Abstract
The enormous amount of biomolecule measurement data generated from high-throughput technologies has brought an increased need for computational tools in biological analyses. Such tools can enhance our understanding of human health and genetic diseases, such as cancer, by accurately classifying phenotypes, detecting the presence of disease, discriminating among cancer sub-types, predicting clinical outcomes, and characterizing disease progression. In the case of gene expression microarray data, standard statistical learning methods have been used to identify classifiers that can accurately distinguish disease phenotypes. However, these mathematical prediction rules are often highly complex, and they lack the convenience and simplicity desired for extracting underlying biological meaning or transitioning into the clinic. In this review, we survey a powerful collection of computational methods for analyzing transcriptomic microarray data that address these limitations. Relative Expression Analysis (RXA) is based only on the relative orderings among the expressions of a small number of genes. Specifically, we provide a description of the first and simplest example of RXA, the k-TSP classifier, which is based on k pairs of genes; the case k = 1 is the TSP classifier. Given their simplicity and ease of biological interpretation, as well as their invariance to data normalization and parameter-fitting, these classifiers have been widely applied in aiding molecular diagnostics in a broad range of human cancers. We review several studies which demonstrate accurate classification of disease phenotypes (e.g., cancer vs. normal), cancer subclasses (e.g., AML vs. ALL, GIST vs. LMS), disease outcomes (e.g., metastasis, survival), and diverse human pathologies assayed through blood-borne leukocytes. The studies presented demonstrate that RXA—specifically the TSP and k-TSP classifiers—is a promising new class of computational methods for analyzing high-throughput data, and has the potential to significantly contribute to molecular cancer diagnosis and prognosis.
Keywords: relative expression, classification, microarray analysis, computational biology
Introduction
High-throughput measurements in biology (e.g., transcriptomics, proteomics, metabolomics) provide an enormous amount of information, but only implicitly, in the form of raw expression values. Harnessing this information means converting it to knowledge and, for the purposes of classification, useful decision rules; this conversion can enable a greater understanding of cancer and drive advances in personalized medicine. A systems-level approach, which employs computational and statistical tools to reveal and evaluate patterns with diagnostic or prognostic value, is critical to fully exploiting these new technologies. In particular, molecular signatures derived from patterns in gene expression microarray experiments have great potential to detect the presence of disease, to discriminate among cancer sub-types, to predict clinical outcomes, and to provide insight into specific changes that occur during disease progression.
Perhaps the most evident challenge for developing useful molecular signatures is to identify classifiers that are accurate for a specific study or platform, and that are also robust across a wide range of settings. Previous studies have aimed to identify sets of individual genes (“signatures”) whose differential expression is highly correlated with phenotypic changes (e.g., genes that may be over- or under-expressed in cancer relative to normal). In these cases, increased or decreased absolute mRNA concentration levels above some threshold (i.e., more than would be statistically expected by chance for a gene on the microarray) are put forth as candidates for disease-induced (or causing) perturbations. Unfortunately, the statistically significant genetic changes often depend largely on the context of the microarray experiment. Even when thresholds are tuned to produce statistically significant results, findings can depend heavily on a number of factors, such as the experimental design and the type of data normalization. Consequently, there may be little to no overlap in the molecular signatures identified from one platform to another, or by extension, from one clinical setting to another.
A less evident, but equally important, challenge for phenotype classification using gene expression data is to develop techniques that not only yield accurate and robust decision rules, but also provide rules that are easy to interpret and might contribute to biological understanding. Advanced statistical learning and pattern recognition methods are routinely applied to transcriptomics and other high-throughput data types. These include neural networks (1-3), decision trees (4-6), boosting (5, 7) and support vector machines (8, 9). In many cases, these methods achieve good classification performance, with sensitivities and specificities above ninety percent. However, they generally result in extremely complex decision rules based on nonlinear functions of many gene expression values. Therefore, whereas advanced methods may be more accurate than those based on the patterns of individual genes, they usually produce decision rules which are virtually impossible to interpret. Furthermore, as the number of variables (transcripts) far exceeds the number of observations in most microarray studies, building more complex classifiers entails a greater risk of over-fitting the training data and poor generalization.
An important potential benefit of simple and interpretable decision rules is to provide insight into the underlying biological differences between phenotypes. Notably, malignant phenotypes in cancer arise from the net effect of interactions among multiple genes and other molecular agents within biological networks. Genes in networks operate in a combinatorial manner—the actions of one gene greatly influence the actions of other genes. This often limits the information that can be gleaned from the expression patterns of individual genes. As an alternative approach, studying gene expression in the context of networks may yield greater insight into mechanisms and functional changes associated with disease. Recently, methods for analyzing microarray data have focused not on individual genes, but instead on biologically meaningful pathways or networks (10-13). These frameworks have been applied to diverse cancer systems and serve as a robust source of biological discovery (12, 14).
At scales smaller than biological networks or even pathways, assessing the relationships among a small number of genes—for example, the patterns of interactions among just two or three genes—can provide useful information about biomolecular processes. One way to probe the interactions among several genes is to study their relative expression, i.e., the ordering among the expression values, rather than their absolute expression values. One then searches for characteristic perturbations in this ordering from one phenotype to another. The simplest form of such an interaction is the ordering of expression among two genes, in which case one seeks to identify typical “reversals”—pairs of genes for which one of the two possible orderings is usually present in one phenotype and rarely present in the other. We refer to the family of such rank-based methods as Relative Expression Analysis (RXA). This methodology is characterized by replacing each expression level across all genes by its corresponding rank within a single microarray profile.
