COMPUTER programs that simulate genetics results can play a useful role in teaching the principles of genetics, but some people have argued that such programs have generally not fulfilled their initial promise to engage students. I discuss some of the reasons for this shortfall on the basis of my experience in writing and distributing the “Hands On Genetics” programs (http://www.handsongenetics.com).
It is well accepted that the principles of genetics cannot easily be learned only from lectures, even with excellent descriptive textbooks. Although the evidence for this conclusion is largely anecdotal, there are some objective studies that support this view (e.g., Stewart et al. 1992; Hickey et al. 2003; Pukkila 2004). The question thus becomes: What additional activities are required to transmit the basic concepts of genetics in an introductory course?
Two activities can be invaluable supplements to lectures: direct experience from practical classes and problem solving. Computer simulation should assist implementation of both activities. Some experiments, particularly those involving Mendelian genetics, are amenable to simulation. Complete crossing programs that are impossible in live organisms can be carried out rapidly at almost no cost. Problems can also be posed in connection with such simulations in such a fashion that that each student can receive a different set of questions. Although there has been some acceptance of instructional software for genetics, it is not clear that it has transformed learning. Here I review my experiences with computer-based genetics education.
Simulation programs can be of many types. Here I describe two different types of simulation from the “Hands On Genetics” programs (http://www.handsongenetics.com). These programs run under Windows and Macintosh (Classic OS only). Further technical details about the programs are included in the supporting information, File S1.
My vision for these programs was to achieve interactivity between the user and the software. Having learned genetics in the precomputer age, my hope was that the old-fashioned way of learning and thinking—with a pencil in hand—might be replaced by creativity with a mouse in hand, moving objects (concepts?) around the screen.
Mendelism:
The program allows the instructor to implement genetic exercises that require students to solve a range of problems. It does this by providing the student a genetic setup, generally a pair of individuals that differ in one or more phenotypes, together with a posed question.
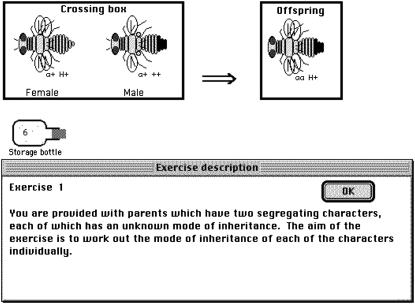
Figure 1 shows how a problem is presented to the student, in this case using Drosophila crosses. In the exercise, there are two segregating loci, one of which controls the presence or absence of the aristae and the other the conversion of the haltere into a second wing pair. The rest of the exercise is left up to the student, who must determine the crosses needed to work out the pattern of inheritance for each trait. To solve the problem, the student drags flies into the “crossing box,” which leads to the production of offspring whenever the “offspring box” is empty. Bottles are available for storage to simplify the counting.
Figure 1.—
Example of what the student sees when a Mendelism exercise is presented (colors not shown).
Genotypes are shown in the example of Figure 1 because the student has chosen to run a “trial” exercise. Repeated running of trials allows a student to begin to understand the relationship between genotype and phenotype in preparation for the awkward moment when only phenotypes are shown and genotypes must be deduced.
In its standard mode, the software checks the genotypes entered by the student and assigns a grade. This mode has utility in allowing the student to carry out multiple experiments with self-monitoring and in grading large classes where student–teacher interaction is limited. However, the process is not continuously monitored by the instructor and thus does not provide ongoing feedback on student progress. A more satisfactory way of checking student answers is one in which the instructor knows the correct answer, which is not available to the student. For the software to be used in this way, it is necessary to perform a preliminary run of the program in which access is password-authenticated. The program then displays and prints the correct answers. This mode also allows for a written report to be part of the course requirement.
