Abstract
The interior of cells is highly crowded with macromolecules, which impacts all physiological processes. To explore how macromolecular crowding may influence cellular protein folding, we interrogated the folding landscape of a model β-rich protein, cellular retinoic acid-binding protein I (CRABP I), in the presence of an inert crowding agent (Ficoll 70). Urea titrations revealed a crowding-induced change in the water-accessible polar amide surface of its denatured state, based on an observed ca. 15% decrease in the m-value (the change in unfolding free energy with respect to urea concentration), and the effect of crowding on the equilibrium stability of CRABP I was less than our experimental error (i.e., ≤1.2 kcal/mol). Consequently, we directly probed the effect of crowding on the denatured state of CRABP I by measuring side chain accessibility using iodide quenching of tryptophan fluorescence and chemical modification of cysteines. We observed that the urea-denatured state is more compact under crowded conditions, and the observed extent of reduction of the m value by crowding agent is fully consistent with the extent of reduction of the accessibility of the Trp and Cys probes, suggesting a random and nonspecific compaction of the unfolded state. The thermodynamic consequences of crowding-induced compaction are discussed. In addition, over a wide range of Ficoll concentration, crowding significantly retarded the unfolding kinetics of CRABP I without influencing the urea dependence of the unfolding rate, arguing for no appreciable change in the nature of the transition state. Our results demonstrate how macromolecular crowding may influence protein folding by effects both on the unfolded state ensemble and on unfolding kinetics. (end of abstract)
Introduction
The intracellular environment is highly crowded with macromolecules; total macromolecular concentrations range from 50 to 400 g/l.1–3 Due to excluded volume effects, cellular crowding increases the chemical potential of macromolecules, and hence the thermodynamic driving force for them to react.4 Macromolecular crowding also affects the nature of diffusion within the cell. Hence, ligand binding events, protein-protein interactions, enzymatic activities, protein folding, and essentially all other biochemical processes are expected to be modulated relative to their propensities in dilute solution by the high concentrations of macromolecules in cells. We have been studying how protein folding energy landscapes are altered by the cellular environment. Our results point to striking differences in the nature of a urea denaturation process inside E. coli cells from that in dilute solution.5,6 Among the many factors that differ between typical in vitro environments and those encountered in vivo, macromolecular crowding would seem to be a potentially major one that may influence the folding/unfolding equilibrium in vivo.
The impact of macromolecular crowding on protein folding has been explored both theoretically and experimentally.7–10 Yet despite numerous studies, fundamental gaps remain in our understanding of the effects of crowding. Theoretical prediction of quantitative crowding effect on protein stability varies largely depending on how the unfolded state is modeled.11–13 It is crucial that we gather direct experimental data on the impact of macromolecular crowding on the nature of the unfolded state. There are many experimental studies examining how crowding affects protein folding energetics,14–22 and the results mostly point to a modest stability effect. Experimental studies on the effects of crowding on kinetics of folding or unfolding do not form a clear picture as yet, with reports of retarded unfolding,21,23 no effect on unfolding kinetics,14,24 acceleration of refolding,22,24–26 or retardation of refolding.21,26 Studies characterizing crowding effects on ensemble distributions of the unfolded state are sparse and inconclusive.27,28 Crowding has been observed to favor aggregation and misfolding,25,29,30 potentially complicating experimental investigation of folding under crowded conditions.
In this study, we have used an inert macromolecular crowding agent (Ficoll 70) to probe the impact of crowding on the folding landscape of a model β-sheet rich protein, cellular retinoic acid-binding protein (CRABP I), a monomeric 18-kDa protein with a well studied folding mechanism in dilute solution.31–35 Three native tryptophans (Trp 7, Trp 87, Trp 109) all contribute to the intrinsic fluorescence of CRABP I and provide probes that are sensitive to local structure and environment. Our initial finding based on equilibrium urea denaturation experiments was that crowding caused a decrease in the m-value, the dependence of unfolding free energy on [urea]. The observed m-value change is most likely a result of compaction of the denatured state in the presence of crowding agent. Thus, we directly investigated how macromolecular crowding modulates the properties of the fully populated denatured state by measuring tryptophan accessibility using iodide quenching of Trp fluorescence and cysteine susceptibility to chemical modification. We obtained compelling experimental evidence that crowding leads to unfolded state compaction. Crowding significantly increased the Cm, the urea concentration for half-maximal unfolding, although the effect on protein stability at zero [urea] (ΔG°) was not significant given the experimental uncertainty (i.e., less than 1.4 kcal/mol). In addition, our results from investigating urea-induced unfolding over a wide range of Ficoll concentration also provide a clear picture of how crowding affects the unfolding rate of CRABP I. We observe significant crowding-induced retardation without significant change in urea dependence of unfolding rate, implying no appreciable change in the nature of the transition state. In this work, we propose a correction for the concentration of small molecules to account for volume exclusion by the crowding agent. Application of this correction enabled a better dissection of the effect of crowding on the m-value and how unfolding kinetics changed with crowding agent concentration.
Materials and Methods
Chemicals
Ultrapure urea (>99% pure, FW 60.06) was obtained from MP Biomedicals, LLC (Solon, Ohio). Ficoll 70 (FW 74 kDa) was from Fluka, Biochemika (Sweden) and O-(2-Maleimidoethyl)-O’-methyl-polyethylene glycol 5000 (Mal-PEG with FW 5 kDa, ≥90% purity (NMR)) was obtained from Fluka (Buchs, Switzerland). KI and KCl (certified ACS) were from Fisher Scientific Company (Fair Lawn, New Jersey). Na2S2O3 (99%) was from Aldrich (Milwaukee, WIS). N-Acetyl-L-Tryptophan-Amide (NATA) was from Sigma-Aldrich (St. Louis, MO). All other chemicals are reagent or molecular biology grade.