Here we focus on RXA methods which involve a small number of gene pairs, each exhibiting a characteristic “relative expression reversal” between the phenotypes or classes of interest. Aggregating the decisions from a few such pairs, even just one, is surprisingly powerful. Basing decisions on one pair is called the top-scoring pair (TSP) classifier (15) and on k pairs is called the k-TSP classifier (16). Thus, in TSP, a sample is classified based on a decision rule which only involves comparing the ranks, hence the relative expression levels, of two genes within a profile. For the k-TSP classifier, the decision rule combines a disjoint set of TSPs by simple majority voting. Other RXA methods include those based on the six possible orderings among three genes (the top-scoring triplet classifier (17)) and comparing the average ranks in two groups of genes (18). Herein we review the TSP and k-TSP computational methods, focusing on their utility for aiding molecular diagnostics in a broad range of human cancers. Our review is largely restricted to applications with transcriptomic data, since this is the most plentiful and has been the most used to date. However, RXA is generally applicable to any ordinal data type, such as protein expression, DNA copy number, chromosomal position, and so forth.
Relative Expression Analysis
Microarray Data and Analysis
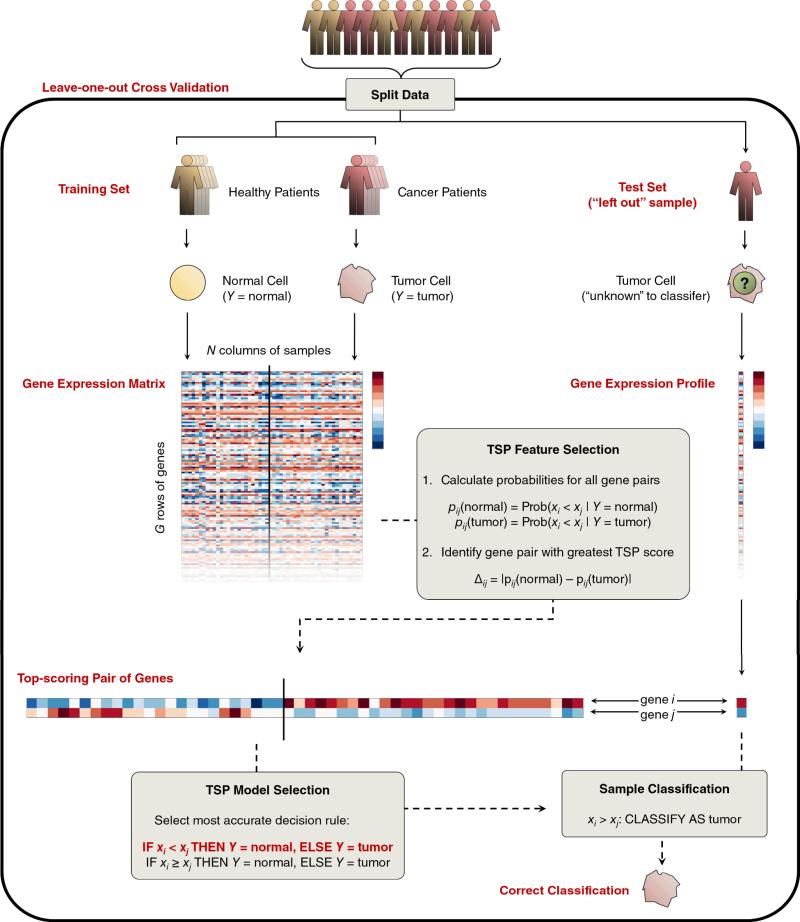
For those readers less familiar with computational approaches to microarray analysis, we first describe the typical features of microarray data and common procedures relevant to the results presented here. Whereas we discuss computational analysis of microarray data in the context of RXA (Figure 1), the notation, representation of the data, and basic steps are the same for other approaches. Computational analysis of microarray data typically involves two steps. First, a classifier is trained on a collection of microarray profiles (samples) referred to as the training set. This involves selecting a subset of genes and choosing a mathematical algorithm (decision rule) to apply to the selected genes in order to determine the phenotype of a new sample. Of course the goal is to identify an algorithm that is predicted to work well on a new data set, and the second step is then to evaluate the performance of the classifier on held-out data. Usually, the algorithm works quite well on the training data and hence validation is essential.
Figure 1.
Schematic overview of phenotype classification with the top-scoring pair (TSP) algorithm in cross validation.
Microarray data are typically represented as a matrix of G rows of genes and N columns of samples (e.g., different tumors, tissues, patients, time points). The nth column of this matrix is therefore a G × 1 vector representing the expression profile xn of the nth sample. Each profile contains expression values for gene one (g1) through gene G (gG). The expression level of gene gi is denoted by Xi. In addition, each sample is labeled by a phenotype Y ∈ {A, B, ...}. For example, yn = A indicates that the nth sample belongs to phenotype A. The labeled data set to be used for classifier training is F = {(x1, y1), ..., (xN, yN)}.
As mentioned above, the simplest method for classifying expression profiles based on the relative ordering of expression values is the top-scoring pair (TSP) algorithm for distinguishing between two phenotypes A and B. In TSP, a particular pair of genes i and j is selected during training and the decision rule is simple maximum likelihood: for the sample to be classified, choose the class, A or B, for which the observed ordering between the expression values of gi and gj is the most likely. Notice that the observed ordering is either Xi < Xj or Xi > Xj (we can assume at this point that ties are broken at random). The pair which is chosen is the one that achieves the highest “score” among all pairs of genes. This score is a quantitative measure of the degree of relative expression reversal estimated from the data and used for classifier training, as explained in the following section. For the k-TSP classifier, the decision rules are conceived in the same manner as in the TSP classifier, but use a combination of gene-pair markers to obtain potentially better classification accuracy. There are currently two software implementations available for researchers who wish to apply these methods: one in Perl (16) and one in R (19).
