrupts the binding of the opposing phosphatase PP1 to CENP-E, thereby establishing a bistable phospho-switch for regulation of CENP-E (see schematic in Figure 7).
Figure 7.
A model of CENP-E regulation by Aurora kinases and PP1. (A) Unattached chromosomes are translocated poleward along a single astral microtubule in a dynein-dependent manner (Rieder and Alexander, 1990). (B) Aurora A phosphorylates CENP-E at T422 near the spindle poles, releasing PP1 from CENP-E. Phosphorylation of CENP-E provides selectivity toward bundles of kinetochore microtubules (K-fiber) for CENP-E to glide along. (C) Our evidence here demonstrates that CENP-E powers the congression of polar chromosomes to the spindle equator along the K-fiber of an already bioriented chromosome, as earlier proposed (Kapoor et al., 2006). (D) As chromosomes congress, kinetochores move away from the Aurora A gradient concentrated at the spindle poles and CENP-E is dephosphorylated. (E) Dephosphorylated CENP-E recruits a high local concentration of PP1 to the outer kinetochores of chromosomes it has translocated away from a pole. CENP-E delivered PP1 is essential for stable, microtubule capture by the kinetochores of these towed chromosomes through its dephosphorylation of key components at the kinetochore-microtubule interface (e.g. Ndc80 and KNL1), therby increasing their affinity for microtubules. (F) Upon biorientation, kinetochore substrates become separated from the inner centromeric Aurora B and kinetochore-recruited PP1 through its direct binding to KNL1 and CENP-E stabilizes kinetochore-microtubule interactions.
The Aurora phosphorylation site on CENP-E is adjacent to its coiled-coil neck, next to several conserved positively charged amino acids. Phosphorylation at T422 diminishes the basic charge of what we propose to be an electrostatic tether directly involved in microtubule binding [analogous to similar domains identified in other kinesin family members (Okada and Hirokawa, 1999; Ovechkina et al., 2002; Thorn et al., 2000)]. Consistently, phosphorylation at T422 reduces CENP-E’s affinity for microtubules and allows the motor to dissociate more readily during processive runs. Phosphorylation of CENP-E 422 is highest on the kinetochores close to the spindle poles. Since Aurora A is concentrated at the poles, it is likely to be responsible for phosphorylation of T422 on such polar-oriented chromosomes. Aurora phosphorylation reduces the proportion of time that each motor molecule is bound unproductively to the many dynamic astral microtubules nucleated near the pole (Figure 7B). Phosphorylation-dependent reduction in CENP-E residence time on an individual microtubule of a kinetochore fiber, on the other hand, will be of little consequence, as rapid rebinding to an adjacent microtubule is likely, given the high local concentration of parallel microtubules that comprise the fiber (Figure 7C). Thus, Aurora-mediated destabilization of CENP-E tethering to individual spindle microtubules yields a variant of kinetic proofreading (Hopfield, 1974), with local, destabilized attachment as a means to eliminate incorrect initial attachments, while allowing productive CENP-E-powered movement along a kinetochore microtubule bundle.
A requirement for Aurora A in modulating CENP-E offers a mechanistic explanation for prior reports that Aurora A inhibition causes chromosome misalignment with a few chromosomes found close to the spindle poles (Hoar et al., 2007; Kunitoku et al., 2003; Marumoto et al., 2003). Although Aurora A-mediated phosphorylation of the centromere-specific histone H3 variant CENP-A has previously been proposed to promote chromosome congression (Kunitoku et al., 2003), we conclude that CENP-E is the kinetochore substrate whose Aurora A-dependent phosphorylation is directly required for chromosome congression. For Aurora B, the absence of tension exerted on mono-oriented polar kinetochores and the juxtaposed position of sister kinetochores on syntelically attached chromosomes would bring it in close proximity to the highly elongated and flexible CENP-E, allowing Aurora B phosphorylation to modulate processivity of CENP-E attached to kinetochores with reduced tension (Figure 7A, B). Further, Aurora B-dependent phosphorylation in and around the inner centromeres of sister kinetochores would also be expected to preferentially destabilize any incorrect attachments made by the 230 nm long CENP-E to microtubules that reach across the inter-kinetochore space.
