Abstract
We have examined the effects of surface nanotopography on in vitro osteogenesis of human mesenchymal stem cells (hMSCs). UV-assisted capillary force lithography was employed to fabricate a scalable (4 cm × 5 cm), well-defined nanostructured substrate of a UV curable polyurethane polymer with dots (150-, 400-, 600-nm diameter) and lines (150-, 400-, 600-nm width). The influence of osteogenic differentiation of hMSCs was characterized at day 8 by alkaline phosphatase (ALP) assay, RT-PCR, and real time PCR analysis. We found that hMSCs cultured on the nanostructured surfaces in osteogenic induction media showed significantly higher ALP activity compared to unpatterned PUA surface (control group). In particular, the hMSCs on the 400-nm dot pattern showed the highest level of ALP activity. Further investigation with real-time quantitative RT-PCR analysis demonstrated significantly higher expression of core binding factor 1 (Cbfa1), osteopontin (OP), and osteocalcin (OC) levels in hMSCs cultured on the 400-nm dot pattern in osteogenic induction media. These findings suggest that surface nanotopography can enhance osteogenic differentiation synergistically with biochemical induction substance.
Introduction
Cell fate is dictated in part by adhesive, mechanical interactions between cells and surrounding extracellular matrix (ECM) substrates as well as the milieu of soluble, diffusible factors. 1-2 In particular, surface nanotopography has been shown to exert influence over adhesion, proliferation, and gene expression in many cell types. Recent advances in micro- and nanofabrication techniques have great potential to exercise a high degree of control over the physical properties of biomaterials such as elastic modulus, 3-4 roughness, 5-6 size, 7-8 and topography 9-12 at cell/tissue-implant interface, allowing for the study of mechanical interactions between cells and their local environment on the sub-cellular scale. 13-18 In the past few years, for example, exploration of synthetic nanometer-scale features to mimic natural matrices has revealed a significant influence of surface nanotopography on cellular behaviors including changes in gene expression, cell proliferation, migration, adhesion, and differentiation. 7, 9, 18-25 Although the influence of the substratum microtopography has been extensively studied, the effect of substratum nano-features, particularly on differentiation of adult stem cells has rarely been investigated.
Engineered nanometer-scale scaffolds hold great promise for stem cell differentiation and transplantation. 6, 12, 26-27 Arguably, to facilitate this research, more facile and efficient fabrication methods need to be developed to direct differentiation of stem cells into a particular lineage for clinical application. Recent studies have shown that mammalian cells including hMSCs are capable of responding to the substratum nano-topography. 22, 28-33 In particular, cues arisen from nanotopographically-defined surfaces apparently directed differentiation into neuronal lineage 30 or fibroblast of human bone marrow stromal cells. 34-36 In addition, hMSCs cultured on the nanograting of 350-nm width lines showed significant neuronal and muscular gene expression. 30
The hMSCs have been isolated and established from several sources including bone marrow, adipose tissue, and umbilical cord blood. Of these, the bone marrow derived hMSCs, with the surface markers of CD105+, CD166+, CD29+, CD14-, CD34-, CD45-, have self limited renewal and differentiation capability into diverse cells of mesodermal origin, e.g., bone, cartilage, muscle, and connective tissues. 37-39 It has been revealed that the differentiation of hMSCs into osteogenesis is influenced by Runx2 (Runt-related transcription factor 2) gene. 40 The Runx2 gene regulates bone development by osteogenesis using G protein-coupled signaling pathway, promoting up-regulation of bone specific extracellular matrix, such as alkaline phosphatase (ALP), osteopontin (OP), osteocalcin (OC), and bone sialoprotein (BSP). Runx2 expression and its activity are influenced by external signal, cell to cell interaction, and growth regulatory factor. 22, 41-42
In the present study, we investigated the osteogenic differentiation of bone marrow-derived hMSCs by culturing these cells on a range of diverse nanostructured surfaces. To achieve this goal, well-defined, large-area (>3×3 cm2) nanopatterns (dots and lines) were fabricated on glass coverslips using UV-assisted capillary force lithography (CFL), which allowed for a simple and scalable approach to creating a structured biomaterial interface for cell adhesion studies. 15, 20, 43 This technique was developed by combining the crucial element of soft lithography – using an elastomeric or soft mold – and the use of capillarity for polymer molding. 44 This method allows one to exploit topographic definition of the substratum while eliminating the need to use an extremely high pressure that is typically needed in nanoimprint lithography. CFL has proven to be effective in fabricating various complex nanopatterns using a mold material with differences in permeability, mechanical modulus, and surface tensions.15
With UV-assisted CFL technique, various scalable (>3×3 cm2) nanopatterns were fabricated such as nanoscale dots (150-, 400-, 600-nm diameter) and lines (150-, 400-, 600-nm width). For the patterning material, a UV curable polyurethane polymer functionalized with acrylate groups (polyurethane acrylate, PUA) was used for its biocompatibility, fast curing time (< 20 s), and easy release from the mold. 45-46 Using these nanopatterns, the effects of surface nanotopography on in vitro osteogenesis of human mesenchymal stem cells (hMSCs) were examined. We found that osteogenic differentiation of hMSCs was much enhanced on the nanostructured surfaces of 150-nm width lines, 150-nm diameter dots, and 400-nm diameter dots in osteogenic induction media (OM), as verified by alkaline phosphatase (ALP) assay, RT-PCR analysis. Further investigation with real-time quantitative RT-PCR analysis showed that the significantly higher Cbfa1 level of hMSCs was expressed from culture on the nanostructured surface of 400-nm diameter dots (p<0.05) compared with the unpatterned PUA surface (control group). These results suggest that surface nanotopography, in the presence of suitable biochemical signals, can exert significant influence on regulating stem cell differentiation.