The TSP and k-TSP classifiers are parameter-free methods that are invariant to all normalization techniques that are monotonic transformations of the original expression values within each chip or microarray. That is, if the data are processed in such a way that if gene gi is expressed more than gene gj before normalization (original data) and it is still expressed more after normalization (processed data), then the TSP and k-TSP classifiers derived from the original and processed data are the same. It is in this sense that these classifiers are “invariant” to normalization. Moreover, the TSP and k-TSP classifiers are especially favorable in terms of the simplicity of the decision rule and the small number of genes involved in classification. They are easy to implement in practice since the classifier only requires measurement of the expression of small number (at most 2k) of genes using techniques such as RT-PCR. They also remain context-independent by not requiring any parameter-tuning or data pre-processing based on genes outside of the pairs involved. Furthermore, since data normalization is not required, RXA classifiers have been shown to be useful in the integration of data across different studies and platforms for the purpose of increasing sample size and facilitating meta-analysis of microarray data (20).
Training RXA Classifiers
In relative expression approaches, the features selected are pairs of genes. Consider first TSP. Because only gene pairs are considered, it is possible to completely enumerate all possible pairs and select the “best” ones using the training data. The natural criterion is performance, which anticipates how the pair of genes will be used for classification. As a result, one then selects the pair of genes gi and gj for which the difference |Prob(Xi < Xj | Y = A) – Prob(Xi < Xj | Y = B)| is maximized. This can be shown to be equivalent to choosing the pair of genes which achieves the lowest error rate on the training set. In many cases, there is a single pair of genes achieving the top score. Otherwise, in order to select a unique pair of genes, a secondary score is applied which is based on the average difference in expression values over all samples. An important feature of the top-scoring pair of genes is that it may not be the case that both genes are highly differentially expressed on the basis of their individual t-statistics; in fact, one gene may serve as a “pivot” for the other.
Depending on which of the two probabilities Prob(Xi < Xj | Y = A) or Prob(Xi < Xj | Y = B) is larger, the decision rule is either:
Rule 1: If expression gene i < expression gene j, THEN class A, ELSE class B.
Rule 2: If expression gene j < expression gene i, THEN class A, ELSE class B.
In the case of k-TSP, the classifier is constructed from the k top-scoring pairs of genes. Each pair votes for class A or class B the same way as in TSP and the class with the majority vote is chosen. Effectively, this is the maximum likelihood rule: choose the class for which the k observed orderings are the most likely. Usually, the pairs are constrained to be “disjoint,” meaning that a gene cannot appear in more than one pair, and the number of pairs (k) is determined by cross-validation up to some limit (e.g., kmax = 10) in order to keep the total number of genes manageable. Consequently, the size of the gene “signature” is two for TSP and 2k for k-TSP. Unlike other methods, once the signature is determined so is the classifier. That is, there are no parameters to tune, which reduces over-fitting the training data.
Testing RXA Classifiers
Classifier training is followed by performance evaluation on a test dataset. The gold standard for testing any predictive method is to use an independent dataset collected solely for testing. However, due to the scarcity of data, the test set usually consists of samples collected from the original training dataset and set aside. Even in this case, repeated training and testing, known as cross-validation, is preferred due to small sample sizes. Such procedures involve splitting the original training dataset F into two smaller sets: the set of samples on which the classifier is trained, Ftrain; and the set of samples on which the classifier is tested, Ftest. Importantly, no information from Ftest can be utilized when learning the classifier on Ftrain. The data is repeatedly split into training and test groups, and the cross-validated accuracy is the average classifier performance across all test groups. Leave-one-out cross-validation (LOOCV) is commonly used, in which the total of N samples is divided into a training set of size N – 1 with the test set consisting of the single remaining sample. While error estimation with LOOCV is known to have high variance relative to the true error (21), it is particularly useful for TSP and k-TSP because there is a technique (16) which yields a very significant reduction in the computation involved in looping over all pairs of genes in each loop of cross-validation.
A number of different metrics can be used to measure the performance of classifiers. Particularly common measures include sensitivity, specificity, and overall accuracy. These metrics are most easily understood for experiments with a case (e.g., cancer) and a control (e.g., normal), but can be extended to any binary phenotype comparison as well as to multiclass problems by decomposing them into sets of binary comparisons. If a classifier correctly predicts that a cancer profile belongs to the cancer class this is known as a true positive (TP), and the probability of correctly labeling future cancer samples is the sensitivity of the classifier (also known as the true positive fraction). Similarly, a true negative (TN) is when a classifier correctly labels a normal sample and the probability of doing this on new samples is the specificity of the classifier. Importantly, the sensitivity and specificity computed on the samples used for training are upwardly biased and not predictive of cross-validated rates. Finally, overall accuracy can be defined in several ways; perhaps the simplest is the average of sensitivity and specificity.
