Abstract
Polysaccharides constitute a major component of bacterial cell surfaces and play critical roles in bacteria/host interactions. The biosynthesis of such molecules, however, has mainly been characterized through in vivo genetic studies, thus precluding discernment of the details of this pathway. Accordingly, we present a chemical approach which enabled reconstitution of the E. coli O-polysaccharide biosynthetic pathway in vitro. Starting with chemically prepared N-Acetyl-D-galactosamine-diphospho-undecaprenyl, the E. coli O86 oligosaccharide repeating unit was assembled via sequential enzymatic glycosylation. Successful expression of the putative polymerase Wzy via a chaperone co-expression system then allowed demonstration of polymerization in vitro using this substrate. Analysis of additional substrates revealed a defined mode of recognition for Wzy towards the lipid moiety. Specific polysaccharide chain length modality was furthermore demonstrated to result from the action of Wzz. Collectively, polysaccharide biosynthesis was chemically reconstituted in vitro, providing a well-defined system for further underpinning molecular details of this biosynthetic pathway.
Bacterial cell surface polysaccharides, such as lipopolysaccharides (LPSs) and capsular polysaccharides (CPSs), play important roles in processes critical for bacterial pathogenicity such as bacterium-host interactions, resistance to serum-mediated killing, and regulation of the host immune response1,2. This property has been exploited to develop polysaccharide-based vaccines against infectious diseases such as pneumonia, meningitis and sepsis3–9.
LPS typically consists of three structural parts: lipid A (endotoxin), core oligosaccharide10 and O-polysaccharide11. Of these, O-polysaccharide is the most surface-exposed region and is an important component in resistance to serum-mediated killing, phagocytosis and killing by cationic peptides12,13. While the biogenesis of LPS is a very complex process involving a large number of enzymes/complexes11, all steps in lipid A biosynthesis have been well characterized14. It is also known that the core oligosaccharide is assembled in a step-wise manner onto lipid A in the cytoplasm11. In contrast, O-polysaccharide is assembled separately onto an Undecaprenyl phosphate (Und-P, 1) carrier and then ligated to the core-Lipid A structure11,15.
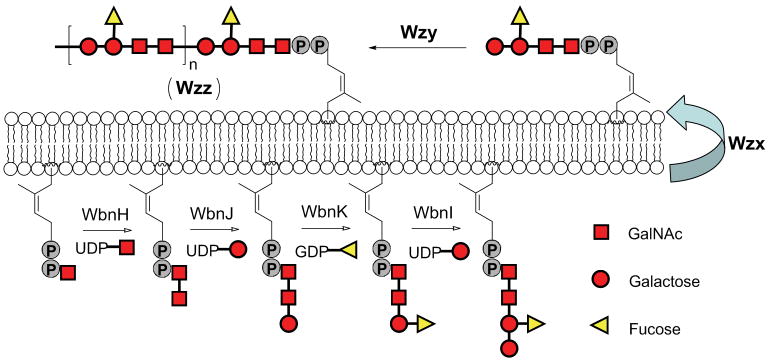
Increasing efforts have been put forth in the past several decades to better understand the biosynthesis of O-polysaccharides. Three major models have been proposed based on the genetic investigations of an increasing number of sequenced gene clusters dedicated to polysaccharide biosynthesis11,16. These models include the wzy-dependent pathway, ABC-transporter-dependent pathway and synthase-dependent pathway. Among them, the wzy-dependent pathway accounts for the synthesis of the vast majority of heteropolysaccharides17. Specifically, the pathway is initiated by the addition of a sugar phosphate to diphospho-Und to form monosaccharide-PP-Und, a precursor for oligosaccharide repeating units assembled by a series of dedicated glycosyltransferases on the cytoplasmic face of the inner membrane18,19 (Fig. 1). The flippase Wzx then translocates these repeating units to the periplasmic side of the membrane20, where the putative polymerase Wzy catalyzes their polymerization from the non-reducing end via a block-transfer mechanism21,22. The final component of the wzy-dependent pathway, the Wzz protein, is postulated to work as a chain length regulator, enabling generation of strain specific polysaccharide chain lengths11.
Figure 1.
wzy-dependent pathway of O-polysaccharide biosynthesis (E. coli O86:B7 O-polysaccharide as an example). Biosynthesis is initiated with sequential assembly of repeating units on the cytoplasmic face of the inner membrane by glycosyltransferases (WbnH, WbnJ, WbnK and WbnI), after which translocation to the periplasmic face occurs. Polymerization of repeating units on the periplasmic face of the inner membrane then follows through the action of Wzy polymerase in a block transfer mechanism which is regulated by Wzz.
Despite extensive genetic studies, this wzy-dependent model has yet to be biochemically verified in vitro. This has hampered the mechanistic studies of polysaccharide polymerization as well as investigations concerning export and assembly of polysaccharides into the outer membrane. Specifically, the following difficulties have impeded efforts towards reconstituting the wzy-dependent pathway in vitro: 1) structurally-defined repeating unit substrates are not readily available, 2) over-expression and isolation of the purified Wzy membrane protein has been unsuccessful and 3) development of a straightforward and sensitive in vitro assay to monitor polymerization has proven challenging.