Experimental section
Fabrication of nanostructured substrates with UV-assisted CFL
A small amount (∼0.1 – 0.5 mL) of a UV curable PUA prepolymer was drop-dispensed on silicon master having positive patterns (features sticking out) and a supporting poly(ethylene terephthalate) (PET) film was carefully placed on top of the surface to make conformal contact. The PET film used in this study was surface modified with urethane groups to increase adhesion to the acrylate-containing monomer (Minuta Tech., Korea). The silicon masters had been prepared by photolithography or electron-beam lithography. To cure, the resin was exposed to UV (wavelength: 250 ∼ 400 nm) for 17 s at an intensity of 100 mW/cm2, and the cured 1st replica was peeled off from the master using a sharp tweezer. The 1st replica was additionally exposed to UV overnight to remove any uncured active groups on the surface. The 2nd replica was prepared by using a capillary molding process on glass coverslips with the over-cured 1st PUA as a mold, resulting in the same pattern as the silicon master. After curing, the 1st replica was removed from the surface using a sharp tweezers. The fabricated PUA nanopatterns were sterilized by rinsing with IPA (isopropyl alcohol) and D.I. water, and coated with 0.1 % gelatin for 1 hr at room temperature prior to cell culture. A schematic diagram of the fabrication procedure is illustrated in Figure 1 along with the self-replication characteristic of the PUA material.
Figure 1.
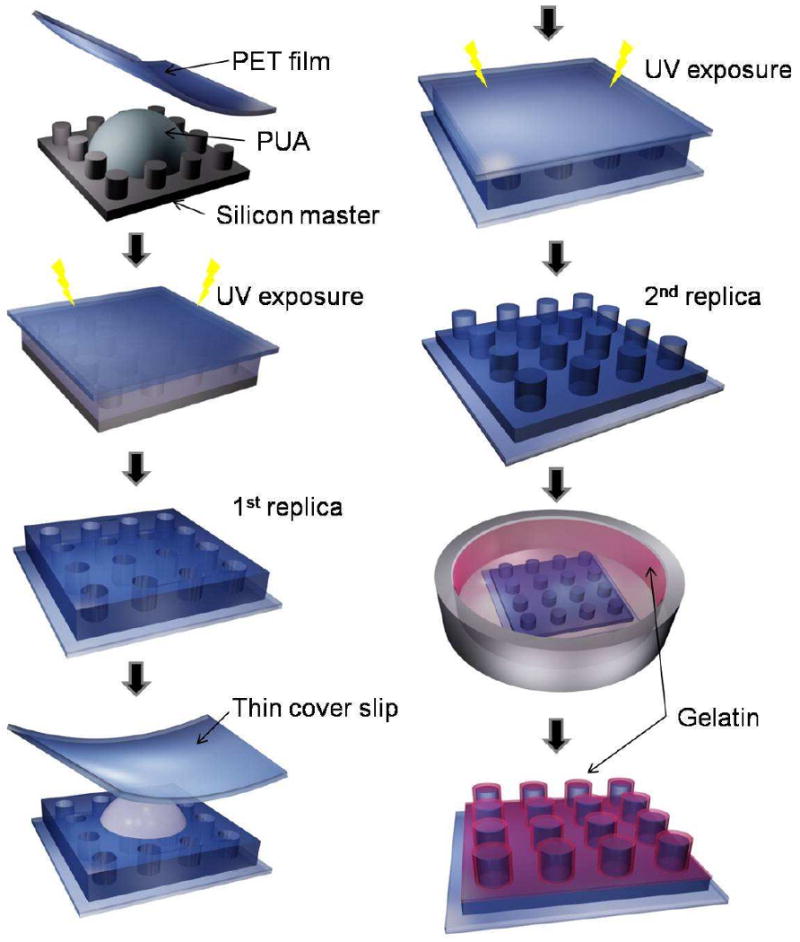
A schematic diagram for the fabrication of PUA nanopatterns by self-replication and capillary force lithography (CFL). As shown, the PUA patterns (2nd replica) have the same geometry with the silicon master after double replications.