RXA in the Study of Cancer
Cancer Studies Using Relative Expression Values Before TSP and k-TSP
Gene-pair relative expression markers, specifically in the form of a two-gene expression-level ratio, have been previously used for disease classification and prognosis. Gordon et al. (22) successfully distinguished between malignant pleural mesothelioma (MPM) and adenocarcinoma (ADCA) of the lung based on ratios of expression. Although genetically disparate, the tissues of MPM and ADCA can be difficult to distinguish based on established histopathological methods. Gorden et al. (22) tested the fidelity of ratio-based diagnosis in differentiating between the two tissue types in 181 samples (31 MPM and 150 ADCA). First, the investigators used a training set of 32 samples (16 MPM and 16 ADCA) to identify differentially expressed genes based on various methods (fold changes, standard t-tests, expression cutoffs, etc.). They then formed 15 ratios using individual or combinations of those genes that showed the highest significance in inversely correlated expression levels. Any single ratio of the 15 examined was at least 90% accurate in predicting diagnosis for the remaining 149 samples (e.g., test set). They then examined (in the test set) the accuracy of multiple ratios combined to form a simple diagnostic tool. Using two and three expression ratios, the investigators found that the differential diagnoses of MPM and lung ADCA were 95% and 99% accurate, respectively. Whereas, in this study, these gene-pairs are not combined in the same way as TSP, they are sensitive to normalization and parameter choices. Still, their work illustrates the utility and discriminatory power of gene pairs in important clinical diagnoses.
Ma et al. (23) found that a two-gene expression ratio derived from a genome-wide, oligonucleotide microarray analysis of estrogen receptor (ER)-positive, invasive breast cancers predicts tumor relapse and survival in patients treated with tamoxifen. Tamoxifen is one of the most commonly used medications in the treatment of early-stage and metastatic ER-positive breast cancer (24, 25). When administered to women with surgically treated ER-positive breast cancer, tamoxifen therapy reduces the annual risk of recurrence by 40-50%, leading to a 5.6-10.9% improvement in 10-year survival (26). However, 25-66% of women diagnosed with ER-positive breast tumors fail to show a prolonged response or develop early resistance to adjuvant therapy (24, 27). Currently, there are no markers that reliably predict clinical outcome of cancer patients treated with tamoxifen. Therefore, a reliable means to accurately predict tamoxifen treatment outcome is crucial for early-stage breast cancer management.
In the tamoxifen study conducted by Ma et al. (23), a set of 60 patients with receptor-positive primary breast cancers were treated with tamoxifen alone. The results from gene expression profiling of the extracted tumor tissues before therapy indicated that the homeobox gene (HOXB13) was over-expressed in patients who experienced disease recurrence, whereas the interleukin-17B receptor gene (IL-17BR) and EST gene were over-expressed in those with no evidence of recurrence after a 5-year treatment period. The investigators evaluated the prognostic utility of each of these three genes by itself and in combination with genes that have opposing patterns of expression between the two classes. Results from t-test and ROC analyses revealed that a two-gene ratio of HOXB13 over IL-17BR had a stronger correlation with treatment outcome than any of the genes alone with AUC values reaching 0.84, and was able to accurately predict tumor recurrence in adjuvant tamoxifen-treated patients
This observation was also confirmed in real-time quantitative PCR analysis, where the predictive accuracy of the two-gene ratio was 81%. Furthermore, the expression ratio of HOXB13 over IL-17BR outperformed existing biomarkers for prognosis of breast cancer, such as patient age, tumor size, grade, and lymph node status. In this study pre-dating any formal RXA classification approaches, Ma et al. (23) demonstrated the utility of a two-gene expression biomarker in identifying a subset of patients with early-stage ER-positive breast cancer who are at a risk for tumor recurrence even with tamoxifen therapy. Such a biomarker provides a potential means to identify patients appropriate for alternative therapeutic regimens in early-stage breast cancer.
Comparative Analysis of TSP and k-TSP Performance in Cancer Classification
Geman et al. (15) introduced the TSP method and demonstrated its efficacy on several gene expression data sets involving breast, prostate and leukemia cancers. The phenotype classification problems considered were: (i) predicting the status of lymph nodes (affected vs. non-affected) in patients with breast tumors using data from (28); (ii) classifying the sub-types of leukemia (AML vs. ALL) using data from (29); and (iii) distinguishing prostate tumors from normal profiles using data from (30). The reported accuracies for TSP results were based on LOOCV, and comparison to randomly permuted data was made to estimate the statistical significance for each classifier.
In predicting the status of lymph nodes in the breast cancer data set, a cross-validation classification rate of 79% was achieved from 49 patient samples. The authors also mention a separate study where estimated error rates for these data—based on LOOCV and using a wide variety of common machine learning techniques—are summarized for varying numbers of pre-filtered genes (28). Other methods, more complex than TSP and using many more genes, did not result in better classification rates, and the low accuracy observed in all methods applied to date is probably a function of the complexity and similarity of the phenotypes being separated. In the case of separating AML from ALL, the TSP classifier correctly classified 68 samples out of 72 samples in cross-validation. In comparison, the study in Golub et al. (29) used a fifty-gene classifier to predict 65 samples correctly out of 72.
In addition to demonstrating improved performance in classifying breast cancer and leukemia samples, Geman et al. (15) also investigated the ability of TSP to detect the presence of prostate cancer. In a previous study, Singh et al. (30) found a strong correlation between patterns of gene expression of prostate cancer and various clinical and pathological aspects of the disease. The top-scoring gene pair using the TSP algorithm on their data could discriminate non-tumor versus prostate tumor samples at a prediction rate of 95%. Hence, the classification rates using TSP were comparable to the best results reported previously in the literature, often incorporating hundreds of genes or more in complex decision rules.