Protein
For expression and purification, we followed the published protocol31 with only minor change. Instead of sonication, a microfluidizer with 16 k-atm of h pressure was used to rupture the cells. Purified protein was lyophilized and stored at −20 °C. CD spectroscopy and fluorescence emission scans were detected for each batch of purified protein. Proteins were expressed with a His-tag for purification, which was not removed; previous studies show no influence of the His-tag on stability or folding/unfolding kinetics.32 Two CRABP I mutants were utilized in this study: (i) P85A, in which a stabilizing R131Q mutation has also been introduced, was used in all studies, and (ii) WT*, with only the R131Q mutation, was used only in unfolding kinetics. CD and fluorescence emission spectra show that there are no structural differences between the CRABP I variants.33,36
Equilibrium urea denaturation
Urea denaturation was performed to determine m-value, ΔG° (extrapolated protein stability in the absence of denaturant), and Cm (half-maximal urea concentration). Urea samples for denaturation titrations were prepared by mixing urea stock (in Tris buffer: 10 mM Tris·HCl, pH 8.0, with temperature dependence taken into account) with Tris buffer in a series of specified ratios, with both having the same concentration of Ficoll 70. Protein was then added to each sample from a single stock in Tris buffer/Ficoll 70, to a final concentration of 0.8–6.4 μM, with thorough and gentle mixing. Urea, Ficoll solution, and protein stock were always made fresh for each measurement, and each had 2 mM TCEP present as reductant. Urea denaturation samples were equilibrated in a closed incubator (to avoid water condensation) at 20 or 37 °C for a time determined to be sufficient to reach equilibrium at a given temperature (at least 8.5 and 5.5 hours for 20 and 37 °C, respectively), and then monitored by fluorescence emission at 350 ± 3 or 4 nm with excitation of 280 ± 2 nm using a PTI QM-1 fluorimeter with Peltier temperature control (Photon Technology International, New Brunswick, NJ) with emission scan (300–380 nm) also collected. A limited number of titrations were also monitored by far-UV CD signal (210–260 nm) to insure that consistent results were obtained and thus to check the two-state behavior of the system. Background for urea or Ficoll solutions was always subtracted as blank. All samples were centrifuged before measurements.
Urea denaturation curves were analyzed using an equilibrium two-state model and assuming validity of linear extrapolation of unfolding free energy with respect to urea concentration.37,38 Global fitting was performed on multiple data sets (usually 3–4, and 6 for 37 °C/150 g/l Ficoll) to obtain parameter values and standard errors. Besides conventional fitting of the ΔG° and m-value together, we also fitted the Cm and m-value together to estimate the error of Cm, otherwise there was no difference for the two kinds of fitting. Origin 8 software was used for regression analysis with the default ‘asymptotic-symmetry’ method used for standard error estimation.
Iodide quenching of tryptophan fluorescence
Dynamic quenching of tryptophan fluorescence by iodide was used to probe Trp accessibility.39,40 Quenching measurements were performed for a series of samples of increasing [KI] at 37 °C, basically as previously described.39 To insure constant ionic strength as the iodide concentration was increased, the total KI plus KCl concentration was held constant at 300 mM (330 mM also checked with no differences observed). Fresh KI and KCl stocks, in a solution of 8.0 M urea, 0 or 150 g/l Ficoll 70, with Tris buffer plus 2 mM TCEP, were added to protein or NATA stock dissolved in the same solution and incubated for 4 hours. Final protein concentrations were 3–5 μM, in the same range as used in urea denaturation experiments. A small amount of S2O32− (1 mM) was added to minimize I3− formation. Fluorescence emission scans (300–380 nm) were collected using the PTI QM-1 fluorimeter with excitation at 295 ± 3 nm. Background signal from the same solutions without protein or NATA were subtracted.
Cysteine PEGylation
PEGylation, i.e., reaction with polyethylene glycol bis-maleimide (Mal-PEG), has been used to map topological accessibility of Cys residues in native proteins,41 and was used in this study to probe Cys accessibility in the unfolded state. P85A CRABP I samples were unfolded in 8.1 M urea/Tris buffer with 0 or 150 g/l Ficoll 70 and 2 mM TCEP for over 4.5 hours at 20 °C. For PEGylation, 2 μl Mal-PEG stock (50-fold excess over the total Cys concentration, given that each protein has three cysteines) were added to 200 μl unfolded protein solution (with protein concentration of 5 μM), and the sample was thoroughly mixed. After 30 s of PEGylation reaction, 1 μl Cys-HCl was added at 40-fold excess of [Mal-PEG] to quench the reaction. After 2 minutes, the reaction solution was combined with 5X gel loading buffer and boiled for 7 min before running 10% SDS-PAGE. PEGylation time was tested in the presence or absence of Ficoll to insure complete reaction.
Protein unfolding kinetics
Native protein stock was added to urea solutions (both in Tris buffer/Ficoll 70 with 2 mM TCEP) to a final protein concentration of 3–5 μM to initiate unfolding at 20 or 37 °C. The unfolding traces were monitored by fluorescence emission at 350 ± 3–4 nm with excitation of 280 ± 2 nm using the PTI QM-1 fluorimeter. Quartz cuvettes (750 μl) equipped with a magnetic stirring bar were used to mitigate possible photobleaching effects due to slow diffusion in crowded solutions. An emission scan from 300 to 380 nm was collected at the end of each kinetic trace to confirm that the protein was completely unfolded. Single exponentials were found to provide good fits to the kinetic traces.
