SUMMARY
Genetic conservation allows ancient features of fat storage endocrine pathways to be explored in C. elegans. Multiple studies have used Nile red or BODIPY-labeled fatty acids to identify regulators of fat mass. When mixed with their food, E. coli bacteria, Nile red, and BODIPY-labeled fatty acids stain multiple spherical cellular structures in the C. elegans major fat storage organ, the intestine. However, here we demonstrate that, in the conditions previously reported, the lysosome-related organelles stained by Nile red and BODIPY-labeled fatty acids are not the C. elegans major fat storage compartment. We show that the major fat stores are contained in a distinct cellular compartment that is not stained by Nile red. Using biochemical assays, we validate oil red O staining as a method to assess major fat stores in C. elegans, allowing for efficient and accurate genetic and functional genomic screens for genes that control fat accumulation at the organismal level.
INTRODUCTION
Many pathways regulating energy homeostasis are conserved between humans and the nematode C. elegans. Genome-wide investigation of genes that alter fat storage in whole-living animals is possible in C. elegans using classical forward genetic approaches (Jones et al., 2009; Mak et al., 2006; Soukas et al., 2009) or functional genomic approaches such as RNAi (Ashrafi et al., 2003; McKay et al., 2007). Thus, unlike in higher metazoans, a comprehensive identification of the signals exchanged between tissues that mediate energy storage and mobilization and the centers of appetite control is possible in C. elegans.
Nile red is a phenoxazone, lipophylic dye derived from Nile blue that is concentrated in hydrophobic environments when used in vitro or in cell lines (Fowler and Greenspan, 1985; Greenspan et al., 1985). In a hydrophobic environment, Nile red undergoes an increase in yellow-gold fluorescence, making it a useful indicator of lipid droplets in cells or TLC fractionated lipids (Bonilla and Prelle, 1987; Fowler et al., 1987). BODIPY-labeled fatty acids have similar properties when used in cell culture (Guo et al., 2008). Both stains have been used as vital dyes on cells, tissues, or, in the case of C. elegans, on living animals. Nile red and BODIPY-labeled fatty acids stain living worms when mixed with their food, E. coli bacteria (Ashrafi et al., 2003; Mak et al., 2006). The C. elegans metabolism field has extensively used these fluorescent dyes to indicate fat stores, as they stain vesicular structures in the C. elegans main fat storage organ, the intestine. More than 45 papers have used Nile red as a proxy for fat mass in C. elegans, ascribing a fat regulatory role to more than 400 genes with an altered Nile red phenotype. However, Nile red poorly stains the germline, eggs, and hypodermis, tissues known to be high in fat content by other measures. In addition, for the insulin-receptor-like mutant daf-2, two recent analyses showed a decreased Nile red or BODIPY-fatty acid staining phenotype that is opposite to the increased fat storage revealed by biochemical analysis (Soukas et al., 2009; Wang et al., 2008). These and other paradoxical findings presented here indicate that Nile red or BODIPY fatty acids are not accurate proxies for major fat stores in the worm.
The C. elegans intestine has several classes of morphologically and functionally distinct ‘granules’ or vesicles. One class of large granule is autofluorescent and birefringent; is acidic; stains with Nile red and acridine orange; expresses PGP-2, GLO-3, FUS-1, and other markers; and is likely to be lysosome-related organelles (LROs) (Hermann et al., 2005; Nunes et al., 2005; Schroeder et al., 2007; Treusch et al., 2004). By virtue of this granule staining with Nile red, LROs were thought to be a site of major lipid storage in C. elegans (Hermann et al., 2005; Schroeder et al., 2007; Rabbitts et al., 2008). Other classes of granules are neutral, not autofluorescent, are suggested to be Nile red negative, and express endosomal markers such as RME-1 and RAB-5 or the late-endosomal markers LMP-1 and RAB-7 (Nunes et al., 2005; Rabbitts et al., 2008; Treusch et al., 2004). Electron microscopy in some mutants with defective LRO formation suggests that there are additional, distinct, small granules that may be classical lysosomes (Ruaud et al., 2009).