Chemical approaches using homogenously synthesized substrates and purified enzymes offer a powerful complement to genetic studies, and can provide unambiguous biochemical evidence so as to help delineate the molecular details of the pathway. Previously, we initiated the investigation of O-polysaccharide biosynthesis using an E. coli O86 model system23–26. In this study, we demonstrate in vitro reconstitution of E. coli O-polysaccharide biosynthesis using chemically-defined substrates and purified biosynthetic enzymes. This study lends direct biochemical evidence to the roles of Wzy and Wzz as polymerase and chain length regulator, respectively. It also provides groundwork for further delineating molecular details of polysaccharide biosynthesis.
Results
Reconstitution of Repeating Unit Substrates
Chemical synthesis of GalNAc-PP-Und and other substrates
It is widely accepted that Und-P is the predominant lipid carrier of oligosaccharide intermediates for the biosynthesis of various bacterial polysaccharides such as peptidoglycans, LPSs, CPSs, teichoic acids, colanic acids and other carbohydrate polymers27, as well as for bacterial protein N- and O-glycosylation28–31. In nature, one of the common precursors for polysaccharide biosynthesis, N-acetyl-D-galactosaminyl/glucosaminsyl linked PP-Und (GalNAc-PP-Und, 2; GlcNAc-PP-Und, 3) is synthesized from UDP-GalNAc/GlcNAc and Und-P by the inner membrane protein WecA11. Recently, a WecA homolog from Thermotoga maritima was over-expressed and purified to homogenous form on a milligram scale32. The enzymatic synthesis of GalNAc/GlcNAc-PP-Lipid analogs nonetheless still presents a challenge given that WecA appears specific in regards to the Lipid region of the substrate19. Accordingly, we turned to chemical synthesis to obtain GalNAc-PP-Und and related compounds with different lipid moieties. Full synthetic procedures and product characterization can be found in the Supplementary Methods online.
Enzymatic assembly of repeating unit-PP-Und
Gal-α1,3-(Fuc-α1,2)-Gal-β1,3-GalNAc-α1,3-GalNAc-PP-Und, the repeating unit pentasaccharide-PP-Und (RU-PP-Und, 4), was obtained from GalNAc-PP-Und via the sequential addition of sugar residues through use of four glycosyltransferases: WbnH, WbnJ, WbnK and WbnI (Fig. 1 and 2). The intermediate formed in each enzymatic step was analyzed using LC-MS or labeled with 2-aminobenzamide and analyzed with HPLC coupled with MALDI-MS (Fig. 2). In previous studies, our lab has characterized the function of each glycosyltransferase 24,26. We were thus able to directly utilize the purified enzymes to synthesize the authentic substrate, RU-PP-Und, in a step-wise manner.
Figure 2.
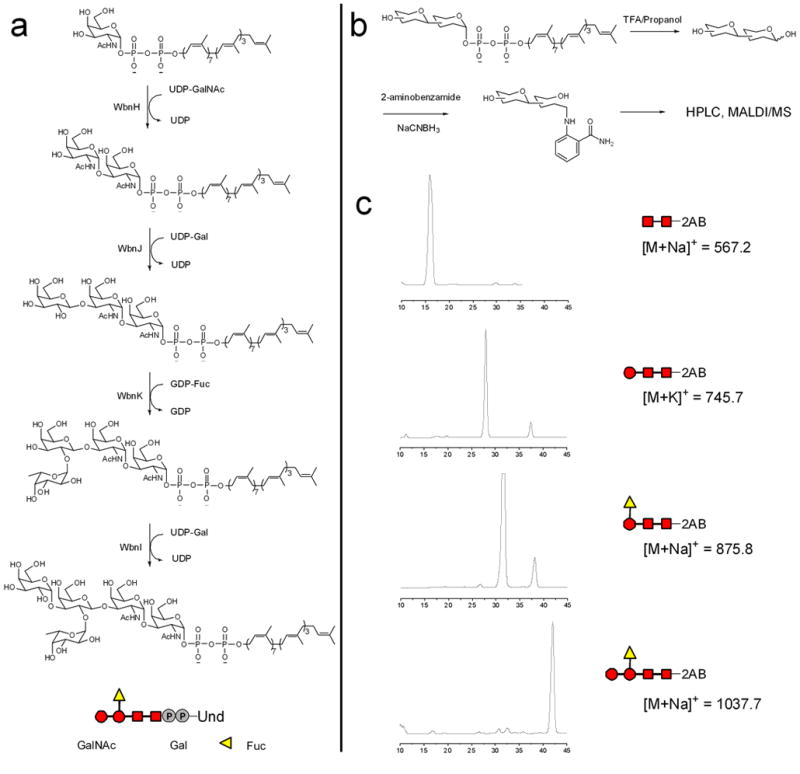
In vitro reconstitution of E. coli O86 polysaccharide repeating unit biosynthesis and associated product characterization. (a) Enzymatic synthesis of GalNAc-α1,3-GalNAc-PP-Und (5), Gal-β1,3-GalNAc-α1,3-GalNAc-PP-Und (6), Fuc-α1,2-Gal-β1,3-GalNAc-α1,3-GalNAc-PP-Und (7), and the E. coli O86 repeating unit substrate RU-PP-Und. (b) Hydrolysis and reductive amination labeling using 2-aminobenzamide (2AB, 8) to form the labeled disaccharide (9), trisaccharide (10), tetrasaccharide (11) and pentasaccharide (12). (c) HPLC profile and MALDI-MS of labeled products.