Results
Ficoll 70 is an inert, non-perturbing crowding agent
Consistent with previous report that Ficoll 70 (74 kDa) is an inert crowding agent,10 we found no significant soft interaction of Ficoll with urea by vapor pressure osmometry (see Supporting Information and Figure S1), and we found the partial specific volume of Ficoll 70 to be independent of urea concentration (see Supporting Information). We saw no perturbation to the structure of CRABP I, native or unfolded, upon addition of crowding agent (see Supporting Information and Figure S2), in contrast to recent reports.19,27,42 Our characterization of unfolded state properties was mostly performed at 150 g/l Ficoll, while our kinetics study spanned a wider range of [Ficoll]. The Ficoll concentrations studied clearly revealed trends from addition of crowding agent and fell into the ranges of macromolecular crowding reported for various cellular environments. 1–3,43
Urea concentration correction
The effect on small molecule concentration of the substantial excluded volume in solutions containing high concentrations of macromolecular species has been widely neglected, with a few exceptions (for example, see references 44–46). Comparing urea denaturation and urea-induced kinetic studies in the presence and absence of an inert crowding agent, we noticed that crowding agent is not the only factor that is varying between the two systems and that urea activity coefficient also varies at the same apparent [urea]. An inert crowding agent increases the activity coefficient of any species by decreasing its available volume through pure steric repulsion (γi = 1/favailable,i, where favailable,i is the fractional volume available to species i).8,47 Therefore, to be able to attribute observed thermodynamic/kinetic effects to crowding agent alone, we corrected the effect of the excluded volume of crowding agent on the urea activity coefficient. We propose a simple way to take into account this effect by subtracting the solvent-excluded volume of crowding agent (calculated using its partial specific volume) from the total solution volume to approximate the volume available to small molecules considering their negligible size. The activity coefficient, 1/favailable, determined in this way is then used to correct the apparent [urea] measured by refractive index48 (we verified that this measurement is not affected by Ficoll). Unless specified, urea concentration in this paper denotes the corrected concentration ([urea]eff).
Urea denaturation of P85A CRABP I in Ficoll follows a reversible two-state transition
To be able to attribute any observed effects of macromolecular crowding to altered properties or shifts in population of the two end-states, folded or denatured, we needed to insure that our conditions were consistent with a two-state model; i.e. that there were no complications due to protein aggregation or misfolding, both of which are likely to be enhanced under crowding conditions.25,29,30 CRABP I is a well-behaved protein that reversibly refolds in dilute solution without detectable equilibrium intermediates, although it visits kinetic intermediates upon refolding from urea.31–34,36 Under the conditions investigated (0, 75, 150 g/l Ficoll, 0.8–6.4 μM protein), two-state equilibrium behavior was confirmed by observation of a single transition in urea denaturation curves monitored by intrinsic fluorescence at 350 nm and by CD at either 218 (dominated by backbone contributions) or 232.5 nm (dominated by aromatic packing) (Figure S3). Denaturation data from all three measurements are well fit by a two-state transition, with, importantly, the same Cm and m-value. Folding reversibility under crowded conditions was demonstrated by the agreement of fluorescence emission spectra of protein unfolded and refolded to the same urea concentration (in the transition region) (Figure S3), confirming the absence of aggregation, which is consistent with the observation of no protein concentration dependence of urea denaturation. The single transition and reversibility justifies the applicability of an equilibrium two-state model to analyze urea denaturation curves.
Ficoll increases Cm and decreases the M-value
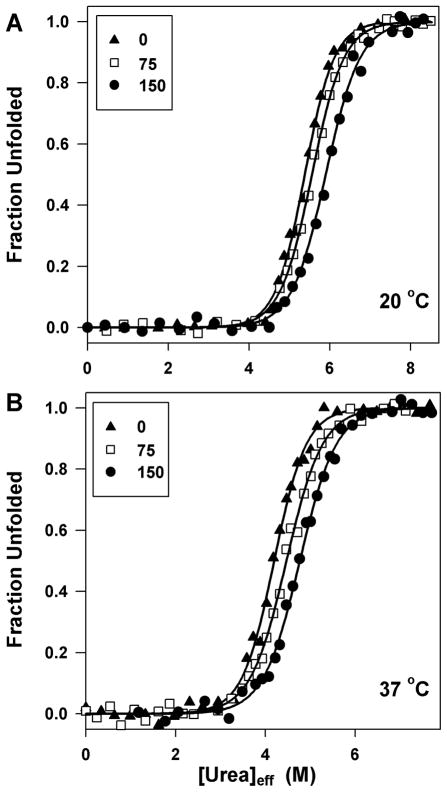
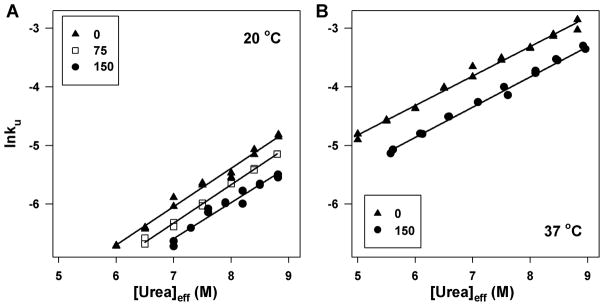
Urea denaturation of P85A CRABP I has been performed in 0, 75, 150 g/l Ficoll at either 20 or 37 °C (Figure 1). The data were analyzed in an unbiased way, and the resulting parameter values plus standard errors of m-value, Cm, and ΔG° from global fitting (with m-value and Cm or ΔG° shared, and parameters for native and unfolded state base lines floated for each data set) are reported in Table 1. Parameter values obtained from fitting individual curves at each condition yield mean values no different from the globally fitted parameter values and have significantly smaller standard deviations than the uncertainties obtained from global fitting (most are ≤50% the latter), indicating high reproducibility. A few conclusions can be drawn from these data: First, as revealed by an increased Cm, crowding makes protein more resistant to urea denaturation, but the effect of crowding on protein stability (ΔG°) is less than the experimental uncertainties of our fits (≤ 1.2 kcal/mol). Second, we find that crowding reduces the m-value to a small but statistically significant extent (by 13–15% between 0 and 150 g/l Ficoll). Although an apparent decrease in m-value can arise from accumulation of intermediate in the transition zone, we find the evidence for two-state equilibrium unfolding of CRABP I in crowded solution (see above) to be strong. Finally, protein stability decreases significantly as temperature increases from 20 to 37 °C whether in the presence of Ficoll or not, but the effect of Ficoll is comparable at either temperature. Correcting urea concentration for excluded volume enabled both the trend in m-values and in unfolding kinetics to be observed. Figure S3A, as an example, compares a denaturation curve in the presence of crowding agent as a function of uncorrected [urea] with one plotted versus [urea]eff and shows that plotting against [urea]eff leads to a higher Cm and a smaller m-value with negligible difference in ΔG°.