This two-locus unknown exercise has been used in introductory genetics courses at the University of Sydney for many years, preceded by its mainframe precursor Drosim (Sved 1980). While there has been no systematic evaluation of student understanding or performance with these programs, feedback from students and experienced tutors has generally been positive. The majority of students cope well with the two-locus exercise, with some students even complaining that it is too easy. The exercise simply asks the student to consider the inheritance of the two loci individually. The best students sometimes go on to consider the joint segregation, to show, in this case, that the two alleles are inherited independently. The initial cross presents the loci “out of phase”; therefore, to make a double heterozygote genotype to test the joint segregation requires a considerable amount of thought. Other, more advanced exercises are provided to deliberately test this aspect of the problem.
Although the Mendelism program requires the student to work out the underlying mode of inheritance, it does not help the student to think through the inheritance process, other than to allow trials where genotypes are shown. In many cases, students need a pencil and paper to follow what is going on. Such thinking may more easily be done away from the computer, and students are encouraged to do that.
SexLink:
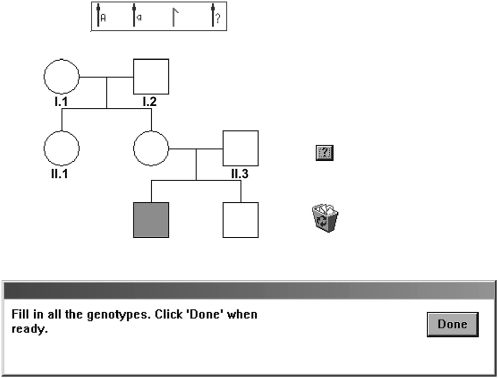
By contrast, another type of simulation lends itself to a direct solution on the computer screen and aids thinking about inheritance. An example is the SexLink program, shown in Figure 2.
Figure 2.—
Example of a pedigree presented to students in the SexLink program.
In this program, students directly drag gene/chromosome symbols into individuals. The top box of Figure 2 shows the genetic ingredients: X chromosomes with dominant and recessive alleles, the Y chromosome, and the “?” symbol for use where the genotype cannot be unambiguously determined. The pedigree contains three founder individuals: I.1, I.2, and II.3. The chromosomes for these individuals need to come from the top box. Other individuals in the pedigree need to inherit chromosomes by dragging them from their parents.
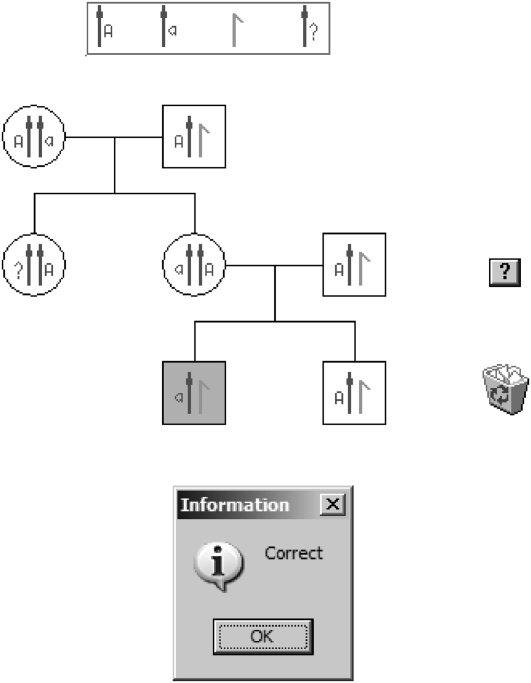
Figure 3 shows the completed exercise. Note that the chromosomal constitution of one individual, II.1, cannot be determined, and for whom the “?” chromosome must be selected from the top box.
Figure 3.—
The completed pedigree of Figure 2.
This appears to be one exercise where learning can be achieved at the computer. Furthermore, with repetition of the exercise, students come to appreciate that there is a limited set of pedigree types and eventually absorb the principles of sex-linked inheritance.
Simulation and problem solving in molecular genetics:
Other programs in the Hands On Genetics set include an elementary simulation of DNA replication, transcription, and translation; a PCR simulation; and a restriction enzyme site mapping exercise. A meiosis simulation program is also included.