The performance of TSP and k-TSP classifiers were compared with those of other machine learning methods on 19 gene expression datasets involving human cancers in a study by Tan et al. (16). The study investigated a number of publicly available datasets, with sample sizes ranging from 33 to 327 for each disease phenotype within a particular dataset. The collection of datasets comprised various studies of human cancer, including colorectal, leukemia, lung, prostate, breast, central nervous system, lymphoma, bladder, melanoma, renal, uterus, pancreas, ovary, and mesothelioma. The classification performance of TSP and k-TSP was compared to that of decision trees (DT), Naïve Bayes (NB), k-nearest neighbor (k-NN), support vector machines (SVM), and prediction analysis of microarrays (PAM), which is essentially linear discriminant analysis. The TSP and k-TSP techniques were also extended beyond binary classification to the multi-class setting, where several well-known aggregation strategies, such as “one-vsall” and “one-vs-other,” were applied to combine the results of binary sub-problems into one final decision rule.
In this study, LOOCV was used in order to estimate the classification rate. The best classifier based on the average accuracy for the binary classification problems used in this study was k-TSP (92.01%), followed by SVM (91.18%), PAM (88.91%) and TSP (88.26%). The differences in accuracies were small, so it was concluded that all four methods perform classification similarly. The authors also elucidate the biological meaning of the classifiers by showing the connections between the genes in the markers and their corresponding cancer types. For the multi-class problems, TSP achieved an average accuracy of 85.12% over 10 problems, somewhat less than PAM (88.50%) and SVM (88.10%), which performed the best overall but used hundreds or thousands of genes.
In the initial variant of RXA, Geman et al. (15) showed that the TSP classifier provides decision rules that are highly accurate in binary classification problems and involve very few genes. Tan et al. (16) compared the TSP and k-TSP approach to other machine learning techniques on a broad source of human cancer gene expression data. The performance of TSP and k-TSP on both binary and multi-class problems were comparable to those of the other techniques, while no single method was found to have the best performance across all datasets. TSP and k-TSP were thus shown to have comparable accuracy to state-of-the-art methods, involve fewer genes and yield transparent, context-independent classifiers which are invariant to most forms of data normalization.
Specific Cancer Studies Using TSP or k-TSP
TSP-based classification methods have been applied to a number of specific cases of predictive studies in cancer. These studies can be broadly divided into those that identify classifiers for disease diagnosis and studies that develop relative expression classifiers for disease prognosis. Specifically, diagnosis can refer to determination of the presence or absence of disease, the particular sub-type of a disease, or in some cases the stage of disease. In contrast, prognosis aims to predict the outcome of patients with the disease. Examples of disease prognosis include response to treatment, survival time, and tumor metastasis. Importantly, a number of the studies presented here demonstrate not only the power of TSP methods to accurately classify microarray profiles, but also their utility for integrating microarray datasets from different sources and even across different measurement technology platforms.
Gene-pair Classifiers for Diagnosis
Gastrointestinal stromal tumor (GIST) and leiomyosarcoma (LMS) are common mesenchymal tumors with similar phenotypic features. A whole-genome gene expression study of 68 well-characterized tumor samples identified a two-gene relative expression classifier using TSP that distinguished GIST and LMS with 99.3% accuracy on microarray samples and 97.8% accuracy in cross validation (31). The classifier, which predicts GIST when OBSCN > C9orf65 and LMS otherwise, was validated using RT-PCR on 20 samples from the original dataset and on 19 independent samples, achieving 100% accuracy. Immunostaining for the Kit protein marker is currently the best test to differentiate GIST and LMS. Using expression of c-Kit to classify samples (with a cutoff determined by 1D linear discriminate analysis) achieved only 87.3% accuracy. That is, as some GIST samples have low Kit expression and some LMS samples have high Kit expression, testing for levels of the protein marker was more prone to error than predictions based on the OBSCN/C9orf65 expression ratio.
The TSP classification method is invariant to standard procedures for monotonic data normalization, as it relies only on the ranks of gene expression values within the microarray. As such, using TSP for classification enables the integration of microarray profiles from multiple datasets, thereby increasing the sample size of the training data and the predictive potential of the classifiers. Xu et al. (20) identified a TSP marker for prostate cancer (HPN > STAT6) that achieves high accuracy, sensitivity, and specificity on two datasets from different platforms. Performance of the HPN/STAT6 TSP marker—trained on integrated microarray data—was better than other TSP classifiers trained on individual datasets. In training the classifier, three microarray datasets from different prostate cancer studies were integrated and TSP was applied to analyze both individual and integrated datasets. It was found that TSP markers vary between individual datasets, but as more samples are added to the integrated training dataset, TSP selection becomes consistent. Stability analysis was also performed to calculate the appearance frequency of markers (i.e., how often the same TSP markers were selected) when samples were randomly removed from the dataset. The TSP marker was tested on an independent cross-platform dataset, comprising prostate tumor expression values from both Affymetrix and spotted cDNA platforms. Samples in the independent test set were classified with 93.8% accuracy, 91.7% sensitivity, and 97.7% specificity.
Gene-pair Classifiers for Cancer Prognosis
Xu et al. (32) integrated three independent microarray datasets containing 358 total samples for prediction of distant metastases in breast cancer. All samples in the integrated dataset were obtained from lymph-node-negative patients who had not received adjuvant systemic treatment. Gene expression data was directly merged using 22,283 probe sets on the Affymetrix HG-U133A microarray, and the top 200 “features” were selected as gene pairs with the highest TSP scores. In accordance with clinical treatment guidelines defined by the St. Gallen (Switzerland) expert consensus and the NIH, the goal of the authors was to achieve the highest possible specificity while maintaining high sensitivity (~90%). The optimal signature size (80 pairs, 112 distinct genes) was determined in k-fold cross-validation, and a likelihood ratio test (LRT) for classification based on this signature achieved 88.6% sensitivity and 54.6% specificity in an independent external test set of 154 samples. Since the LRT assumes statistically independent gene pairs, the decision rule amounts to weighted voting among the gene pair classifiers and hence is very similar to k-TSP.