Synthesis of disaccharide GalNAc-GalNAc-PP-Und with WbnH
WbnH, an α-1,3-N-acetyl galactosaminyl transferase, has been previously shown to transfer GalNAc from UDP-GalNAc to a GalNAc-PP- C11OPh acceptor26. When WbnH was incubated with GalNAc-PP-Und and UDP-GalNAc, LC/MS analysis revealed a peak at m/z = 665.5, which corresponded to GalNAc-α1,3-GalNAc-PP-Und [M-2H]2−. To further confirm product formation, the Und lipid portion was hydrolyzed, after which the free saccharide was fluorescently labeled with 2-aminobenzamide by reductive amination for HPLC/MS analysis. A peak with a retention time of 16 min was observed (Fig. 2). When this peak was collected and subjected to ESI-MS analysis, a peak with m/z = 567.2 was identified that corresponded to the sodium ion adduct of labeled disaccharide GalNAc-GalNAc-2AB (Supplementary Fig. 1 online). These results confirm that disaccharide GalNAc-GalNAc-PP-Und was successfully synthesized in vitro.
Synthesis of tri-, tetra- and pentasaccharides
Preparation of the trisaccharide, tetrasaccharide and pentasaccharide proceeded in a manner analogous to that described above using the glycosyltransferases WbnJ, WbnK and WbnI, respectively (Fig. 2 and Supplementary Fig. 1 online).
In summary, the authentic repeating unit substrate, RU-PP-Und, was reconstituted by step-wise enzymatic reactions in vitro. This not only provides biochemical evidence for the sequential assembly model of repeating units in the wzy-dependent pathway, but also affords access to homogenous synthetic substrates.
Expression and purification of the membrane protein Wzy
The wzy gene of E. coli O86 is proposed to encode a sugar polymerase. Specifically, a mutant strain depleted of the wzy gene displays a semi-rough LPS phenotype in which only one repeating unit is linked to the Lipid-A-core23. The in vitro confirmation of polymerase activity for Wzy, however, has been hampered by the difficulty of obtaining practical amounts of purified Wzy.
Wzy of E. coli O86 is a membrane protein with 10 predicted transmembrane segments, a fact that poses a significant challenge for over-expression and purification. In this study, we constructed a recombinant plasmid, pBAD-wzy-his10, which encoded an optimized, truncated Wzy (nineteen N-terminal amino acids were removed due to the high percentage of rare codons with no effect on activity) with a C-terminal His10 epitope for western-blot detection and purification via a Ni2+ affinity column. The pBAD-wzy-his10 plasmid was co-transformed with the GroEL/GroES chaperone expression vector into an E. coli O86 mutant strain depleted of wecA and waaL for co-expression. Membrane fractions were isolated, purified by chromatography, and analyzed by SDS-PAGE with coomassie blue staining and Western blotting. Coomassie staining showed a strong purified protein band with an apparent molecular weight of 36 kDa (Supplementary Fig. 2 online), corresponding to the monomer of Wzy. Two bands at higher molecular weights were also observed. One of these bands, with an apparent molecular weight of 58 kDa, was proposed to be the dimer of Wzy, while the band showing a molecular weight of more than 250 kDa was thought to be the aggregate. While the monomer and aggregate showed positive signals on the Western blot using an antibody against the His tag, no significant band was observed for dimeric Wzy (Supplementary Fig. 2 online). Such a result likely stems from the apparent small amount of dimeric Wzy seen in the SDS-PAGE as well as the low transfer efficiency of such membrane proteins to the nitrocellulose membrane. To further confirm the identity of Wzy, the respective bands corresponding to monomer, dimer and aggregate were subjected to in-gel trypsin digestion and analysis by capillary-liquid chromatography-nanospray tandem mass spectrometry (nano-LC/MS/MS). Expectedly, multiple peptides were identified for each sample, all of which gave a credible match score to E. coli O86 Wzy (Supplementary Fig. 3 online). Such analysis thus conclusively shows that Wzy has been expressed and purified in vitro. This is, to our knowledge, the first demonstration of Wzy being isolated in purified form in vitro.
In vitro Polymerization of Repeating Units by Wzy
To investigate if purified Wzy catalyzes the polymerization of repeating units, we carried out an in vitro reaction which was monitored using a radioactivity-based assay 33,34. RU-PP-Und was labeled with 3C in the final enzymatic reaction through use of WbnI in the presence of UDP-[3C] Gal. The radio-labeled substrate was then incubated for 4 hr at room temperature with purified Wzy. The reaction was quenched with cold methanol and analyzed using paper chromatography and SDS-PAGE. The control reaction, which contained the Ni-NTApurified product from expression of an empty pBAD vector, had the highest radioactivity at an Rf of 0.75 on the paper. However, in the reaction containing Wzy, a significant amount of radioactivity appeared at a position with a low Rf value (Supplementary Fig. 4 online), indicating the formation of higher molecular weight compounds. To further verify this result, we subjected the reaction mixture to SDS-PAGE and detection by autoradiography (Fig. 3, A1 and A2). The radioactive signal on the gel gave a ladder-like pattern, consistent with the formation of compounds containing multiple repeating units. Up to 13 repeating units can be resolved under the optimized experimental conditions. It is thus evident from the PAGE assay that Wzy is both necessary and sufficient to catalyze polymerization of repeating units.