Animation programs:
The most common type of software might be described as “animation,” rather than simulation software, as in the Hands on Genetics programs. Perhaps the best example of this is the comprehensive and informative “DNA from the Beginning” (http://dnaftb.org), a free set of modules offered by Cold Spring Harbor Laboratory. Although these modules have a molecular emphasis, they provide much information on Mendelian genetics.
Animation software falls somewhere between textbooks and interactive simulation programs in the presentation of material. A key advantage of animation software is that a greater breadth of material can be presented compared to simulation programs. Animation development is expedited by programs such as Authorware; simulation programs are typically more laborious to write. Small simulations can be embedded in animation programs (as in the Cold Spring Harbor programs).
Despite its breadth and comparative ease of delivery, it is unclear whether computer animation improves on a good series of lectures backed up by a textbook. On the other hand, for a student unable to take advantage of traditional instruction, such software is an advance.
Difficulties in the production of simulation programs:
The development of simulation programs has been limited by the complexities of the production process. In contrast to commercial programs, developed by teams of managers, producers, designers, programmers, quality assurance personnel, and more, most teaching software is written by teachers in their spare time. My experience has been that the time necessary to develop programs is incompatible with actually using them in classes; the majority of the software development was done after my retirement from teaching. In terms of the learning experience, problem-solving simulations do not necessarily require a complex presentation. Low budgets and time constraints, however, pose a challenge when trying to develop software designed to engage the attention of students used to the sophisticated computer animation applications developed by the entertainment industry.
One area in which a simulation program has no obvious advantage is in the presentation of new material. Unless students have some familiarity with the basic terms and concepts, they usually have difficulty solving the problem. Optimal use of simulation programs therefore depends on judicious presentation of material prior to use of the program.
One size does not fit all:
Genetics is taught at many levels—from primary or early secondary school to the graduate level. This breadth creates problems in designing software suitable for each level. The Mendelism program provides elementary- and advanced-level exercises. But it has not been easy to determine what levels are appropriate for particular classes. and for teachers to appreciate which sections are relevant to their goals, they must devote considerable time and effort in examining the programs.
Feedback on the software:
I have found it difficult to get feedback from users of the software at other institutions (other than word of mouth from colleagues). A mechanism for feedback, perhaps via a website or “Wiki” where teachers could report their experiences using the software, would be useful. I hope this Letter will stimulate implementation of such a scheme.
Editor's Note:
The above letter highlights both the difficulty in the creation of software packages designed for teaching genetics by individual investigators and the need for a much more interactive method of sharing and supporting these programs. The creation of a Teaching Committee as a standing Committee of the GSA Board and the hiring of Teaching Coordinator as a member of the GSA staff is intended to address precisely this issue, as well as other teaching-related concerns.
Acknowledgments
I am indebted to the many generations of students at the University of Sydney who suffered through various versions of these programs. Bill Engels of the University of Wisconsin provided great help in my initial attempts at Graphic User Interface programming. Discussion with Patricia Pukkila was instrumental in the writing of this Letter to the Editors. During development of the Hands On Genetics programs, I made a conscious effort to avoid being influenced by other teaching programs, and I therefore apologize for my limited knowledge and acknowledgment of other programs in the area.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.116640/DC1.
References
- Hickey, D. T., A. C. H. Kindfield, P. Horwitz and M. T. Christie, 2003. Integrating curriculum, instruction, assessment, and evaluation in a technology supported genetics learning environment. Am. Educ. Res. J. 40 495–538. [Google Scholar]
- Pukkila, P. J., 2004. Introducing student inquiry in large introductory genetics classes. Genetics 166 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, J., R. Hafner, S. Johnson and E. Finkel, 1992. Science as model building: computers and high-school genetics. Educ. Psychol. 27 317–336. [Google Scholar]
- Sved, J. A., 1980. A computer program which saves on cooking and washing up. Drosoph. Inf. Serv. 55 171–172. [Google Scholar]