Recent evidence has demonstrated that KNL1, one of the core microtubule binding components thought to be responsible for end-on attachment at metazoan kinetochores (Cheeseman et al., 2006), binds PP1 on chromosomes aligned at metaphase. Binding is through a motif for PP1 docking with an overlapping Aurora phosphorylation site (Liu et al., 2010), a situation similar to what we now report for CENP-E. Thus, the vertebrate kinetochore has evolved multiple modules for recruiting PP1, with recruitment by KNL1 and CENP-E each providing different functions. Blocking KNL1 recruitment of PP1 increased the number of kinetochores without cold stable microtubules and decreased the level of PP1 recruited to kinetochores. Nevertheless, it did not affect congression or chromosome alignment, but did lead to an unexplained inhibition of cell growth (Liu et al., 2010). In contrast, we have now shown that once CENP-E tows initially misoriented chromosomes to the cell center, its subsequent dephosphorylation and rebinding of PP1 is essential for stable microtubule attachment to the kinetochores on these chromosomes (Figure 7D). Thus, we propose a model in which CENP-E powers chromosome movement away from the high Aurora activity at poles and then exploits its flexible coiled-coil and plus end-directed motility to deliver PP1 phosphatase activity within its ~230 nm reach at the outer kinetochore (Figure 7E, F). For the kinetochores on these chromosomes, our evidence implicates dephosphorylation of the core microtubule-binding proteins (e.g., KNL1 and the four member Ndc80 complex) by CENP-E-bound PP1 as an essential step in reversing their prior inactivation by Aurora-dependent phosphorylation.
Finally, the spatial regulation of CENP-E by Aurora kinases and PP1 may provide an insight into the classic observation that phosphorylation controls the directionality of two opposing kinetochore motors on isolated chromosomes (Hyman and Mitchison, 1991). To coordinate prometaphase chromosome movement, this phosphorylation-dependent switch must turn off the minus end-directed motor and turn on the plus end-directed motor at the spindle poles. Here we have shown that the plus-end directed motor properties of CENP-E (and binding of PP1) are altered by a gradient of Aurora kinase activity emanating from the spindle poles. This provides spatial information within the mitotic spindle to regulate CENP-E activity according to the position of chromosome.
Experimental Procedures
Constructs
The full-length human CENP-E open reading frame was cloned into a pcDNA5/FRT/TO based vector (Invitrogen) modified to contain an amino-terminus Myc-LAP epitope tag. The LAP tag consists of GFP-TEV-S-peptide as previously described (Cheeseman et al., 2004). TagRFP-T (a kind gift from Roger Tsien, UCSD) was cloned into pET23d vector (Novagen) containing Xenopus CENP-E (aa1-473). This cloning strategy generates a 16 amino acid-long linker (MASMTGGGGMGRLR) between CENP-E and TagRFP-T. All human and Xenopus CENP-E mutants were generated by site-directed mutagenesis (QuickChange, Stratagene).
Cell culture and siRNA treatment
Cells were maintained at 37 °C in a 5% CO2 atmosphere in Dulbecco’s modified medium (DMEM) containing 10% tetracycline free fetal bovine serum (Clontech), 100 U/ml penicillin, 100 U/ml streptomycin and 2 mM L-glutamine. For siRNA treatment, 1.5 × 105 cells were plated in a 6 well plate and duplexed siRNAs were introduced using Oligofectamine (Invitrogen). siRNAs directed against CENP-E (5′-CCACUAGAGUUGAAAGAUA-3′) and GAPDH (5′-UGGUUUACAUGAUCCAAUA-3′) were purchased from Dharmacon. Cells were processed for immunofluorescence microscopy or live cell imaging 48 hours after transfection.