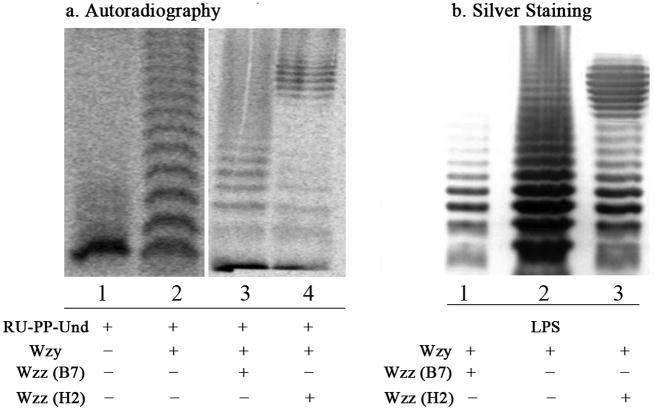
Figure 3.
Analysis of the Wzy polymerization reaction with the Und-based donor via SDS-PAGE and visualization with auto-radiography. a1. Control reaction containing Ni-affinity elution from expression of empty pBAD vector; a2. Reaction containing Ni-affinity elution of Wzy; a3. Polymerization reaction containing Wzy and Wzzfrom the B7 strain; a4. Polymerization reaction containing Wzy and Wzz from the H2 strain; b1. LPS profile of E. coli O86:B7; b2. LPS profile of E. coli O86:B7 Δwzz; b3. LPS profile of E. coli O86:B7 Δwzz complemented with wzzH2 (plasmid pTR-102). The O-polysaccharide-PP-Und generated by in vitro Wzy-Wzz reconstitution (a3 and a4) exhibits a modality similar to that of LPS regulated by WzzB7 and WzzH2 (b1 and b3), respectively. It should also be noted that the O-polysaccharides from these two strains share the same branched pentasaccharide repeating unit but differ in the anomeric configuration of the linkage25.
Generation of polysaccharide chain length modality
Cell surface polysaccharides generally have a strain-specific pattern of chain lengths termed modality that has been shown to be closely associated with the virulence of pathogens. Genetic studies suggest that modality is conferred by the Wzz protein, possibly through concerted action with Wzy35–37. To investigate this hypothesis in vitro, we introduced purified Wzz proteins obtained from two different E. coli O86 strains (B7 and H2) into the polymerization system. O-polysaccharides from these two strains share the same branched pentasaccharide repeating unit but differ in the anomeric configuration of the linkage25. Furthermore, although Wzz proteins from these two strains share 90% amino acid identity, the LPS modality of the two strains is markedly different. B7 derived LPS exhibits a very short modal distribution (2 to 5 units) (Fig. 3, B1), while H2 derived LPS exhibits an intermediate modal distribution (9 to 16 units). The same LPS modality (9–16 units) was observed for E. coli O86:B7 Δwzz complemented with plasmid pTR-102 (with wzzH2 gene)23 (Fig. 3,B3). To demonstrate its effect on chain length, purified WzzB7 or WzzH2 was added to the previous polymerization reaction mixture containing the radiolabeled repeating unit substrate and Wzy. The reaction was analyzed using SDS-PAGE/autoradiography. The radioactive signal on the gel indicated the presence of a very short modal distribution (3–6 units) in the case of WzzB7 (Fig. 3, A3), whereas that for WzzH2 showed a pattern of intermediate modality (10–14 units) (Fig. 3, A4). These patterns are similar to those observed for LPS from E. coli O86:B7 and O86:B7Δwzz complemented with wzzH2, respectively. The LPS of E. coli O86:B7 Δwzz was also provided (Fig. 3, B2) and shows a similar modality as the in vitro Wzy reaction without Wzz (Fig. 3, A2). On the other hand, incubation of Wzz with the repeating unit substrate in the absence of Wzy did not produce any detectable polysaccharides (Supplementary Fig. 4 online). Collectively, these results demonstrate that our Wzy-Wzz system is sufficient to generate polysaccharides with specific chain lengths. This observation also opens up an exciting possibility of modulating polysaccharide chain lengths in vitro.
Evaluation of lipid specificity of Wzy
We furthermore evaluated the lipid specificity of Wzy through use of various repeating unit analogs. The preparation of these analogs derived from lipids (Table 1) other than Undecaprenol followed the same sequential enzymatic synthesis as outlined above. Analysis was carried out through use of ESI-MS, the results of which can be found in the product characterization section of the Supplementary Methods.
Table 1.
Comparison of Wzy activity among repeating unit donors with variable lipid moieties. The numbers refer to the radioactivity of O-polysaccharide-PP-lipid against the total radioactivity in the paper chromatography assay [Number/100 = (total radioactivity of the chromatography paper – radioactivity of paper squares where input RU-PP-lipid was located)/total radioactivity of the chromatography paper]. The reactions were allowed to proceed for 4 hours with three replicates being performed for each lipid analog. Percentages shown are the means ± standard error of two independent experiments with triplicate determinations.