Figure 1.
Representative urea denaturation curves of CRABP I in the presence of different amounts of crowding agent at 20 °C (panel A) and 37 °C (panel B); fraction unfolded was determined by monitoring fluorescence at 350 nm. The symbols are the same for both panels: 0 (▲), 75 (□), and 150 (●) g/l Ficoll. The lines through the data are non-linear fits to an equilibrium two-state model.
Table 1.
Effect of macromolecular crowding on folding energetics of P85A CRABP Ia
| 20° C | 37° C | |||||
|---|---|---|---|---|---|---|
| Ficoll 70 (g·l−1) | m-value (cal·mol-1·M−1) | Cm (M) | ΔG° (cal·mol−1) | m-value (cal·mol-1·M−1) | Cm (M) | ΔG° (cal·mol−1) |
| 0 | 1568 ± 109 | 5.36 ± 0.02 | 8401 ± 580 | 1698 ± 127 | 4.27 ± 0.02 | 7252 ± 537 |
| 75 | 1448 ± 143 | 5.61 ± 0.03 | 8123 ± 791 | 1551 ± 92 | 4.48 ± 0.02 | 6951 ± 412 |
| 150 | 1326 ± 118 | 5.99 ± 0.04 | 7942 ± 681 | 1477 ± 90 | 4.81 ± 0.02 | 7100 ± 431 |
The parameter values plus the standard errors at each condition are the result of a global fitting of independent measurements (three to four data sets in general, and six for 37 °C/150 g/l Ficoll).
Macromolecular crowding leads to a more compact ensemble for the unfolded protein
The m-value for chemical denaturation has been correlated with the change in solvent accessible surface area (ΔASA) between the folded and unfolded states.49 Subsequent studies have pointed out that the m-value is actually probing the change in solvent-exposed polar amide surface upon unfolding.50,51 With regard to a local-bulk domain model interpretation of the m-value,52 we conclude that the proportionality between the m-value and the ΔASA remains the same in the presence of crowding agents, so long as the [denaturant] is corrected for the excluded volume of crowding agents (see Supporting Information). Thus, the reduced m-value observed in the presence of crowding agent suggests that crowding either caused the unfolded state to become more compact, or the native state to become more expanded. The former is much more likely, based on our having observed no structural change in crowded conditions and on theoretical grounds7, 53 and other reports.54 Thus, we directly characterized the unfolded state for its compaction in two ways: First, we investigated how crowding affects accessibility of tryptophan residues by assessing their susceptibility to iodide quenching. In contrast to some previous work,27, 28 we were characterizing a fully populated unfolded state with or without crowding agent. Parallel measurements of iodide quenching of unfolded protein in 8.0 M urea in 0 or 150 g/l Ficoll were carried out at 37 °C, with N-acetyl-tryptophanamide (NATA) as a control for fully accessible tryptophan. The Stern-Volmer plots (Figure S4) of both protein and NATA are linear, consistent with a dynamic quenching mechanism over the concentrations of iodide used.39,40,55 To determine the accessibility of Trps, we used a modified Stern-Volmer law,55 according to which, if there are n accessible fluorophores with the same value of Stern-Volmer constant (KQ) and the rest are inaccessible, the following relation exists between the fluorescence in the absence (F0) or presence of quencher (F), the molar quencher concentration X, and the fractional fluorescence arising from accessible fluorophores (fa)
| (1) |
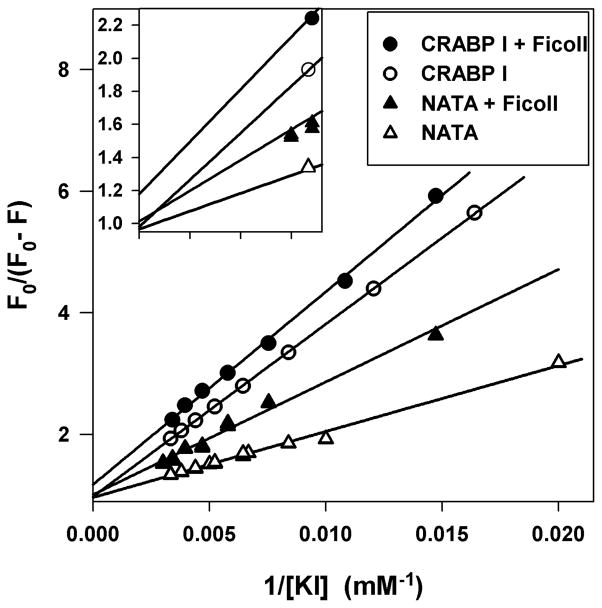
Based on eq. 1, a plot of F0/(F0-F) vs. 1/X yields a straight line with intercept 1/fa. If each exposed fluorophore has a different KQ or if there is static quenching, the plot of F0/(F0-F) vs. 1/X will show curvature. The intercept, 1/fa, for NATA is 1 in the presence or absence of Ficoll, as anticipated for a fully accessible Trp. Our analysis of iodide quenching data for the unfolded protein gives a linear plot (Figure 2), suggesting that the three Trp residues are indistinguishable in terms of iodide quenching. The intercept value of 1.00±0.02 (the standard deviation among three independent measurements) for unfolded CRABP I in dilute solution reveals full accessibility of the three fluorophores to iodide in the absence of macromolecular crowding. The intercept value of 1.16±0.02 for unfolded protein in 150 g/l Ficoll indicates a reduction of 14% (fa is 86%) in Trp accessibility to iodide quencher and, therefore, a more compact unfolded state in the presence of crowding agent.
Figure 2.
Representative modified Stern-Volmer plots for unfolded CRABP I at 37 °C in 150 (●) and 0 (○) g/l Ficoll and NATA (in 8.0 M urea) in 150 (▲) and 0 (△) g/l Ficoll. Lines are fits to eq. 1. The inset is the plot of the region near the intercept.