Here we show that instead of staining C. elegans major fat stores, Nile red and BODIPY-labeled fatty acids, under the conditions previously reported, stain acidified cellular compartments, or LROs. Further, we show that LROs stained with Nile red, BODIPY-labeled fatty acids, or PGP-2::GFP largely do not colocalize with neutral lipid droplets stained with the fixative-based dyes LipidTOX and oil red O. In concert with these findings, we report that mutants with altered Nile red or BODIPY phenotypes often show no lipid phenotype or the opposite phenotype to that indicated by vital dyes when measured by lipid biochemistry. In addition, Nile red levels do not drop upon fasting, whereas oil red O staining does decrease upon fasting. We conclude that Nile red and BODIPY-labeled fatty acids do not stain the major fat stores in C. elegans, and that major fat stores are contained in subcellular compartments independent of the Nile red-stained compartment. Finally, we put forward oil red O staining as a new method to study fat mass in C. elegans, and validate it using quantitative lipid biochemistry.
RESULTS
Mutant Analyses Reveal Inconsistencies between Nile Red and Triglyceride Levels
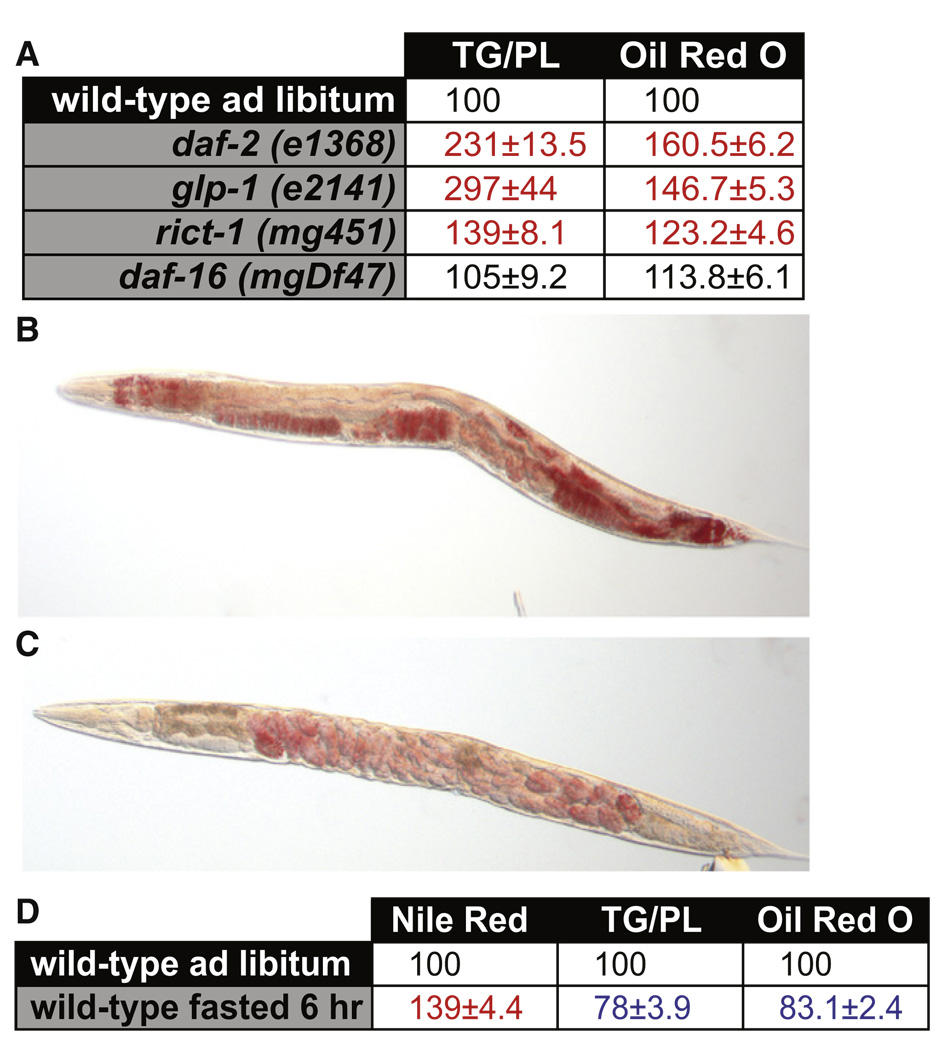
We identified multiple instances where the Nile red or C1-C12-BODIPY-labeled fatty acid staining of C. elegans did not agree with biochemical measurement of triglycerides, the major form of long-term energy stores. daf-2 mutants have been demonstrated to have increased fat mass by fixative-based staining with Sudan black (Kimura et al., 1997) and biochemically (Ashrafi et al., 2003; Perez and Van Gilst, 2008). In our original report of the use of Nile red as a vital dye for fat storage, we reported that, consistent with these biochemical results, animals containing loss-of-function mutations in daf-2, the sole C. elegans insulin/IGF receptor, had increased Nile red staining (Ashrafi et al., 2003).However, in our recent analyses, we were not able to repeat the finding that Nile red staining is increased in daf-2 mutants, finding instead that daf-2 mutants show decreased Nile red staining (Table 1 and Soukas et al., 2009; Wang et al., 2008).