| Lipid | Geometry | Structure | Percent (%) |
|---|---|---|---|
| Undecaprenol | 7 cis, 3 trans | 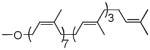 |
51 ± 5 |
| Heptaprenol | 4 cis, 2 trans | 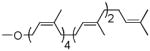 |
34 ± 4 |
| Pentaprenol | 2 cis, 2 trans | 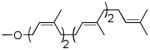 |
15 ± 2 |
| Cis-Pentaprenol | 4 cis, 0 trans | 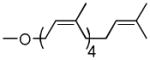 |
29 ± 4 |
| Solanesol | 0 cis, 8 trans | 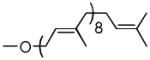 |
0 |
| MS-Pentaprenol | 1 cis, 2 trans | 3 ± 1 |
As can be seen (Table 1, entries 1–3), the amount of repeating unit successfully polymerized decreased as the length of the lipid was shortened in the Heptaprenol (13) and Pentaprenol (14) containing substrates. However, given that the number of cis-double bonds, which are adjacent to the hydroxy terminus, also decreases as lipid length decreases, one must consider that the double bond geometry rather than lipid length is the primary source of decreased polymerization levels. The donor derived from cis-Pentaprenol (15) was thus evaluated. The degree of polymerization observed, in comparison to that seen with Pentaprenol was notably greater (Table 1, entry 4). This indicates that double bond geometry is a more important factor than lipid length in determining polymerization efficiency. To further verify this hypothesis, the Solanesol (16) derived donor (Table 1, entry 5) was tested as it is only two isoprene units shorter than the natural lipid, Und, while containing only trans double bonds. No detectable polymerization was observed with this donor. Finally, it had not escaped our attention that eukaryotic systems utilize Dolichol as the lipid moiety in donor substrates rather than Undecaprenol. The importance of unsaturation in the α-isoprene unit was therefore considered through use of the Monosaturated Pentaprenol (17) based donor (Table 1, entry 6). The lack of activity observed using this donor verifies the expected preference for bacterial-based Und-containing substrates rather than eukaryotic-based Dolichol-containing substrates.
Discussion
Bacteria possess polysaccharides with enormous structural complexity. Efforts to understand their biosynthesis and export have been driven by their importance in host-bacterium interface biology and, in particular, their strong association with bacterial pathogenicity. While such efforts have revealed the wzy-dependent pathway to arguably be the most widely distributed polysaccharide biosynthetic pathway in nature, convincing evidence that Wzy functions as a polymerase has not previously been presented. Rather, assignment of Wzy as the putative polymerase was based on the generation of a semi-rough LPS phenotype upon genetic depletion of the wzy gene. Such assignment can be problematic, however, and warrants caution as deletion of the wzy gene can potentially disrupt critical interactions and propagate changes in the overall biosynthetic pathway. Definitive assignment of polymerase activity has instead awaited a well-defined, reconstituted system that consists of a minimal yet sufficient number of components. We have accordingly demonstrated that purified Wzy is both necessary and sufficient to induce polymerization of chemically synthesized oligosaccharide substrates in vitro. Thus, we present direct and unambiguous biochemical evidence of Wzy as a sugar polymerase.
While not necessary to effect polymerization, Wzz is also proposed to play an interesting, but as of yet mechanistically undefined, role in imparting a defined modal distribution to the polysaccharides. Generation of modal distributions consistent with those of E. coli O86:H2 and O86:B7 upon introduction of the respective Wzz protein into our in vitro system further confirms the role of Wzz in this process. Determination of precisely how Wzz imparts modality, however, remains an even more intriguing topic. Currently, two possible mechanisms have been proposed. One possibility suggests that Wzz functions as a molecular timer, modulating Wzy activity between two states that favor either the polymerization or ligation reaction38. The alternative model, however, suggests that Wzz functions as a molecular ruler, regulating the oligomerization state of a complex between Wzy and the ligase WaaL to define the modality39. Structural studies on periplasmic domain and full length Wzz seem to favor the molecular ruler model. Small-angle X-ray scattering data40, crystallographic evidence41 and the most recent electron microscopy results42 have all suggested that Wzz proteins form oligomers. Nonetheless, no direct evidence illustrates how these oligomers modulate the chain length of O-polysaccharides. Several pieces of evidence do suggest the presence of multi-protein complexes in the wzy-dependent pathway35,36. In addition, it has been shown in vivo that chain length modality is conferred prior to the ligation reaction43, suggesting that regulation occurs on the polymerizing substrate. Our in vitro reconstitution system further confirms such a hypothesis and suggests the existence of a meaningful interaction among Wzy, Wzz and polyisoprenoid lipid-linked polymers. Work is ongoing in our lab to identify Wzy-Wzz interactions using a pull down assay, and to modulate chain length modality by varying the stoichiometry of Wzy-Wzz in the system. We expect our well-defined system will thus provide further insights into the mechanism of action of Wzz.
Apart from Wzy and Wzz, also central to our studies was Und-P, the predominant lipid carrier in bacteria for the assembly of a diverse range of polysaccharides. Unlike typical glycosyltransferase-catalyzed reactions, where the diphospho-lipid moiety is part of the acceptor structure, in the Wzy-catalyzed polymerization, the diphospho-lipid moiety also serves as part of the donor substrate and is directly involved in bond breaking in the reaction. Therefore, it was speculated that the lipid structure might have a more dramatic effect in this instance. This notion garnered support from lipid specificity studies of TGase44 and PglB45,46, which both use similar diphospho-lipid sugar moieties as donors. Our specificity studies with Wzy revealed a preference similar to those observed in those studies, suggesting a defined mode of recognition for Wzy towards lipid structures in the sugar donor. Additionally, it is now apparent that much simpler lipid based donors can be utilized in future studies of this system, thus remedying problems associated with the use of the highly hydrophobic lipid, Und.