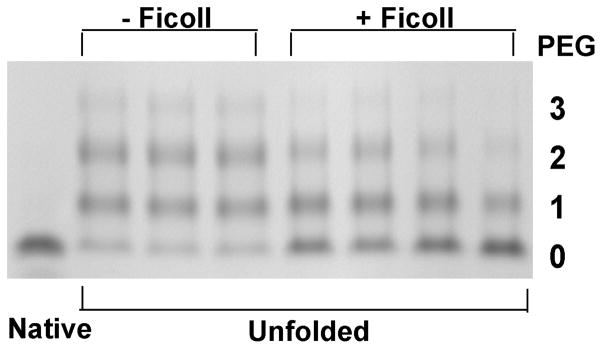
The second method we used to test the possible compaction of the unfolded state in the presence of macromolecular crowding was assessing cysteine side chain accessibility by modification with PEG-maleimide, which reacts with exposed cysteine sulfhydryls and adds approximately 5 kDa to the molecular weight of the modified protein per PEG group transferred. At a urea concentration of 8.1 M, 20 °C, 100% of the CRABP population is unfolded in the presence or absence of 150 g/l Ficoll according to our equilibrium urea denaturation data. CRABP has three Cys residues, all of which are sequestered in the native state and, as shown in Figure 3, readily PEGylated in the unfolded state in the absence of crowding agent. Upon addition of 150 g/l Ficoll, the band for un-PEGylated protein is significantly more intense and bands for PEGylated protein (whether adding one, two or three PEG groups) are weaker. In other words, the Cys residues are significantly less accessible to PEGylation in the presence of macromolecular crowding agent, again providing direct support for a more compact unfolded state. Quantification of the gel bands from several independent reactions in dilute (11) and crowded (14) solutions gives an average 18±3% decrease in PEGylation due to macromolecular crowding. The different extents of PEGylation are not due to crowding effect on PEGylation kinetics (see Supporting Information and Figure S5).
Figure 3.
Extent of PEGylation of unfolded CRABP I in 0 or 150 g/l Ficoll at 20 °C. Native protein was used as a control since all three cysteines are inaccessible in the native state in 0 or 150 g/l Ficoll. The number of PEGylated residues is indicated at right.
Macromolecular crowding substantially retards CRABP unfolding
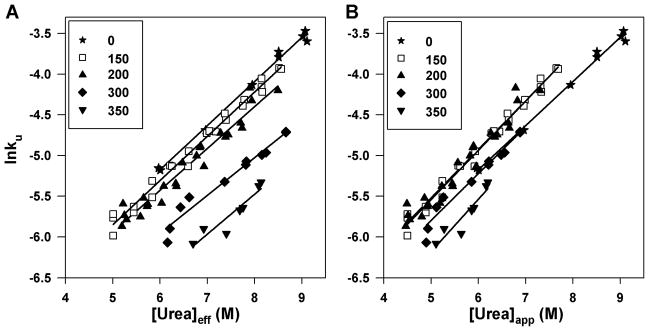
Unfolding kinetics were monitored at urea concentrations where the protein would be fully unfolded at equilibrium (see Figure S6 for a representative raw unfolding trace). We examined unfolding kinetics for CRABP I (WT*) in 0, 150, 200, 300, 350 g/l Ficoll at 37 °C (Figure 4) and to insure that observed kinetics were not affected by a slow cis-trans isomerization around the Leu84-Pro85 peptide bond previously found to contribute a minor phase to folding kinetics,36 we also measured kinetics for P85A CRABP I in 0, 75, 150 g/l Ficoll at 20 and 37 °C and saw no appreciable differences in the trends (Figure 5). We report here a distinct retarding effect on protein unfolding rate by crowding agent, and we are able to assess this kinetic crowding effect as a function of crowding agent concentration. For both variants of CRABP I, the urea dependence of unfolding rate (from which the mu-value is derived from the slope of ln(unfolding rate) as a function of [urea]) does not change significantly with crowding agent concentration, arguing that macromolecular crowding does not appreciably alter the nature of the transition state ensemble. Again, we observed marked impact of urea concentration correction on unfolding kinetics under crowding conditions. When plotted as a function of apparent [urea], the unfolding kinetics show no systematic trend as Ficoll concentration increases. Some rates are observed to be accelerated, contrary to predictions based on excluded volume effects, at and below 200 g/l Ficoll. However, when the unfolding rate is plotted as a function of [urea]eff, a clear trend emerges, with the rate becoming smaller with increased concentration of crowding agent (Figure 4).
Figure 4.
Unfolding rate of WT* CRABP I at 37 °C plotted as a function of [urea]eff (A) and of apparent [urea] (B) in 0 (★), 150 (□), 200 (▲), 300 (◆), 350 (▼) g/l Ficoll. Straight-line fits of the data are shown.
Figure 5.
Unfolding rate of P85A CRABP I as a function of [urea]eff at 20 °C (A) and 37 °C (B) in 0 (▲), 75 (□), 150 (●) g/l Ficoll. Straight-line fits of the data are shown.
Discussion
In this study, we interrogated the folding landscape of the β-rich monomeric protein, CRABP I, in the presence of an inert macromolecular crowding agent and found it refolds reversibly by a two-state equilibrium process and suffers no competing aggregation under the conditions used. Therefore, observed effects of macromolecular crowding can be attributed to the altered populations or properties of the two end states (native and denatured). We found that the change in CRABP I stability from crowding was less than the error limits of our experiments (viz., 1.2 kcal/mol) for all Ficoll concentrations tested, at either 25 or 37 °C. We observed a significant upward shift in the half-maximal urea concentration for unfolding (Cm) upon addition of crowding agent and conclude that it arose primarily from a crowding-induced decrease in the urea dependence of the equilibrium constant for denaturation (the m-value), arguing for a change in the nature of the unfolded ensemble in the presence of crowding agent. Our direct characterization of a fully populated denatured state, as assured by our equilibrium urea denaturation data, showed that the crowding agent causes a significant (~15%) compaction.