Human mesenchymal stem cell culture
Cultured hMSCs were purchased from FCB-Pharmicell CO., Ltd., Sungnam, South Korea. Initially, hMSCs were obtained from 20 mL of aspirates from the iliac crest of normal human doners. hMSCs isolation and culture procedures were performed under GMP conditions as previously reported. 47-48 hMSCs were maintained in growth medium (GM; DMEM, 10% FBS, 0.3 mg/ml glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin). Fifth passage hMSCs was used for experiments. For osteogenic differentiation, hMSCs were cultured on diverse nanopatterns (150-, 400-, 600-nm width lines and 150-, 400-, 600-nm diameter dots) in osteogenic differentiation medium (OM, 50 μM ascorbic acid-2-phosphate, 10mM β-glycerophosphate, and 100 nM dexamethasone) for 8 days at a cell density of 3000 cells/cm2. As a negative control, hMSCs were cultured in GM for 8 days at the same density using the same patterns. To ensure reproducibility and biological significance of the data, each experiment was run three times under the identical conditions with subsequent analysis of osteogenic differentiation of hMSCs by using ALP activity assay as well as real time RT-PCR.
ALP activity assay
After 8 days culture, hMSCs (3000 cells/cm2) were harvested, and total lysate was prepared using a RIPA lysis buffer supplemented with a protease inhibitor cocktail (Calbiochem, San Diego). Protein concentrations of the lysate were determined by a Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA), and ALP activity was analyzed by incubating the lysates with p-nitrophenylphosphate (4.5 mg/ml) at 37 °C for 2 hours prior to the measurement of OD 410 by a spectrophotometer (Bio-Rad, Japan). By performing the assay using known concentration of ALP in parallel with the samples, the concentrations of ALP for the samples were calculated and normalized to metabolic activity.
Cell staining
ALP was stained using Sigma kit #85 according to the manufacturer's instructions. In brief, samples were fixed in acetone/citrate, rinsed in water, and stained with Fast Blue RR/naphthol.
RT-PCR analysis
Total RNA was purified from the cells using TRIzol reagent, according to the manufacturer's protocol (Invitrogen). 1 μg of total RNA was used in cDNA synthesis with random hexamers as primers (Promega). PCR amplification was subsequently performed with the following primer sets: for Cbfa1, sense: 5′-AGAGCTGAACAGGAACAACGT-3′ and antisense: 3′- CACCAGCAAGAAGAAGCCTTTG-5′; for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sense: 5′-ATTACAGTCCATGCCATCA-3′ and antisense: 5′- TCCACCACCCTGTTGCTGTA-3′; for alkaline phosphatase(ALP), sense: 5′- TTAGTGCCAGAGAAAGAG-3′ and antisense: 5′-CTTGGCTTTTCCTTCATGGTG-3′; for peroxisome proliferator activator receptor γ 2(PPARγ2), sense: 5′-GCTGTTATGGGTGAAACTCTG-3′ and antisense: 5′-ATAAGGTGGAGATGCAGGTTC-3′; for Osteopontin (OP), sense: 5′-CACATCGGAATGCTCATTGC-3′ and antisense: 5′-ATCACCTGTGCCATACCAGT-3; for Osteocalcin (OC) sense: 5′-ATGAGAGCCCTCACACTCCTC-3′ and antisense: 5′-GCCGTAGAAGCGCCGATAGGC-3; for Sox 9, sense: 5′-ACGTCATCTCCAACATCGAGACC-3′ and antisense: 5′-CTGTAGTGTGGGAGGTTGAAGGG-3 All PCR was carried out with an initial denaturation at 94 °C for 2 minutes followed by 35 cycles at 94 °C for 20 seconds, annealing temperatures of 57.4 °C (Cbfa1), 41.5 °C (ALP), 53.8 °C (GAPDH), 59.3 °C (PPARγ2), 56.0 °C (OP), 55.0 °C (OC), 55.0 °C (Sox 9) for 30 seconds, and a final extension at 72 °C for 5 minutes. All samples were run in triplicates in each experiment.