Since the wzy-dependent pathway had only been studied in vivo prior to the studies described herein, many molecular details are yet to be revealed. For example, how is the activity of Wzy functionally regulated? Does Wzy interact with Wzz as suggested by previous in vivo studies? If so, is the interaction required for chain length regulation? The answers to these critical questions now appear obtainable with our in vitro reconstitution of this pathway. Thus, this system has supplied the groundwork necessary for probing the molecular details of Wzy polymerization and Wzz chain length regulation and represents one step forward towards understanding and re-defining the mechanisms of polysaccharide biosynthesis in general.
Methods
Bacterial strains, plasmids and materials
E. coli O86:K61:B7 (ATCC 12701) was obtained from American Type Culture Collection (Rockville, MD, USA). E. coli DH5α [lacZΔM15 hsdR recA] competent cells were obtained from Gibco-BRL Life Technology. E. coli C43 (DE3) competent cells were obtained from Lucigen Co. (Middleton, WI). Expression plasmid pBAD-myc-His was purchased from Invitrogen (Carlsbad, CA). HiTrap Chelating Ni HP column was obtained from GE Healthcare (Piscataway, NJ). All other chemicals and solvents were purchased from Sigma-Aldrich. GroES/EL expression plasmid, pGro7, was kindly provided by Dr. Jian Liu from the University of North Carolina.
Expression and purification of glycosyltransferases
Four glycosyltransferases (WbnH, WbnJ, WbnK and WbnI) were expressed and purified to homogeneity by Ni-affinity chromatography or GST-affinity chromatography as previously described24,26.
Cloning, expression and purification of Wzy
The optimized wzy with a His10 tag at the C-terminus was obtained from GeneScript (Piscataway, NJ) and inserted into the pBAD vector (Invitrogen) to form the pBAD-wzy-his10 recombinant plasmid. This plasmid was co-transformed with pGro7 vector into E. coli O86 (ΔwecA, ΔwaaL) for protein expression. Cells were cultured in 6 L of low-salt LB medium (10 g tryptone, 5 g yeast extract, 5 g NaCl for 1 L) with the antibiotics ampicillin (100 μg/mL) and chloramphenicol (34 μg/mL). Expression was induced with 0.2% L-arabinose followed by further incubation at 20 °C for 20 hrs. Cells were harvested and washed with 20 mM Tris-HCl (pH 7.5). Cells were disrupted by sonication, followed by centrifugation (10,000 g, 10 min, 4 °C) to remove debris. Membrane fractions were isolated by ultracentrifugation (150,000 g, 60 min, 4 °C) and solubilized in 5 mL buffer A (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5% sodium dodecanoyl sarcosine (SDDS)) overnight at 4 °C. Unsolubilized material was removed by ultracentrifugation (150,000 g, 60 min, 4 °C), after which soluble material was incubated with 1 mL of Ni-NTA resin (Qiagen) pre-equilibrated with buffer B (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM Imidazole, 0.1% n-Dodecyl-β-D-maltopyranoside (DDM), 10% glycerol). After washing with 100 mL of buffer B containing different imidazole concentrations (10 mM, 20 mM and 50 mM), bound proteins were eluted with 5 mL of buffer C (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 250 mM imidazole, 0.1% DDM, 10% glycerol). The fraction was concentrated and desalted using an ultra-filtration tube (Millipore, MA). Further purification by FPLC then used a SP Sepharose Fast Flow column (Pharmacia) equilibrated with buffer D (50 mM Tris-HCl buffer (pH 7.5), 1 mM dithiothreitol (DTT), 5% glycerol, 0.1% DDM). Elution was performed using a 0–100% buffer E (buffer D with 1 M NaCl) gradient over a 30 min interval. The final purification step involved a superdex 200 column (Pharmacia) with an eluent of buffer F (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 0.1% DDM, 10% glycerol).
Cloning, expression and purification of Wzz from E. coli O86:B7 and O86:H2
Details can be found in the Supplementary Methods online.
Chemical synthesis of GalNAc-PP-Lipid Substrates
Full synthetic procedures and product characterization can be found in the Supplementary Methods online.
Enzymatic synthesis of RU -PP-Lipid substrates
To a buffer containing 20 mM Tris-HCl (pH 7.0) and 5 mM MnCl2, 5 mg of GalNAc-PP-Lipid (solubilized in DMSO) was added followed by 1% (v/v) Triton X-100/10 mM UDP-GalNAc. The mixture was vortexed vigorously and sonicated for 10 min in a water bath. The reaction was initiated by the addition of 50 μg of the enzyme WbnH and was incubated at room temperature for 2 hrs or until the reaction was complete. The reaction product was purified using reverse phase chromatography with methanol as the eluent. About 0.01 mg of the dried product was incubated with 50 μL of n-propanol:2 M trifluoroacetic acid (1:1) at 50 °C for 20 min to release the Lipid-PP moiety. After evaporating the mixture to dryness, hydrolyzed sugars were then labeled with 2-aminobenzamide by reductive amination47. Post-labeling clean-up was achieved by ascending the mixture on chromatography paper with acetonitrile overnight. The labeled product was then eluted from the paper using 2 × 500 μL distilled water. The eluted product mixture was further separated on a normal phase analytical HPLC column using 50 mM aqueous ammonium formate (pH 4.5) (solvent A) and acetonitrile (containing 5 mM ammonium formate, solvent B). A gradient of 48–80% solvent B was used at a flow rate of 0.5 mL/min. The peaks were detected with a fluorescence detector (λex=330 nm,λem=420 nm), collected and then characterized by MALDI-MS. Remaining product was re-solubilized in enzyme reaction buffer and elaborated to the pentasaccharide-PP-Lipid product using the above procedures.
