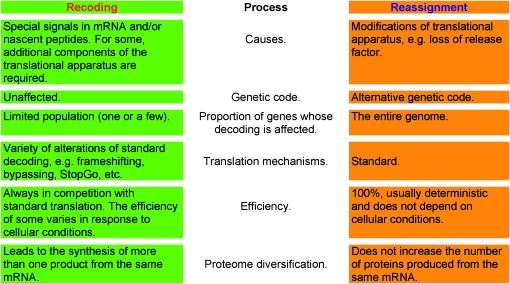
IN several recent reports, the term “recoding” is used to describe the phenomenon of codon reassignment (e.g., Charriere et al. 2006; McCutcheon et al. 2009). McCutcheon et al. (2009) misuse the term in numerous locations, suggesting that confusion is systemic and not inadvertent. This provokes us to clarify the distinction between these two natural phenomena. Reassignment and recoding are different (see Figure 1 for the key distinctive features).
Figure 1.—
Summary of distinctions between recoding and codon reassignment phenomena.
With the term “codon reassignment,” the meaning of a codon is altered irrespective of its mRNA context. The new meaning applies to the entire mRNA repertoire of the organism, and there is generally a complete change of meaning: the genetic code table is altered. Well-known examples include the reassignment of UGA to specify cysteine in the ciliate Euplotes and tryptophan in certain minute symbiont bacteria and specific mitochondrial genomes (Knight et al. 2001). Such reassignments are usually due to changes in the translational apparatus, such as alteration or loss of a release factor.
With “recoding,” the new meaning is site- or mRNA-specific and is dynamic; i.e., it is in competition with the standard meaning, with some of the product reflecting the new meaning and some reflecting the standard meaning. Recoding signals that are separate from the recoded site greatly enhance the efficiency of the nonstandard event (Gesteland et al. 1992; Gesteland and Atkins 1996; Baranov et al. 2002, 2006; Namy et al. 2004; Atkins and Gesteland 2010). Such signals have been characterized by many investigators and are known to act by a variety of mechanisms: through RNA secondary structures, complementary interactions with ribosomal RNA, or alteration of the ribosomal state by the encoded nascent peptide. Some examples of such signals are the following: an important signal for the ribosomal frameshifting that occurs during translation of bacterial release factor 2 mRNA is the pairing between a specific sequence 5′ of the shift site with the rRNA of translating ribosomes; a key signal for the (50% efficient) translation bypass of 50 nucleotides between codons 46 and 47 of bacteriophage T4 gene 60 is a specific sequence in the nascent peptide encoded 5′ of codon 46; a pseudoknot 3′ of the shift site is important for infectious bronchitis virus frameshifting; a stem loop close to the shift site is important for the frameshifting required to synthesize HIV GagPol; distant 3′ signals are involved in some cases of codon redefinition (not to be confused with codon reassignment) and in frameshifting in decoding the Barley Yellow Dwarf virus; and the recoding signal for eukaryotic selenocysteine specification by UGA is an mRNA structural element known as SECIS, located in the 3′ UTR. The context specificity provided by this variety of recoding signals is in marked contrast to context-independent codon reassignment.
Of course, as is common in biology, the boundaries are not sharp. An example of an intermediate situation may be in the specification of pyrrolysine by archael methanogens, where a 3′ RNA structure appears to enhance pyrrolysine specification but is not required for it (Longstaff et al. 2007). Several research groups use the term “recoding” to describe RNA editing that alters the protein product (Nishikura 2010), and here a parallel with translational recoding is evident. However, the existence of a few intermediate situations does not diminish the utility of using different words for situations where overall there is a clear distinction. The term “recoding” should be used for context-dependent alterations of gene readout, such as in the genes collected in the current version of the Recode Database (Bekaert et al. 2010). The term “codon reassignment” should be used to describe a process responsible for the differences between genetic codes, such as those collected in NCBI Genetic Code tables at http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi.
References
- Atkins, J. F., and R. F. Gesteland (Editors), 2010. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer Verlag, New York.
- Baranov, P. V., R. F. Gesteland and J. F. Atkins, 2002. Recoding: translational bifurcations in gene expression. Gene 286 187–201. [DOI] [PubMed] [Google Scholar]
- Baranov, P. V., O. Fayet, R. W. Hendrix and J. F. Atkins, 2006. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 22 174–181. [DOI] [PubMed] [Google Scholar]
- Bekaert, M., A. E. Firth, Y. Zhang, V. N. Gladyshev, J. F. Atkins et al., 2010. Recode-2: new design, new search tools, and many more genes. Nucleic Acids Res. 38 D69–D74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charriere, F., S. Helgadottir, E. K. Horn, D. Soll and A. Schneider, 2006. Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 103 6847–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland, R. F., and J. F. Atkins, 1996. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 65 741–768. [DOI] [PubMed] [Google Scholar]
- Gesteland, R. F., R. B. Weiss and J. F. Atkins, 1992. Recoding: reprogrammed genetic decoding. Science 257 1640–1641. [DOI] [PubMed] [Google Scholar]
- Knight, R. D., S. J. Freeland and L. F. Landweber, 2001. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2 49–58. [DOI] [PubMed] [Google Scholar]
- Longstaff, D. G., S. K. Blight, L. Zhang, K. B. Green-Church and J. A. Krzycki, 2007. In vivo contextual requirements for UAG translation as pyrrolysine. Mol. Microbiol. 63 229–241. [DOI] [PubMed] [Google Scholar]
- McCutcheon, J. P., B. R. Mcdonald and N. A. Moran, 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5 e1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy, O., J. P. Rousset, S. Napthine and I. Brierley, 2004. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell 13 157–168. [DOI] [PubMed] [Google Scholar]
- Nishikura, K., 2010. Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 79 321–349. [DOI] [PMC free article] [PubMed]