Over-expression of the Src tyrosine kinase in pancreatic cancer is thought to play a significant role in tumor development and progression. The in vivo efficacy of an orally active small molecule Src inhibitor AZD0530 was investigated in a collection of pancreatic tumor xenografts (33). The k-TSP algorithm was applied to gene expression profiles from the tumors in order to identify predictive biomarkers of response to AZD0530. Tumor growth index (TGI) was used to morphologically classify xenografts as sensitive (TGI < 50%) or resistant (TGI > 50%) to AZD0530 treatment. In the training set of 16 xenografts (3 sensitive, 13 resistant), the TSP classifier LRRC19 > IGFBP2 most accurately predicted cases as sensitive (and correspondingly predicted cases as resistant when LRRC19 ≤ IGFBP2).
The k-TSP classifier achieved an estimated LOOCV accuracy of 97.8% on the microarray data set. The two-gene predictor was tested and validated on eight independent xenografts not included in the original training set and achieved an overall accuracy of 87.5%, specificity of 83.3%, and sensitivity of 100%. RT-PCR was performed on the two genes in the eight independent xenografts, showing the relative expression of LRCC19 and IGFBP2 was the same as measured by microarray gene expression in all cases. This stability across different measurement platforms is critical for application in the clinic, and represents an advantage of methods based on RXA.
A two-gene expression ratio (RASGRP1/APTX) has been found that accurately predicts response to the drug tipifarnib in patients with acute myeloid leukemia (AML) (34). The TSP algorithm was applied to transcriptional profiles of bone marrow samples from newly diagnosed AML patients—including 13 responders and 13 patients with progressive disease, achieving 92.3% sensitivity and 100% specificity (96% accuracy) in LOOCV. External validation of the two-gene classifier was performed in an independent dataset of 54 samples from patients with relapsed or refractory AML (10 responders, 44 with progressive disease). When applied to the independent test set, the classifier predicted tipifarnib response with sensitivity of 80% and specificity of 52.3%. This reduction in accuracy compared to LOOCV may derive from the initial very small sample set not being sufficient to represent the amount of variance in the population, and thus further data collection and classifier development is needed. Still, the results are encouraging considering the subtle difference of the phenotypes being considered and the small amount of training data.
In another study, Weichselbaum et al. (35) applied k-TSP to a previously determined gene expression signature—the IFN-related DNA damage signature (IRDS)—in order to develop a therapy-predictive marker of adjuvant chemotherapy for metastatic breast cancer. 78 breast cancer patients were divided into two IRDS status groups (IRDS(+) and IRDS(–)) using hierarchical clustering of microarray data. The k-TSP classifier was trained using 49 genes in the IRDS along with 534 previously-defined intrinsic breast cancer genes, with the optimal number of gene pairs determined using 10-fold cross validation. Each of the seven selected gene pairs in the k-TSP classifier contained one IRDS gene and a second gene for comparison. Classification was based on a majority vote, where samples were classified as IRDS(+) if expression of the IRDS gene was higher than the other gene in at least four of the seven pairs.
For the purpose of employing a non-binary measure for survival analysis, the number of positive-scoring gene pairs was used to define a TSP IRDS score. Specifically, the sum of pair-wise comparisons in which the IRDS gene was more highly expressed defined an ordinal scale from zero to seven, with seven representing the most IRDS(+)-like pattern. To examine the IRDS as a predictive marker for therapy outcome, a data set of 295 patients with early stage breast cancer was analyzed based on the TSP IRDS score. A multivariate Cox proportional-hazards model for metastatic risk when an interaction with chemotherapy is considered revealed a hazard ratio of 1.2—signifying a 1.2-fold increased risk of metastasis for each incremental increase in the TSP IRDS score from 0 to 7. These statistically significant results suggested that an association of the IRDS with clinical outcome depends on the use of adjuvant chemotherapy.
Broad Application of TSP in Disease Diagnosis and Prognosis
A more recent study has shown that two-transcript classifiers have the potential to reliably classify diverse human diseases (36). In this study, the investigators sought to assess the effectiveness of the TSP approach in the identification of diagnostic classifiers in a number of human diseases including bacterial and viral infection, cardiomyopathy, diabetes, Crohn's disease, and transformed ulcerative colitis through analysis of both local diseased tissue and the immunological response assayed through blood-borne leukocytes. The results of this study showed that several diseases of solid tissues could be reliably diagnosed through TSP classifiers based on the blood-borne leukocyte transcriptome. The TSP method identified multiple predictive gene pairs for each phenotype, with LOOCV accuracy ranging from 70 to nearly 100 percent. Performance compared favorably with that of pre-existing transcription-based classifiers, and in some cases approached the accuracy of current clinical diagnostic procedures. Thus, this study provided further evidence that the TSP classifier represents a simple yet robust method to differentiate between phenotypic states based on gene expression profiles of diverse human pathologies. The experimental simplicity of this method results in measurements that can be easily translated to clinical practice.