In vitro polymerization of RU-PP-Lipid substrates by Wzy
The 3C-labeled pentasaccharide-PP-Lipid was prepared for the final enzymatic reaction using WbnI and 50 μM UDP-[3C]-Gal. For the paper chromatography assay, the reaction mixture (10 μL) containing 0.03 mM radiolabeled pentasaccharide-PP-Lipid, 0.4% Triton-X100 and 50 μg purified Wzy protein was incubated at room temperature for 4 hr in 20 mM Tris-HCl (pH 7.5), 5 mM MnCl2 and 100 mM NaCl. The reaction was quenched with an equal volume of cold methanol. After centrifugation to remove the precipitate, the mixture was concentrated to < 10 μL and spotted on Whatman 3 MM chromatography paper (1×14 cm). The paper strip was developed in isobutyric acid: 1 M NH4OH (5:3, v/v). The radioactivity was analyzed as previously described44 (Supplementary Fig. 4 online). For the SDS-PAGE assay, the reaction mixture was quenched with an equal volume of SDS loading buffer containing 50% glycerol and 0.001% bromophenol blue. This mixture was electrophoresed in the Bio-Rad Mini-PROTEAN system. The gel was soaked in Amplify Fluorographic Reagent (GE Healthcare) for 45 min, dried and exposed to Hyperfilm MP (GE Healthcare) at −80 °C for 6 days before development.
In vitro generation of chain length modality
10 μg of purified WzzH2 or WzzB7 was added to the aforementioned polymerization reaction mixture. The reaction was allowed to proceed overnight, after which it was quenched and analyzed by SDS-PAGE/autoradiography as described above.
Analysis of LPS
Details can be found in the Supplementary Methods online.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jian Liu for providing the GroES/EL expression vector and helpful discussions of expression conditions. R.W. acknowledges the NIH Predoctoral Trainee Program (TM GM008512). L.L. acknowledges support from the China Scholarship Council (2007102057). P.G.W. acknowledges the National Cancer Institute (R01 CA118208), NIH (R01 GM085267), NSF (CHE-0616892), Bill & Melinda Gates Foundation (51946) for financial support.
Footnotes
Author Contributions: R.W., W.Y. and P.G.W. designed research. R.W., L.L., W.Y., G. Z., H.E., R.S.P., H.G., J.K.S., E.M., L.C., P.K., X.L., W.H., W.Z., Y.D. and M.L. performed research. L.L., W.Y., G. Z., H.E. and P.G.W. analyzed data. R.W., L.L., W.Y., G.Z. and P.G.W. wrote the paper.
Supplementary Information is available as a separate file.
References
- 1.Robbins JB, Schneerson R, Egan WB, Vann W, Liu DT. Virulence properties of bacterial capsular polysaccharides - unanswered questions. Life Sci Res Rep. 1980;16:115–32. [Google Scholar]
- 2.Moxon ER, Kroll JS. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- 3.Finn A. Bacterial polysaccharide-protein conjugate vaccines. Br Med Bull. 2004;70:1–14. doi: 10.1093/bmb/ldh021. [DOI] [PubMed] [Google Scholar]
- 4.Plotkin SA. Vaccines: past, present and future. Nat Med. 2005;11:S5–11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briles DE, Paton JC, Swiatlo E, Crain MJ. Gram-Positive Pathog. 2. 2006. Pneumococcal vaccines; pp. 289–298. [Google Scholar]
- 6.Westphal O, Jann K, Himmelspach K. Chemistry and immunochemistry of bacterial lipopolysaccharides as cell wall antigens and endotoxins. Prog Allergy. 1983;33:9–39. [PubMed] [Google Scholar]
- 7.Costerton JW, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–64. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 8.Jenkinson HF. Adherence and accumulation of oral streptococci. Trends Microbiol. 1994;2:209–12. doi: 10.1016/0966-842x(94)90114-k. [DOI] [PubMed] [Google Scholar]
- 9.Sahly H, Keisari Y, Crouch E, Sharon N, Ofek I. Recognition of bacterial surface polysaccharides by lectins of the innate immune system and its contribution to defense against infection: the case of pulmonary pathogens. Infect Immun. 2008;76:1322–1332. doi: 10.1128/IAI.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holst O. The structures of core regions from enterobacterial lipopolysaccharides -an update. FEMS Microbiol Lett. 2007;271:3–11. doi: 10.1111/j.1574-6968.2007.00708.x. [DOI] [PubMed] [Google Scholar]
- 11.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield C, Amor PA, Koplin R. Modulation of the surface architecture of gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol Microbiol. 1997;23:629–38. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 13.Nesper J, et al. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun. 2001;69:435–45. doi: 10.1128/IAI.69.1.435-445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abeyrathne PD, Lam JS. WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol Microbiol. 2007;65:1345–59. doi: 10.1111/j.1365-2958.2007.05875.x. [DOI] [PubMed] [Google Scholar]
- 16.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 17.Guo H, Yi W, Song JK, Wang PG. Current understanding on biosynthesis of microbial polysaccharides. Curr Top Med Chem. 2008;8:141–51. doi: 10.2174/156802608783378873. [DOI] [PubMed] [Google Scholar]
- 18.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J Bacteriol. 2007;189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Dabbagh B, Mengin-Lecreulx D, Bouhss A. Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J Bacteriol. 2008;190:7141–6. doi: 10.1128/JB.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Cole RA, Reeves PR. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol. 1996;178:2102–7. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbins PW, Bray D, Dankert MA, Wright A. Direction of chain growth in polysaccharide synthesis. Science. 1967;158:1536–42. doi: 10.1126/science.158.3808.1536. [DOI] [PubMed] [Google Scholar]