Our data point to a non-specific and random compaction due to crowding, because the extent of compaction deduced from the reduction in solvent-exposed polar amide surface (13–15%) (based on the m-value decrease), the decrease in Trp accessibility (14%) (from iodide quenching), and the lowered Cys accessibility (18%) (from PEGylation) due to 150 g/l Ficoll is similar. This may account for the lack of change detected in CD or fluorescence spectra. To understand the meaning of this change in solvent-accessible surface, we estimated the magnitude of the reduction in polar amide surface area under crowding condition. Using the proportionality of m-value/ΔASApolar amide51 and neglecting its temperature effect, the observed m-value decrease from 0 to 150 g/l Ficoll of 242 or 221 cal/(mol ·M) [at 20 and 37 °C, respectively] corresponds to 291 or 267 Å2 of polar amide surface, about the size of 11–12 backbone groups.51 The observation of a more compact unfolded state in the presence of a crowding agent is consistent with predictions by Minton.12 We suspect that comparable previous experiments have not revealed this crowding effect on the unfolded state either because they were confounded by coexistence of intermediate and unfolded states, or by formation of aggregates under crowded conditions,27 or because they were not designed to provide results quantitative enough to measure this small but significant effect.28 Compaction of an unfolded state ensemble reflected by a change in the m-value of two-state equilibrium unfolding has been previously reported; in this case, the compaction occurred in response to a pH change.56,57
Compaction of the unfolded state in the presence of macromolecular crowding agents has an important general thermodynamic consequence, as illustrated by Figure 6. Due to their large excluded volume effect, macromolecular crowding agents increase the free energy of all macromolecular solutes; the larger the molecule, the larger the effect.4,58 The differential increase in the free energy of native and unfolded state determines how crowding affects the stability of the native relative to the unfolded state. A more compact unfolded state will suffer a smaller free energy increase due to the excluded volume effect than a more expanded unfolded state. Thus, our data showing compaction of the unfolded state argue for a smaller free energy difference between the unfolded and native states in the presence of Ficoll than one would expect if a more expanded unfolded state were present in the crowded solution. In this argument, we are neglecting the energy effect due to chain entropy reduction in a more compact unfolded state, which we believe will be small, but unfavorable, given the relatively modest extent of chain compaction.
Figure 6.
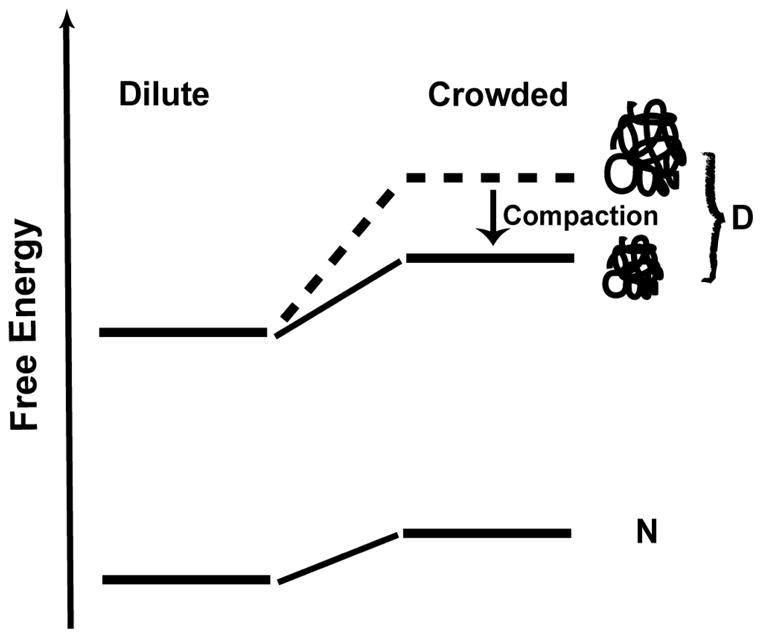
Crowding favors a more compact denatured state (D), which leads to a smaller difference in free energy between native (N) and D state in crowded solution than one would expect if a more expanded unfolded state (indicated as dashed line) were present in the crowded solution.
We observed a distinct retarding effect of crowding agents on the unfolding rate of CRABP I. The retardation cannot be explained simply by elevation of the activation energy due to the differential increase in the free energy of the transition state relative to the native state. For example, the decrease in unfolding rate of P85A by 150 g/l Ficoll at 20 and 37 °C corresponds to an activation energy increase of 330 cal/mol. Using scaled particle theory and modeling all macromolecules as hard spheres58 and using a radius of 2 nm for the native protein (estimated from the crystal structure59) and 5.5 nm for Ficoll,46 we found that to increase activation energy by ~330 cal/mol the transition state must be expanded in radius by 2.5 fold, in which case the transition state would be nearly as expanded as a fully unfolded state.60 This interpretation is inconsistent with the small ratio of mu to m (the Tanford β value61, 62) for P85A CRABP I (0.24 at 20 °C or 0.18 for 37 °C), which argues for a quite native-like transition state. Therefore, there must be other factors affecting unfolding rate. One possibility is a viscosity effect that acts on the pre-exponential factor.63 Protein unfolding is likely to be accompanied by a large local viscosity change due to collision with the macromolecular crowding agent during polypeptide chain rearrangements involved in the unfolding process. The lack of change in the mu-value in the presence of crowding agent implies that crowding has little effect on the compaction of the transition state ensemble, consistent with the observation of a native-like transition state. The lack of change in mu-value and a decrease in m-value implies a reduced urea concentration dependence of the overall refolding rate in Ficoll solution.