Table 1.
Nile Red and Triglyceride Levels Do Not Correlate in Several Metabolic Mutants
| Strain | Nile Red (Percent of Wild-Type) |
Triglyceride/ Phospholipid (mg/mg, Percent of Wild-Type) |
|---|---|---|
| Wild-type | 100 ± 4.9 | 100 ± 16.6 |
| daf-2(e1368) | 79 ± 8.3 | 231 ± 13.5 |
| tph-1(mg280) | 49.7 ± 1.1 | 156.7 ± 4.5 |
| tub-1(nr2004) | 155 ± 5.1 | 103 ± 3.8 |
| kat-1a | 194 ± 10.1 | 127.6 ± 20.6 |
| tub-1(nr2004);kat-1(mg368) | 337 ± 13.7 | 128.7 ± 39.6 |
| glp-1(e2141) | 91 ± 7.1 | 297 ± 44 |
| rict-1(mg451) | 240 ± 11.1 | 129 ± 4.9 |
| daf-16(mgDf47) | 142 ± 5.5 | 110 ± 9.2 |
Nile red mean intensity measured for least 30 animals from at least two biological replicates is shown in the first column. Quantitative triglyceride/phospholipid measurements done with at least two independent biological replicates are shown in the second column. Data are shown as mean ± SEM.
kat-1 analyses were done with two different alleles: kat-1(mg368) for Nile red staining and kat-1(mg447) for lipid biochemistry.
To determine whether Nile red and BODIPY-labeled fatty acids were generally discrepant from biochemical lipid quantification, we examined several other mutants with robust Nile red phenotypes. Mutations in daf-16, the C. elegans FOXO transcription factor ortholog, cause an increase in Nile red; however, lipid biochemistry reveals no difference in fat stores between daf-16 and wild-type worms (Table 1). A 140% increase in Nile red staining is seen in rict-1 mutants, whereas triglyceride levels are only increased 30%. Similarly, mutants in the kat-1 ketothiolase gene, annotated to play a role in fatty acid oxidation, show a 94% increase in Nile red signal (Table 1) and a comparable increase in BODIPY-labeled fatty acid staining (Mak et al., 2006). kat-1 mutants reanalyzed with quantitative lipid biochemistry indicated no significant difference in triglyceride mass (Table 1). Mutants in tub-1, the C. elegans homolog of the mammalian tubby gene, which, when mutated, causes late-onset obesity in rodents (Kleyn et al., 1996), show a 50% increase in Nile red staining (Ashrafi, 2007) but no difference in biochemically measured triglycerides (Table 1). Moreover, double mutants in tub-1 and kat-1, which have a synergistic accumulation of Nile red and BODIPY-labeled fatty acids (Mak et al., 2006), also show no significant increase in triglyceride mass (Table 1). Finally, germline-deficient animals, which have been reported to have reduced fat mass (Wang et al., 2008), show a paradox similar to daf-2, i.e., lower than normal Nile red but more than doubled fat mass measured biochemically (Table 1).