- 22.Kanegasaki S, Wright A. Mechanism of polymerization of the Salmonella O-antigen: utilization of lipid-linked intermediates. Proc Natl Acad Sci U S A. 1970;67:951–8. doi: 10.1073/pnas.67.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H, et al. Molecular analysis of the O-antigen gene cluster of Escherichia coli O86:B7 and characterization of the chain length determinant gene (wzz) Appl Environ Microbiol. 2005;71:7995–8001. doi: 10.1128/AEM.71.12.7995-8001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi W, et al. Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J Am Chem Soc. 2005;127:2040–2041. doi: 10.1021/ja045021y. [DOI] [PubMed] [Google Scholar]
- 25.Yi W, et al. Two different O-polysaccharides from Escherichia coli O86 are produced by different polymerization of the same O-repeating unit. Carbohydr Res. 2006;341:100–8. doi: 10.1016/j.carres.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Yi W, et al. The wbnH gene of Escherichia coli O86:H2 encodes an alpha -1,3-N-acetylgalactosaminyl transferase involved in the O-repeating unit biosynthesis. Biochem Biophys Res Commun. 2006;344:631–639. doi: 10.1016/j.bbrc.2006.03.181. [DOI] [PubMed] [Google Scholar]
- 27.Valvano MA. Undecaprenyl phosphate recycling comes out of age. Mol Microbiol. 2008;67:232–5. doi: 10.1111/j.1365-2958.2007.06052.x. [DOI] [PubMed] [Google Scholar]
- 28.Wacker M, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli Science. 2002;298:1790–3. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 29.Kowarik M, et al. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science. 2006;314:1148–50. doi: 10.1126/science.1134351. [DOI] [PubMed] [Google Scholar]
- 30.Faridmoayer A, Fentabil MA, Mills DC, Klassen JS, Feldman MF. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol. 2007;189:8088–98. doi: 10.1128/JB.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faridmoayer A, et al. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J Biol Chem. 2008;283:34596–604. doi: 10.1074/jbc.M807113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Dabbagh B, Blanot D, Mengin-Lecreulx D, Bouhss A. Preparative enzymatic synthesis of polyprenyl-pyrophosphoryl-N-acetylglucosamine, an essential lipid intermediate for the biosynthesis of various bacterial cell envelope polymers. Anal Biochem. 2009;391:163–165. doi: 10.1016/j.ab.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Barrett D, et al. Analysis of glycan polymers produced by peptidoglycan glycosyltransferases. J Biol Chem. 2007;282:31964–71. doi: 10.1074/jbc.M705440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Synthesis of heptaprenyl-lipid IV to analyze peptidoglycan glycosyltransferases. J Am Chem Soc. 2007;129:3080–1. doi: 10.1021/ja069060g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniels C, Morona R. Analysis of Shigella flexneri wzz (Rol) function by mutagenesis and cross-linking: wzz is able to oligomerize. Mol Microbiol. 1999;34:181–94. doi: 10.1046/j.1365-2958.1999.01591.x. [DOI] [PubMed] [Google Scholar]
- 36.Bengoechea JA, et al. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J Bacteriol. 2002;184:4277–87. doi: 10.1128/JB.184.15.4277-4287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J Bacteriol. 2006;188:5124–5135. doi: 10.1128/JB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bastin DA, Stevenson G, Brown PK, Haase A, Reeves PR. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–34. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 39.Morona R, van den Bosch L, Manning PA. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol. 1995;177:1059–68. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang KH, Guo H, Yi W, Tsai MD, Wang PG. Investigation of the conformational states of Wzz and the Wzz. O-antigen complex under near-physiological conditions. Biochemistry. 2007;46:11744–52. doi: 10.1021/bi701181r. [DOI] [PubMed] [Google Scholar]
- 41.Tocilj A, et al. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol. 2008;15:130–8. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- 42.Larue K, Kimber MS, Ford R, Whitfield C. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J Biol Chem. 2009;284:7395–403. doi: 10.1074/jbc.M809068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels C, Griffiths C, Cowles B, Lam JS. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ Microbiol. 2002;4:883–97. doi: 10.1046/j.1462-2920.2002.00288.x. [DOI] [PubMed] [Google Scholar]
- 44.Ye XY, et al. Better substrates for bacterial transglycosylases. J Am Chem Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 45.Glover KJ, Weerapana E, Numao S, Imperiali B. Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chem Biol. 2005;12:1311–5. doi: 10.1016/j.chembiol.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen MM, Glover KJ, Imperiali B. From peptide to protein: comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry. 2007;46:5579–85. doi: 10.1021/bi602633n. [DOI] [PubMed] [Google Scholar]
- 47.Bigge JC, et al. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230:229–38. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.