Beyond TSP and k-TSP
Top-Scoring Pair of Groups
In an effort to identify a robust common cancer signature, Xu et al. (18) performed a large-scale meta-analysis of cancer gene expression datasets in order to identify a universal cancer signature, and validated their signature using a variant of RXA to separate cancer from normal samples across a wide range of cancers. More specifically, the authors integrated nearly 1,500 microarray gene expression profiles from 26 published cancer data sets across 21 major human cancer types using two different Affymetrix microarray platforms. Michiels et al. (37) had shown that molecular signatures are strongly dependent on the samples in the training data and advocated the use of repeated random sampling for signature validation. In (18), the authors applied an RXA method, referred to as the top-scoring pair of groups (TSPG) classifier, combined with a repeated random sampling strategy to identify of a common cancer signature consisting of 46 genes. The TSPG classifier is an extension of the TSP classifier from two individual genes to two groups of genes. Being an RXA method, it is based entirely on the internal ranking of the genes in the signature. The signature is divided into two disjoint groups, and the average rank is computed for each group and two averages are compared. The decision rule is again maximum likelihood; to choose the class for which the observed ordering between the two rank averages is most likely. It can also be shown that TSPG is a special case of k-TSP, where k is the product of the two group sizes. Given a new expression profile, the classifier was found to discriminate most human cancers from normal tissues, including a validation on six different independent test datasets generated from different Affymetrix microarray platforms. Upon further validation, this cancer signature may be used to improve understanding of cancer pathogenesis and therapeutic targets, and hence lead to the development of effective treatment regimens.
Top-Scoring Triplets
Lin et al. (17) proposed an extension of TSP which bases prediction entirely upon the relative expression ordering among three genes, referred to as the “top-scoring triplets” (TST). The decision rule is to select the class which makes the observed ordering the most likely. In many cases, one gene serves as a “reference” whose expression falls between the expressions of two differentially expressed genes. The objective is to achieve a more discriminating decision mechanism than TSP but without sacrificing interpretability. The investigators explored the different roles the three genes play in the decision mechanism from previous cancer studies, and also applied this methodology to two problems in breast cancer: a cross study validation based on predicting ER status and a clinically relevant application to predicting germ-line BRCA1 mutations. Further analysis on protein-protein interactions among the triplets of genes aided in understanding the biological roles of the classifiers.
Conclusions and Future Directions
The advent of high-throughput measurement technologies for the comprehensive, rapid, and inexpensive detection of biomolecular signatures in human cells, tissues, and serum has led to the generation of a tremendous amount of raw, unprocessed information. However, analyzing and interpreting these data in order to enhance our understanding of human health and genetic diseases (i.e., cancer) continues to be a challenge in the scientific community. In the case of gene expression microarray data, standard statistical learning methods have been used to identify decision rules that can accurately distinguish disease phenotypes. These techniques have been shown to produce accurate classifiers, but still lack the convenience and simplicity desired for extracting any underlying biological rationale for the decision rules.
In this review, we have provided a detailed description of the concepts and methodologies of the TSP and k-TSP classifiers, two bioinformatics techniques for gene expression-based molecular classification based on the analysis of relative expression values. Due to the simplicity of the classifier and ease of biological interpretation, as well as its independence to data normalization and parameter-fitting, the TSP and k-TSP methods have been applied in several studies to perform molecular classification of various pathologies, primarily cancer. These methods, as we have shown above, display highly accurate classification performance in distinguishing a broad range of disease phenotypes (e.g., cancer vs. normal), cancer subclasses (e.g., AML vs. ALL, GIST vs. LMS), disease outcomes (e.g., metastasis, survival), and diverse human pathologies assayed through blood-borne leukocytes. We have also shown that natural extensions of the basic TSP and k-TSP methods can incorporate more genes and allow for indirect microarray data integration and hence large-scale meta-studies. Further work on RXA includes the use of biological network information for phenotype classification and biological discovery as well as decision tree-based strategies for classification of multiple disease phenotypes.
Acknowledgments
We gratefully acknowledge funding for this work from an NIH-NCI Howard Temin Pathway to Independence Award in Cancer Research (NDP), the Grand Duchy of Luxembourg-Institute for Systems Biology Program (PI: Leroy Hood), and a Developmental Research Grant from the Pacific Ovarian Cancer Research Consortium (NIH P50 CA83636, PI: Nicole Urban). The work of DG was partially supported by NIH-NCRR Grant UL1 RR 025005 and by NSF CCF-0625687.