Nile Red Does Not Stain the Major C. elegans Fat Stores
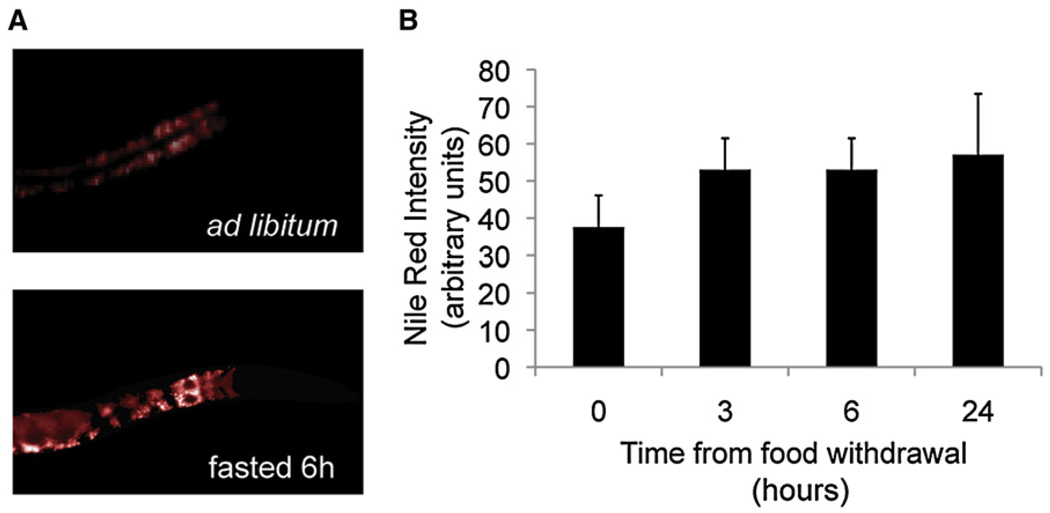
Major animal fat stores primarily take the form of triglycerides and are utilized during periods of food deprivation. However, Nile red signal increases in animals fasted for 6 or 12 hr in the presence of Nile red (Figure 1), an unexpected result if Nile red were indicating triglyceride mass. As expected, biochemically measured triglycerides fall with fasting (Figure 3). This indicates first that the increase in Nile red signal is deceptive, and second that Nile red does not accurately indicate C. elegans triglyceride stores. Supporting this, Nile red poorly stains the germline, eggs, and hypodermis, tissues known to be high in fat content by other histochemical methods. Moreover, neutral lipid droplets stained postfixation with the fluorescent neutral lipid dye LipidTOX green or red mostly do not colocalize with Nile red (Figure 2A) or C1-C12-BODIPY fatty acid-stained granules (see Figure S1 available online), respectively. A 2.2% ± 0.79% overlap was seen between Nile red and LipidTOX green vesicles in doubly stained L3 animals imaged by confocal or apotome Z stacks (n = 5 animals, three 1000× fields per animal were scored; a total of 2163 Nile red and 2024 LipidTOX green vesicles were counted).
Figure 1. Nile Red Signal Increases upon Fasting.
(A) Nile red signal redistributes and becomes more intense upon fasting. Representative images of 1 day adult worms fasted or fed ad libitum for 6 hr are shown.
(B) Nile red intensity increases upon fasting. Quantitative analysis of Nile red intensity after food removal is depicted. Nile red mean intensity was measured for least 30 animals from at least two biological replicates. Data are shown as mean ± SEM.
Figure 3. Oil Red O Stains the Major C. elegans Fat Stores.
(A) Oil red O intensity serves as a proxy for C. elegans triglyceride levels. Quantitative triglyceride/phospholipid measurements done with at least two independent biological replicates are shown in the first column. Oil red O signal measured for least 30 animals from at least two biological replicates is shown in the second column. Data are shown as mean ± SEM.
(B) Oil red O stains fat droplets in all major C. elegans fat storage organs. Oil red O signal is observed in C. elegans intestine, hypodermis, gonad, and eggs.
(C) Oil red O signal drops upon fasting.
(D) Oil red O, but not Nile red, stains long-term C. elegans energy stores. Quantitative analysis of Nile red, triglycerides measured by lipid biochemistry, and oil red O levels upon fasting are presented. Data are shown as mean ± SEM.