Generation of stable cell lines
Stable DLD-1, H2B-RFP cell lines expressing CENP-E were generated using the FRT/Flp-mediated recombination as described previously (Holland et al., 2010). Small molecules were used at the following final concentrations: nocodazole, 0.2 μg/ml; taxol, 10 μM; monastrol, 20 μM; S-Trityl-L-cysteine, 5 M; MG132, 20 μM; ZM447439 (Tocris Bioscience), 3 M; VX-680 (Selleck) 0.5 M and MLN8054 (a kind gift of Patrick Eyers), 0.25 M. All small molecules were from Sigma-Aldrich unless otherwise specified.
Immunofluorescence microscopy
Cells were pre-extracted for 90 sec in MTSB (100 mM PIPES pH 6.8, 0.1% triton X-100, 0.1 mM CaCl2, 1 mM MgCl2) and fixed in 2% formaldehyde in MTSB. Cells were blocked in 2.5% FBS, 0.2 M glycine, 0.1% Triton X-100 in PBS for 1 h. For the pT422 staining, cells were extracted and fixed in the presence of 500 nM Microcystin-LR (EMD). Antibody incubations were conducted in blocking solution for 1 h. DNA was detected using DAPI and cells were mounted in ProLong (Invitrogen). Images were collected using a DeltaVision Core system (Applied Precision) controlling an interline charge-coupled device camera (Cooldsnap, Raper). Kinetochore signal intensity was determined using MetaMorph (Molecular Devices), by measuring integrated fluorescence intensity with a 10 × 10 pixel square. Background signal was subtracted from an area adjacent to the kinetochore. The mean integrated fluorescence intensity of at least 10 kinetochore pairs per cell was calculated. Antibodies used are specified in the Extended Experimental Procedures.
Single molecule assays
CENP-E single molecule assays were performed as previously described (Kim et al., 2008) with the following modifications. Slides and 22 × 22-mm square coverslips were silanized as described (Helenius et al., 2006). A flow chamber was incubated with 50 μg/ml of a rat monoclonal tubulin antibody (YL1/2, Serotec) for 5 min, followed by 1% Pluronic F-127 (Invitrogen) in BRB80 for 15 min and Oregon Green 488-labeled GMPCPP microtubules for 10 min. ~0.2 mg/ml of Xenopus CENP-E1-473-RFP was incubated with 50 μg/ml of Aurora A in 20 mM Tris pH 7.5, 25 mM KCl, 1 mM MgCl2, 1mM DTT, 0.1 mM MgATP for 15 min at room temperature and diluted to ~0.5 nM before imaging in motility buffer (25 mM K-PIPES, pH 6.9, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.25% Brij35, 0.5 mg/ml casein, 4.5 mg/ml glucose, 0.2 mg/ml glucose oxidase, 0.35 mg/ml catalase, 0.5% bME) containing either 3 mM MgATP or 3 mM MgADP. Frames were captured every 500 ms with 200 ms exposure, and the typical duration of imaging was 2~3 min. Note, that since imaging was performed at an elevated temperature (33 °C) and in higher MgCl2, the speed of CENP-E movement was faster than that measured at room temperature in our previous study (Kim et al., 2008).