We observed that crowding agent leads to an increase in Cm, which together with the decrased m-value indicates that the protein becomes more resistant against urea denaturation in crowded solution. We are limited by experimental error in our efforts to determine the effect of crowding on stability and can conclude only that the crowding-induced stability change is < 1–1.2 kcal/mol. Indeed, the macromolecular crowding effect on the stability of CRABP I is not expected to be large given the small size of CRABP I relative to Ficoll 70 (ratio of molecular weights approximately 1:4)12 and the compaction of the unfolded state (see above). Scaled particle theory provides an estimate of the upper bound for the stability effect of crowding, based on modeling all macromolecules in all states as hard spheres:58 Using a radius of 2 nm for the native protein, 5 nm for the unfolded protein (assuming a 2.5-fold increase with respect to native60), and 5.5 nm for Ficoll,46 we estimate a maximal increase in stability of 326 (345) cal/mol at 20 (37) °C from addition of 150 g/l Ficoll. Consistent with this work, other studies involving proteins that are small relative to the crowding agent have also reported only modest stabilizing effects at similar concentration of crowding agent.16,18 When the protein and crowding agent are comparable in size, the effect of crowding on protein stability (ΔG°) is expected to be significantly larger and the stabilizing effect to increase greatly as crowding agent concentration increases.12
The observation that crowding makes the unfolded state more compact represents one way that a protein’s folding landscape in the crowded intracellular environment is markedly different from that sampled in dilute solution in the test tube. Interestingly, the retarding effect on unfolding kinetics by crowding agent suggests that macromolecular crowding alone can make a protein kinetically more stable. Moreover, our data say that crowding reduces the response of a protein to destabilizing conditions such as chemical denaturation. However, revisiting our urea denaturation experiments using FlAsH-labeled CRABP in E. coli cells, where the urea melting isotherm was characterized by a significantly lower Cm and a higher m-value than the dilute solution isotherm,6 we conclude that factors apart from macromolecular crowding, such as chaperone binding, heterogeneity of the cellular environment, weak protein-protein interactions, physiological responses to urea addition, different salt concentrations, and so on, must cause the markedly different urea melt. In addition, we saw more rapid unfolding kinetics and equilibration times in vivo than in vitro, which is the opposite of our observations in the presence of inert macromolecular crowding agents. We are currently pursuing the other factors that contribute to the complexity of the in vivo folding process and how they may remodel the energy landscape.
Conclusion
We provide here compelling experimental evidence that macromolecular crowding makes the unfolded state of a protein more compact. As discussed, such compaction has potential influence on protein folding energetics, and reduces the response of protein to chemical denaturation. We also provide strong evidence that crowding retards protein unfolding but without changing the urea dependence of unfolding rate, implying little change in the nature of unfolding transition state. Lastly, these observations do not account for in-cell urea denaturation studies we have carried out, emphasizing the complexity and multitude of factors that influence folding in the cell.
Supplementary Material
Acknowledgments
We thank Allen Minton for valuable suggestions and thank Anne Gershenson for careful reading of the manuscript and valuable suggestions. This work was supported by an NIH Director’s Pioneer Award (OD000945).
Footnotes
Supporting Information Available: Figures S1–S6. VPO determination of soft interaction of urea with Ficoll. CD and fluorescence data showing that Ficoll 70 does not perturb the structure of CRABP I. Justification that the proportionality between m-value and the change of water accessible surface area (ΔASA) upon protein unfolding is the same in the presence or absence of crowding agent when using corrected urea molar concentration. Control experimental data showing that reduced PEGylation in crowded solution is not due to a crowding effect on PEGylation kinetics. Determination of partial specific volume of Ficoll. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fulton AB. Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerman SB, Trach SO. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 3.Cayley S, Lewis BA, Guttman HJ, Record MT., Jr J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 4.Minton AP. Mol Cell Biochem. 1983;55:119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- 5.Ignatova Z, Gierasch LM. Proc Natl Acad Sci U S A. 2004;101:523–528. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ignatova Z, Krishnan B, Bombardier JP, Marcelino AM, Hong J, Gierasch LM. Biopolymers. 2007;88:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman SB, Minton AP. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 8.Hall D, Minton AP. Biochim Biophys Acta. 2003;1649:127–139. doi: 10.1016/s1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]
- 9.Ellis RJ. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhou HX, Rivas G, Minton AP. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minton AP. Biophys J. 2000;78:101–109. doi: 10.1016/S0006-3495(00)76576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minton AP. Biophys J. 2005;88:971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou HX. J Mol Recognit. 2004;17:368–375. doi: 10.1002/jmr.711. [DOI] [PubMed] [Google Scholar]
- 14.Homouz D, Perham M, Samiotakis A, Cheung MS, Wittung-Stafshede P. Proc Natl Acad Sci U S A. 2008;105:11754–11759. doi: 10.1073/pnas.0803672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPhie P, Ni YS, Minton AP. J Mol Biol. 2006;361:7–10. doi: 10.1016/j.jmb.2006.05.075. [DOI] [PubMed] [Google Scholar]