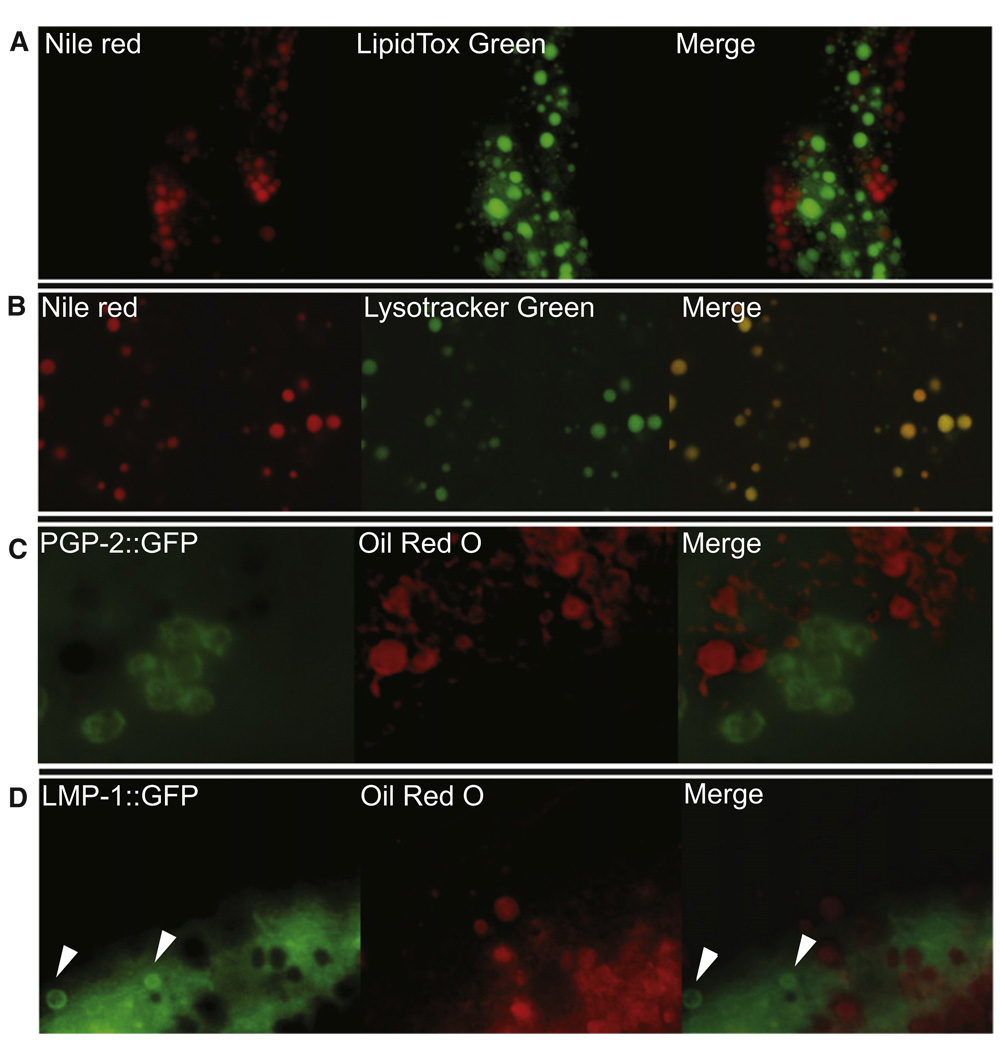
Figure 2. Lysosome-Related Organelles and Neutral Lipid Vesicles Are Mostly Nonoverlapping Compartments.
(A) Neutral lipid droplets stained postfixation with LipidTOX green do not colocalize with Nile red-positive organelles (see also Figure S1).
(B) Nile red colocalizes with the lysosome-specific fluorescent dye LysoTracker Green.
(C) Oil red O-positive vesicles do not colocalize with the PGP-2-positive LRO compartment.
(D) Oil red O-positive vesicles do not colocalize with the LMP-1 LRO compartment. (Note that the oil red O images were pseudocolored in order to allow merging with green fluorescent images.)
Nile Red Stains C. elegans Lysosome-Related Organelles, and Oil Red O Stains Major C. elegans Fat Stores
Given that the Nile red signal distribution and appearance resembles the compartment known as ‘gut granules’ and that Nile red staining is absent in mutants with defective lysosomal vesicles (Schroeder et al., 2007), we hypothesized that Nile red or BODIPY-labeled fatty acids may be treated as xenobiotics and partitioned into a degradative compartment. We tested this hypothesis by simultaneously staining with the vital dyes and the lysosome-specific fluorescent dye LysoTracker. The perfect overlap between Nile red and C1-C12 BODIPY and the lysosome-specific dyes indicates that in the conditions used here and previously reported, Nile red and C1-C12 BODIPY stain a lysosome-like compartment (Figure 2B and Figure S1).
Because Nile red and BODIPY-labeled fatty acids do not reveal major fat stores, we sought an alternative histological technique to determine fat distribution and levels in C. elegans. We tested four fixative-based neutral lipid dyes: fluorescent dyes LipidTOX neutral-lipid green and red, and the nonfluorescent dyes Sudan black and oil red O. LipidTOX green and red, even after extensive permeabilization of the cuticle, stain a very small proportion of animals (less than 10%).Additionally, the signal is very sensitive to photobleaching, an unattractive characteristic for quantification of lipid stores. Sudan black staining was highly variable when used to stain adult animals due to the required final destaining wash with ethanol. Slight variation in the timing of this destaining step greatly affects the final intensity of the Sudan black signal, making the technique error prone. In contrast, oil red O staining, as presented here, allows preservation of worm anatomy and low sample-to-sample variability.