Supplementary Material
Acknowledgments
The authors would like to thank Stephen Taylor, Patrick Eyers, Todd Stukenberg, and Arshad Desai for providing reagents. This work was supported by a grant (GM29513) from the National Institutes of Health to D.W. Cleveland, who receives salary support from the Ludwig Institute for Cancer Research. A.J.H. is supported by a European Molecular Biology Organization (EMBO) Long-Term Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. PNAS. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The Directional Preference of Kinesin Motors Is Specified by an Element outside of the Motor Catalytic Domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J-s, Yates JR, Chan CSM, Drubin DG, Barnes G. Phospho-Regulation of Kinetochore-Microtubule Attachments by the Aurora Kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes & Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The Conserved KMN Network Constitutes the Core Microtubule-Binding Site of the Kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cimini D, Degrassi F. Aneuploidy: a matter of bad connections. Trends Cell Biol. 2005;15:442–451. doi: 10.1016/j.tcb.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED. Aurora Kinase Promotes Turnover of Kinetochore Microtubules to Reduce Chromosome Segregation Errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and Kinetochores: From Epigenetics to Mitotic Checkpoint Signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CSM, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J, Gaussen A, Bieling P, Morin V, Prieto S, Fesquet D, Surrey T, Abrieu A. Phosphorylation Relieves Autoinhibition of the Kinetochore Motor Cenp-E. Mol Cell. 2008;29:637–643. doi: 10.1016/j.molcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Francisco L, Wang W, Chan CS. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Kinesin ATPase: rate-limiting ADP release. PNAS. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking Motif-Guided Mapping of the Interactome of Protein Phosphatase-1. Chem Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Hoar K, Chakravarty A, Rabino C, Wysong D, Bowman D, Roy N, Ecsedy JA. MLN8054, a Small-Molecule Inhibitor of Aurora A, Causes Spindle Pole and Chromosome Congression Defects Leading to Aneuploidy. Mol Cell Biol. 2007;27:4513–4525. doi: 10.1128/MCB.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. PNAS. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard MJ, Cohen P. Regulation of protein phosphatase-1G from rabbit skeletal muscle. 1. Phosphorylation by cAMP-dependent protein kinase at site 2 releases catalytic subunit from the glycogen-bound holoenzyme. FEBS. 1989;186:701. doi: 10.1111/j.1432-1033.1989.tb15263.x. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Mitchison TJ. Two different microtubule-based motor activities with opposite polarities in kinetochores. Nature. 1991;351:206–211. doi: 10.1038/351206a0. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes Can Congress to the Metaphase Plate Before Biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T. CENP-A Phosphorylation by Aurora-A in Prophase Is Required for Enrichment of Aurora-B at Inner Centromeres and for Kinetochore Function. Dev Cell. 2003;5:853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- Liao H, Li G, Yen TJ. Mitotic regulation of microtubule cross-linking activity of CENP-E kinetochore protein. Science. 1994;265:394–398. doi: 10.1126/science.8023161. [DOI] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, Galvin KM, Hoar KM, Huck JJ, LeRoy PJ, et al. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. PNAS. 2007;104:4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278:51786–51795. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- Marumoto T, Zhang D, Saya H. Aurora-A - A guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. PNAS. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N. A Processive Single-Headed Motor: Kinesin Superfamily Protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- Ovechkina Y, Wagenbach M, Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J Cell Biol. 2002;159:557–562. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW. Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sassoon I, Severin FF, Andrews PD, Taba MR, Kaplan KB, Ashford AJ, Stark MJR, Sorger PK, Hyman AA. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora Kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD. Engineering the Processive Run Length of the Kinesin Motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andersen J, Lam YW, Moorhead G, Mann M, Lamond AI. Repo-Man recruits PP1γ to chromatin and is essential for cell viability. J Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andrews PD, Wickramasinghe S, Sleeman J, Prescott A, Lam YW, Lyon C, Swedlow JR, Lamond AI. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol Biol Cell. 2003;14:107. doi: 10.1091/mbc.E02-07-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Weaver BAA, Bonday ZQ, Putkey FR, Kops GJPL, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule Interaction Site of the Kinesin Motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LSB, Cleveland DW. CENP-E Is a Plus End-Directed Kinetochore Motor Required for Metaphase Chromosome Alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Yucel JK, Marszalek JD, McIntosh JR, Goldstein LSB, Cleveland DW, Philp AV. CENP-meta, an Essential Kinetochore Kinesin Required for the Maintenance of Metaphase Chromosome Alignment in Drosophila. J Cell Biol. 2000;150:1–12. doi: 10.1083/jcb.150.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic M, Catling AD, Eblen ST, Renzi L, Hittle JC, Yen TJ, Gorbsky GJ, Weber MJ. Active MAP Kinase in Mitosis: Localization at Kinetochores and Association with the Motor Protein CENP-E. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.