References
- 1.Bicciato S, Pandin M, Didonè G, Di Bello C. Pattern identification and classification in gene expression data using an autoassociative neural network model. Biotechnol. Bioeng. 2003;81:594–606. doi: 10.1002/bit.10505. [DOI] [PubMed] [Google Scholar]
- 2.Bloom G, Yang IV, Boulware D, Kwong KY, Coppola D, Eschrich S, Quackenbush J, Yeatman TJ. Multi-platform, multi-site, microarray-based human tumor classification. Am. J. Pathol. 2004;164:9–16. doi: 10.1016/S0002-9440(10)63090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan J, Wei JS, Ringnér M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat. Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulesteix AL, Tutz G, Strimmer K. A CART-based approach to discover emerging patterns in microarray data. Bioinformatics. 2003;19:2465–2472. doi: 10.1093/bioinformatics/btg361. [DOI] [PubMed] [Google Scholar]
- 5.Dettling M, Buhlmann P. Boosting for tumor classification with gene expression data. Bioinformatics. 2003;19:1061–1069. doi: 10.1093/bioinformatics/btf867. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Yu CY, Singer B. Cell and tumor classification using gene expression data: construction of forests. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4168–4172. doi: 10.1073/pnas.0230559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu Y, Adam BL, Yasui Y, Ward MD, Cazares LH, Schellhammer PF, Feng Z, Semmes OJ, Wright GL., Jr. Boosted decision tree analysis of surface-enhanced laser desorption/ionization mass spectral serum profiles discriminates prostate cancer from noncancer patients. Clin. Chem. 2002;48:1835–1843. [PubMed] [Google Scholar]
- 8.Peng S, Xu Q, Ling XB, Peng X, Du W, Chen L. Molecular classification of cancer types from microarray data using the combination of genetic algorithms and support vector machines. FEBS Lett. 2003;555:358–362. doi: 10.1016/s0014-5793(03)01275-4. [DOI] [PubMed] [Google Scholar]
- 9.Yeang CH, Ramaswamy S, Tamayo P, Mukherjee S, Rifkin RM, Angelo M, Reich M, Lander E, Mesirov J, Golub T. Molecular classification of multiple tumor types. Bioinformatics. 2001;17:S316–322. doi: 10.1093/bioinformatics/17.suppl_1.s316. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 12.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring Pathway Activity toward Precise Disease Classification. PLoS Comput. Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auffray C. Protein subnetwork markers improve prediction of cancer outcome. Mol. Syst. Biol. 2007;3:141. doi: 10.1038/msb4100183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geman D, d'Avignon C, Naiman DQ, Winslow RL. Classifying gene expression profiles from pairwise mRNA comparisons. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1071. Article 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan AC, Naiman DQ, Xu L, Winslow RL, Geman D. Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics. 2005;21:3896–3904. doi: 10.1093/bioinformatics/bti631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin X, Afsari B, Marchionni L, Cope L, Parmigiani G, Naiman D, Geman D. The ordering of expression among a few genes can provide simple cancer biomarkers and signal BRCA1 mutations. BMC Bioinformatics. 2009;10:256. doi: 10.1186/1471-2105-10-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu L, Geman D, Winslow RL. Large-scale integration of cancer microarray data identifies a robust common cancer signature. BMC Bioinformatics. 2007;8:275. doi: 10.1186/1471-2105-8-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leek JT. The tspair package for finding top scoring pair classifiers in R. Bioinformatics. 2009;25:1203–1204. doi: 10.1093/bioinformatics/btp126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu L, Tan AC, Naiman DQ, Geman D, Winslow RL. Robust prostate cancer marker genes emerge from direct integration of inter-study microarray data. Bioinformatics. 2005;21:3905–3911. doi: 10.1093/bioinformatics/bti647. [DOI] [PubMed] [Google Scholar]
- 21.Braga-Neto UM, Dougherty ER. Is cross-validation valid for small-sample microarray classification? Bioinformatics. 2004;20:374–380. doi: 10.1093/bioinformatics/btg419. [DOI] [PubMed] [Google Scholar]
- 22.Gordon GJ, Jensen RV, Hsiao LL, Gullans SR, Blumenstock JE, Ramaswamy S, Richards WG, Sugarbaker DJ, Bueno R. Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res. 2002;62:4963–4967. [PubMed] [Google Scholar]
- 23.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O'Brien K, Wang Y. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 25.Jordan C. Historical perspective on hormonal therapy of advanced breast cancer. Clin. Ther. 2002;24:3–16. doi: 10.1016/s0149-2918(02)85031-7. [DOI] [PubMed] [Google Scholar]
- 26.Clarke MJ. Tamoxifen for early breast cancer. Cochrane Database of Systematic Reviews (Online) 2008:CD000486. doi: 10.1002/14651858.CD000486.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson RI, Gee JMW, Knowlden J, McClelland R, Madden TA, Barrow D, Hutcheson I. The biology of antihormone failure in breast cancer. Breast Cancer Res. Treat. 2003;80:29–34. doi: 10.1023/a:1025467500433. [DOI] [PubMed] [Google Scholar]
- 28.Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–140. [Google Scholar]
- 29.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 30.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 31.Price ND, Trent J, El-Naggar AK, Cogdell D, Taylor E, Hunt KK, Pollock RE, Hood L, Shmulevich I, Zhang W. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3414–3419. doi: 10.1073/pnas.0611373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, Tan AC, Winslow RL, Geman D. Merging microarray data from separate breast cancer studies provides a robust prognostic test. BMC Bioinformatics. 2008;9:125. doi: 10.1186/1471-2105-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajeshkumar NV, Tan AC, De Oliveira E, Womack C, Wombwell H, Morgan S, Warren MV, Walker J, Green TP, Jimeno A, Messersmith WA, Hidalgo M. Antitumor effects and biomarkers of activity of AZD0530, a Src inhibitor, in pancreatic cancer. Clin. Cancer Res. 2009;15:4138–4146. doi: 10.1158/1078-0432.CCR-08-3021. [DOI] [PubMed] [Google Scholar]
- 34.Raponi M, Lancet JE, Fan H, Dossey L, Lee G, Gojo I, Feldman EJ, Gotlib J, Morris LE, Greenberg PL, Wright JJ, Harousseau JL, Lowenberg B, Stone RM, De Porre P, Wang Y, Karp JE. A 2-gene classifier for predicting response to the farnesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood. 2008;111:2589–2596. doi: 10.1182/blood-2007-09-112730. [DOI] [PubMed] [Google Scholar]
- 35.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, Roizman B, Bergh J, Pawitan Y, van de Vijver MJ, Minn AJ. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edelman LB, Toia G, Geman D, Zhang W, Price ND. Two-transcript gene expression classifiers in the diagnosis and prognosis of human diseases. BMC Genomics. 2009;10:583. doi: 10.1186/1471-2164-10-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. The Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]