As in the case of vesicles stained with the neutral lipid dye LipidTOX Green (Figure 2A), oil red O-positive vesicles are mostly nonoverlapping with the LRO and late endosomal compartments. More than one gut granule compartment has been described in C. elegans. The PGP-2-positive compartment perfectly overlaps with Nile red (Schroeder et al., 2007), but it mostly does not overlap with oil red O-positive vesicles (Figure 2C). The nonacidic LMP-1-positive compartment (Nunes et al., 2005) is also mostly independent of the neutral lipid vesicles (Figure 2D). Accurate quantification of the level of overlap between oil red O and the GFP markers is impaired by the negative effect of oil red O staining on GFP signal. Nevertheless, as shown in Figures 2C and 2D, careful examination of multiple fields of well-preserved GFP-expressing cells shows that the neutral lipid and the PGP-2 or LMP-1 compartments are nearly completely independent. As in the case of costaining of the neutral lipid vesicles and the LROs with LipidTOX and vital dyes (Figure 2A and Figure S1), rare overlap between oil red O- and GFP-positive organelles is observed; the overlap is restricted to very few, small vesicles that may represent immature lipid droplets or lipid droplets undergoing degradation (data not shown).
Furthermore, unlike Nile red or BODIPY-labeled fatty acids, oil red O shows clear increases in fat mass for daf-2 mutants (Figure 3A). To further validate oil red O as a proxy for fat mass in C. elegans, we quantified whole-body oil red O signal in several metabolic mutants. In all conditions tested, oil red O quantification correlates with biochemical measurement of triglycerides by solid-phase chromatography followed by gas chromatography/mass spectrometry (Figure 3A). In addition, unlike Nile red or BODIPY labeled fatty acids, oil red O highlights lipid stores in the gonad and eggs, and unlike Nile red, oil red O stains the hypodermis (Figure 3B). Finally, similar to biochemically measured triglycerides, oil red O levels drop upon fasting (Figures 3C and 3D).
DISCUSSION
We here demonstrate that the Nile red-positive LROs do not contain the major C. elegans fat stores. Moreover, we show that the major fat stores are contained in independent specialized neutral lipid-containing vesicles. Nile red and BODIPY-labeled fatty acids do not colocalize with postfixative dyes for neutral lipids in more than 97% of the vesicles but perfectly colocalize with molecular probes for LROs, supporting the idea that vital dyes may be treated as xenobiotics and partitioned into a degradative compartment. Consistent with this hypothesis, mutants deficient in lysosomal biogenesis have reduced Nile red staining (Ashrafi et al., 2003; Schroeder et al., 2007). Moreover, the genome-wide RNAi screen for decreased Nile red staining in C. elegans identified five genes, apt-6, glo-3, glo-4, pgp-2, and vps-16 (Ashrafi et al., 2003), which were subsequently shown to have defective LRO biogenesis (Hermann et al., 2005).
The observation that Nile red- and BODIPY-positive LROs are mostly nonoverlapping with the neutral lipid vesicles in wild-type animals does not rule out a role for the LRO compartment in C. elegans metabolism, or even that LROs might contain physiologically relevant fats. Human genetic diseases, including Tay-Sachs and Niemann-Pick, result from defective lipid degradation in, or lipid trafficking from, lysosomes (Maxfield and Tabas, 2005). The dozens of C. elegans gene inactivations that dramatically decrease Nile red staining may reveal the complex genetic control of the LRO compartment (Ashrafi et al., 2003). Studies of altered vital dye staining in C. elegans, which indicate LRO defects, may therefore contribute to better understanding of these serious metabolic diseases. It is also possible that under certain pathological conditions or with different experimental procedures, Nile red and BODIPY may stain neutral lipids; however, under standard conditions and in the majority of genetic backgrounds, Nile red and BODIPY do not indicate C. elegans major fat stores.
Another interesting aspect is that, as is the case for other cellular byproducts accumulating in the lysosome-like compartment (Samuelson et al., 2007), increased Nile red or BODIPY-labeled fatty acid staining seems to be a predictor of decreased resistance to stress and shortened life span. Lipofucsin, or age pigment, and Nile red and BODIPY-labeled fatty acids levels are lower in long-lived mutants such as daf-2 and glp-1 and accumulate to a greater extent in progeric mutants such as daf-16 and rict-1. It is unclear if a higher ability to recycle cellular byproducts or to mobilize the right molecules to the LRO compartment is a cause or a consequence of the increased longevity phenotype, but it would be interesting to test the degree of correlation between vital dye staining and longevity.