- 16.Qu Y, Bolen DW. Biophys Chem. 2002;101–102:155–165. doi: 10.1016/s0301-4622(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 17.Sasahara K, McPhie P, Minton AP. J Mol Biol. 2003;326:1227–1237. doi: 10.1016/s0022-2836(02)01443-2. [DOI] [PubMed] [Google Scholar]
- 18.Spencer DS, Xu K, Logan TM, Zhou HX. J Mol Biol. 2005;351:219–232. doi: 10.1016/j.jmb.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 19.Stagg L, Zhang SQ, Cheung MS, Wittung-Stafshede P. Proc Natl Acad Sci U S A. 2007;104:18976–18981. doi: 10.1073/pnas.0705127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlton LM, Barnes CO, Li C, Orans J, Young GB, Pielak GJ. J Am Chem Soc. 2008;130:6826–6830. doi: 10.1021/ja8005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee S, Waegele MM, Chowdhury P, Guo L, Gai F. J Mol Biol. 2009;393:227–236. doi: 10.1016/j.jmb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladurner AG, Fersht AR. Nat Struct Biol. 1999;6:28–31. doi: 10.1038/4899. [DOI] [PubMed] [Google Scholar]
- 23.Yuan JM, Chyan CL, Zhou HX, Chung TY, Peng H, Ping G, Yang G. Protein Sci. 2008;17:2156–2166. doi: 10.1110/ps.037325.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ai X, Zhou Z, Bai Y, Choy WY. J Am Chem Soc. 2006;128:3916–3917. doi: 10.1021/ja057832n. [DOI] [PubMed] [Google Scholar]
- 25.Monterroso B, Minton AP. J Biol Chem. 2007;282:33452–33458. doi: 10.1074/jbc.M705157200. [DOI] [PubMed] [Google Scholar]
- 26.van den Berg B, Wain R, Dobson CM, Ellis RJ. EMBO J. 2000;19:3870–3875. doi: 10.1093/emboj/19.15.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel R, Westphal AH, Huberts DH, Nabuurs SM, Lindhoud S, Visser AJ, van Mierlo CP. J Biol Chem. 2008;283:27383–27394. doi: 10.1074/jbc.M802393200. [DOI] [PubMed] [Google Scholar]
- 28.Ittah V, Kahana E, Amir D, Haas E. J Mol Recognit. 2004;17:448–455. doi: 10.1002/jmr.702. [DOI] [PubMed] [Google Scholar]
- 29.van den Berg B, Ellis RJ, Dobson CM. EMBO J. 1999;18:6927–6933. doi: 10.1093/emboj/18.24.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munishkina LA, Cooper EM, Uversky VN, Fink AL. J Mol Recognit. 2004;17:456–464. doi: 10.1002/jmr.699. [DOI] [PubMed] [Google Scholar]
- 31.Clark PL, Weston BF, Gierasch LM. Fold Des. 1998;3:401–412. doi: 10.1016/s1359-0278(98)00053-4. [DOI] [PubMed] [Google Scholar]
- 32.Clark PL, Liu ZP, Zhang J, Gierasch LM. Protein Sci. 1996;5:1108–1117. doi: 10.1002/pro.5560050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark PL, Liu ZP, Rizo J, Gierasch LM. Nat Struct Biol. 1997;4:883–886. doi: 10.1038/nsb1197-883. [DOI] [PubMed] [Google Scholar]
- 34.Liu ZP, Rizo J, Gierasch LM. Biochemistry. 1994;33:134–142. doi: 10.1021/bi00167a017. [DOI] [PubMed] [Google Scholar]
- 35.Rizo J, Liu ZP, Gierasch LM. J Biomol NMR. 1994;4:741–760. doi: 10.1007/BF00398406. [DOI] [PubMed] [Google Scholar]
- 36.Eyles SJ, Gierasch LM. J Mol Biol. 2000;301:737–747. doi: 10.1006/jmbi.2000.4002. [DOI] [PubMed] [Google Scholar]
- 37.Greene RF, Jr, Pace CN. J Biol Chem. 1974;249:5388–5393. [PubMed] [Google Scholar]
- 38.Santoro MM, Bolen DW. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 39.Lehrer SS, Leavis PC. Methods Enzymol. 1978;49:222–236. doi: 10.1016/s0076-6879(78)49012-3. [DOI] [PubMed] [Google Scholar]
- 40.Maruthamuthu M, Selvakumar G. Proc Indian Acad Sci. 1995;107:79–86. [Google Scholar]
- 41.Lu J, Deutsch C. Biochemistry. 2001;40:13288–13301. doi: 10.1021/bi0107647. [DOI] [PubMed] [Google Scholar]
- 42.Perham M, Stagg L, Wittung-Stafshede P. FEBS Lett. 2007;581:5065–5069. doi: 10.1016/j.febslet.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 43.Luby-Phelps K. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 44.Laurent TC. Eur J Biochem. 1971;21:498–506. doi: 10.1111/j.1432-1033.1971.tb01495.x. [DOI] [PubMed] [Google Scholar]
- 45.Jiang M, Guo Z. J Am Chem Soc. 2007;129:730–731. doi: 10.1021/ja065064+. [DOI] [PubMed] [Google Scholar]
- 46.Wenner JR, Bloomfield VA. Biophys J. 1999;77:3234–3241. doi: 10.1016/S0006-3495(99)77154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Record MT, Jr, Courtenay ES, Cayley S, Guttman HJ. Trends Biochem Sci. 1998;23:190–194. doi: 10.1016/s0968-0004(98)01207-9. [DOI] [PubMed] [Google Scholar]
- 48.Pace CN, Shirley BA, Thompson JA. In: Protein Structure: A Practical Approach. Creighton TE, editor. IRL; Oxford: 1989. pp. 311–330. [Google Scholar]
- 49.Myers JK, Pace CN, Scholtz JM. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Auton M, Holthauzen LM, Bolen DW. Proc Natl Acad Sci U S A. 2007;104:15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong J, Capp MW, Saecker RM, Record MT., Jr Biochemistry. 2005;44:16896–16911. doi: 10.1021/bi0515218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Courtenay ES, Capp MW, Saecker RM, Record MT., Jr Proteins. 2000;(Suppl 4):72–85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 53.Minton AP. J Pharm Sci. 2005;94:1668–1675. doi: 10.1002/jps.20417. [DOI] [PubMed] [Google Scholar]
- 54.Samiotakis A, Wittung-Stafshede P, Cheung MS. Int J Mol Sci. 2009;10:572–588. doi: 10.3390/ijms10020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehrer SS. Biochemistry. 1971;10:3254–3263. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- 56.Pace CN, Laurents DV, Thomson JA. Biochemistry. 1990;29:2564–2572. doi: 10.1021/bi00462a019. [DOI] [PubMed] [Google Scholar]
- 57.Pace CN, Grimsley GR, Thomas ST, Makhatadze GI. Protein Sci. 1999;8:1500–1504. doi: 10.1110/ps.8.7.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebowitz JL, Helfand E, Praestgaard E. J Chem Phys. 1965;43:774–779. [Google Scholar]
- 59.Kleywegt GJ, Bergfors T, Senn H, Le Motte P, Gsell B, Shudo K, Jones TA. Structure. 1994;2:1241–1258. doi: 10.1016/s0969-2126(94)00125-1. [DOI] [PubMed] [Google Scholar]
- 60.Goldenberg DP. J Mol Biol. 2003;326:1615–1633. doi: 10.1016/s0022-2836(03)00033-0. [DOI] [PubMed] [Google Scholar]
- 61.Tanford C. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 62.Tanford C. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 63.Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA. Science. 1992;256:1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.