C. elegans energy reserves are expected to be utilized during periods of food deprivation. Here we show that Nile red levels do not drop but increase upon fasting. Previous studies have demonstrated a decrease in Nile red with fasting (Ashrafi et al., 2003; Jo et al., 2009) but used different conditions. First, we eliminated bias introduced by selection of a narrow focal plane by conducting automated, whole-body quantification of the Nile red signal of more than 30 individual animals per treatment. Second, at time 0, animals were moved from plates containing food and Nile red to plates without food but containing Nile red. In previously reported analyses of Nile red signal upon fasting, animals were transferred to plates without food and without Nile red. Those studies most likely revealed Nile red turnover rather than the actual content of the compartment stained with Nile red. The lysosomal compartment is required to recycle nutrients during food deprivation (Finn and Dice, 2006). Consequently, an increase in the contents of the lysosomal compartment during starvation, even an increase in energetically relevant lipids, is not surprising.
Finally, we put forward oil red O staining as a facile method to study fat mass in C. elegans. Oil red O staining correlates, in all cases tested, with biochemically measured triglyceride mass. However, the magnitude of the changes estimated from oil red O images is less pronounced that the one measured by lipid biochemistry. We propose two possible explanations. First, oil red O stains a subset of the major C. elegans fat species. Second, our quantification algorithms for oil red O images are suboptimal. Supporting the latter, Figure S2 shows the striking difference in oil red O signal between wild-type and glp-1 animals; however, oil red O quantification would suggest a mild fat phenotype. Nevertheless, the recurring correlation between lipid biochemistry and oil red O proves oil red O as a reliable proxy for fat mass in C. elegans. Therefore, oil red O staining represents a validated method to study the regulation of long-term energy stores. Until a vital dye alternative is available, C. elegans fat mass is best studied using this fixative-based dye or quantitative lipid biochemistry (Perez and Van Gilst, 2008; Watts and Browse, 2002). The use of accurate fat-assessing methods will enable identification of genes and signals that regulate energy balance in C. elegans.
EXPERIMENTAL PROCEDURES
Strains Used
N2 Bristol was used as the wild-type strain. The following mutant strains were used: daf-16(mgDf47) I, tph-1(mg280) II, tub-1(nr2004) II, kat-1(mg368) II, kat-1(mg447) II, tub-1(nr2004);kat-1(mg368), daf-2(e1368) III, glp-1(e2141) III, rict-1(mg451) II, pwIs50[lmp-1::gfp] (Treusch et al., 2004), and kxEx74[pgp-2:: gfp;rol-6D] (Schroeder et al., 2007).
Nile Red and BODIPY-Labeled Fatty Acid Staining
Nile red and BODIPY-labeled fatty acid analyses were conducted as previously described (Mak et al., 2006) (see the Supplemental Experimental Procedures). At least 30 animals were imaged on at least two separate occasions, and results were consistent between experiments.
Fasting Analysis
For fasting measurements, adult animals were transferred from plates containing food to plates without food but containing 25 ng/ml of Nile red (for Nile red analyses), or to empty plates for the times indicated for oil red O analysis or lipid biochemistry (see the Supplemental Experimental Procedures).
LysoTracker Staining
Lyso Tracker staining was performed as above for Nile red and BODIPY. Lyso-Tracker Green (Invitrogen) was included at 1 µM (final concentration) in E. coli OP50 plates containing Nile red. LysoTracker Red (Invitrogen) was included at 1 µM (final concentration) together with C1-BODIPY 500/510-C12 fatty acids. Animals were allowed to feed on labeled E. coli for 1–2 days in the dark and imaged between L1 and young adult stages; similar results were obtained irrespective of developmental stage.
LipidTOX and Oil Red O Staining and Quantification
For LipidTOX staining, animals that had been fed on E. coli bacteria containing Nile red or BODIPY-labeled fatty acids as above were washed and fixed as for oil red O staining (see below). However, instead of dehydrating in 60% isopropanol, as in the case for oil red O, animals were washed free of paraformaldehyde with 1× PBS. 1:1000 of LipidTOX Red neutral lipid stain (for BODIPY-labeled animals) or LipidTOX green neutral lipid stain (for Nile red-labeled animals) was added directly (LipidTOX DMSO 1000× stocks, Invitrogen). Animals were incubated for 1 hr in the dark with gentle rocking and imaged directly thereafter using a Zeiss Axioimager/Apotome or spinning-disk confocal microscopy.
Oil red O staining was conducted by as previously reported (Soukas et al., 2009) but omitting the freeze-thaw steps (see the Supplemental Experimental Procedures). Animals were mounted and imaged with a Leica color camera outfitted with DIC optics. Oil red O was quantified from color images using the level of excess red intensity in the red channel in comparison to the blue and green channels (see the Supplemental Experimental Procedures).
Quantitative Lipid Biochemistry
Lipid extracts from 7500 worms at day 1 of adulthood were analyzed by solid-phase chromatography followed by GCMS as previously reported (Perez and Van Gilst, 2008; Soukas et al., 2009) (see the Supplemental Experimental Procedures). For all measurements, at least two biological replicates were performed, with data shown as mean ± SEM.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Sean Curran and Buck Samuel for creative input and reading of the manuscript. We also thank other members of the Ruvkun, Kaplan, and Ausubel labs for creative exchanges, and especially Jihong Bai for taking confocal microscopy images. We thank Mason Freeman for the use of his GCMS for lipid analysis. We want to thank Ho Yi Mak and Marc Van-Gilst for sharing their experiences with lipid histochemistry and biochemistry. Thanks to Kaveh Ashrafi and Kevin Jones for sharing their experience with the neutral lipid dye LipidTOX. Thanks to the Caenorhabditis Genome Center and Greg Hermann for providing strains. This work was supported by Award Numbers R01DK070147 (to G.R.) and F32DK080607 (to A.A.S.) from the National Institute of Diabetes and Digestive and Kidney Diseases, and the Human Frontiers Science Program (to E.J.O.).
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include two figures and Supplemental Experimental Procedures and can be found with this article online at http://www.cell.com/cell-metabolism/supplemental/S1550-4131(09)00301-5.
REFERENCES
- Ashrafi K. WormBook, The C. elegans Research Community. 2007. Obesity and the regulation of fat metabolism. ed. doi/10.1895/wormbook.1.130.1 http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Bonilla E, Prelle A. Application of nile blue and Nile red, two fluorescent probes, for detection of lipid droplets in human skeletal muscle. J. Histochem. Cytochem. 1987;35:619–621. doi: 10.1177/35.5.3559182. [DOI] [PubMed] [Google Scholar]
- Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–844. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Fowler SD, Greenspan P. Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J. Histochem. Cytochem. 1985;33:833–836. doi: 10.1177/33.8.4020099. [DOI] [PubMed] [Google Scholar]
- Fowler SD, Brown WJ, Warfel J, Greenspan P. Use of Nile red for the rapid in situ quantitation of lipids on thin-layer chromatograms. J. Lipid Res. 1987;28:1225–1232. [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H, Shim J, Lee JH, Lee J, Kim JB. IRE-1 and HSP-4 contribute to energy homeostasis via fasting-induced lipases in C. elegans. Cell Metab. 2009;9:440–448. doi: 10.1016/j.cmet.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kleyn PW, Fan W, Kovatas SG, Lee JJ, Pulido JC, Wu Y, Berkemeir LR, Misumi DJ, Holmgren L, Charlat O, et al. Identification and characterization of the mouse obestiy gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Suh JM, Avery L, Graff JM. Tripeptidyl peptidase II promotes fat formation in a conserved fashion. EMBO Rep. 2007;8:1183–1189. doi: 10.1038/sj.embor.7401086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes F, Wolf M, Hartmann J, Paul RJ. The ABC transporter PGP-2 from Caenorhabditis elegans is expressed in the sensory neuron pair AWA and contributes to lysosome formation and lipid storage within the intestine. Biochem. Biophys. Res. Commun. 2005;338:862–871. doi: 10.1016/j.bbrc.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Rabbitts BM, Ciotti MK, Miller NE, Kramer M, Lawrenson AL, Levitte S, Kremer S, Kwan E, Weis AM, Hermann GJ. glo-3, a novel Caenorhabditis elegans gene, is required for lysosome-related organelle biogenesis. Genetics. 2008;180:857–871. doi: 10.1534/genetics.108.093534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruaud AF, Nilsson L, Richard F, Larsen MK, Bessereau JL, Tuck S. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic. 2009;10:88–100. doi: 10.1111/j.1600-0854.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder LK, Kremer S, Kramer MJ, Currie E, Kwan E, Watts JL, Lawrenson AL, Hermann GJ. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol. Biol. Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, ’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99:5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.