Real-time quantitative RT-PCR (sybr method)
Total RNA was extracted by using Trizol (Invitrogen). RNA was reverse-transcribed into cDNA. Two microliters of each RT reaction was amplified in a 20 μL PCR assay volume containing 10 μM each primer and power SYBR Green PCR Master Mix (applied Biosystems). Samples were incubated in the ExiCycler for an initial denaturation at 94 °C for 10 min, followed by 40 PCR cycles. Each cycle consisted of the following times and temperatures: 94°C for 40 s, 60°C for 30 s, and 72°C for 30 s. All gene expressions were normalized according to glyceraldehydes 3-phosphate dehydrogenase using the comparative cycle thresholod (ddCt) methods. The Cbfa1, sense: 5′-CACTGGCGCTGCAACAAGA-3′ and antisense: 5′- CATTCCGGAGCTCAGCAGAATAA-3′ (product size 127 bp); for OP, sense: 5′-ATTCTGGGAGGGCTTGGTTG-3′ and antisense: 5′- TGTGGTCCCGACGATGCT-3′; for OC, sense: 5′-CTCCAGGCACCCTTCTTTCC-3′ and antisense: 5′- ATTCCTCTTCTGGAGTTTATTTGGG-3′ for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sense: 5′-GCACCGTCAAGGCTGAGAAC-3′ and antisense: 3′- ATGGTGGTGAAGACGCCAGT-5′ (product size 142 bp).
Data analysis
All data were presented as mean ± SD. The results were analyzed using one-way analysis of variance (ANOVA) test to assess the statistically significant difference of overall differences in ALP activity level and real time PCR results. When one-way ANOVA test revealed p < 0.05, the data were further analyzed by Dunnett's t-test to assess the statistical difference between treatment and control groups. Differences were considered statistically significant, only when p values were less than 0.05 (p < 0.05).
Results and discussion
Nanotopography and substrate characterization
ECM is composed of various protein fibers interwoven with glycosaminoglycan chains. In vivo, cells sense and respond to topographical features of the local microenvironment on multiple length scales such as bone roughness, molecular complexes controlling cell to cell interaction, and collagen banding. Aiming at mimicking the cellular microenvironment, in vitro studies have been centered on modulating the roughness by blasting. 49-51 The previous findings, however, have often been conflicting, hard to reproduce, and performed on the micro- rather than the more relevant nano-meter scale. To overcome these limitations, we developed a well-defined, scalable cell adhesion substratum to direct differentiation of hMSCs into osteoblasts.
For cell culture substrates, various nanoscale dot and line patterns of PUA material were fabricated on optically transparent glass coverslips using UV-assisted CFL and self-replication (Figure 1). Subsequently, the hMSCs were cultured on the nanopatterns, whose feature sizes were at least one or two order of magnitude smaller than that of the cell body. Figure 2 shows representative SEM images of such nanostructured substrates, demonstrating that the patterns exhibited high structural fidelity with well-defined vertical edge profiles. In order to generate uniform patterns, care should be taken during the 1st and 2nd replication steps so as to preserve the original geometry of the silicon master. This is because air cannot easily permeate out of the cured PUA replica and might be trapped in the spaces between nanostructures, resulting in a reduced height or a non-uniform height distribution. To alleviate this problem, a roller was mildly rounded several times on the surface, which helped expel trapped air prior to UV exposure. It is noted that the PUA material has been extensively used as a cell culturing substrate for many cell types with good biocompatibility. 15, 23, 52-54 In the current experiment, no considerable level of cell death was observed during a long culture of < 1 week, as evidenced by tests with a cell toxicity maker with control group (hMSCs cultured in normal 25 flask) (data not shown).
Figure 2.
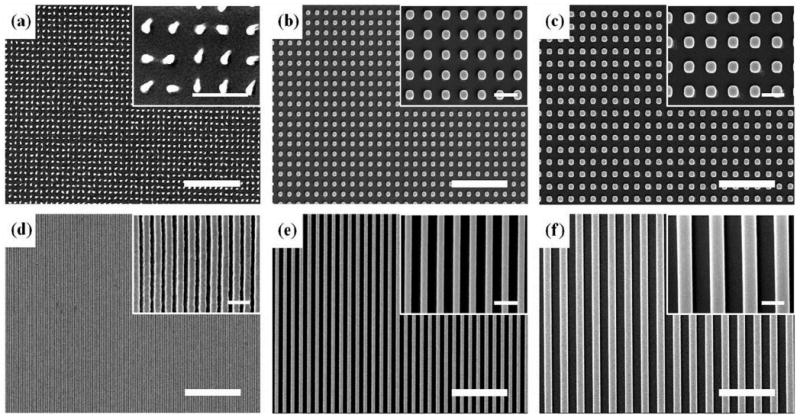
Planar SEM images of the fabricated nanopatterns by UV-assisted CFL. Scale bars represent 5 μm and 1 μm in the panel and inset images, respectively. (a) 150-nm diameter and 500-nm pitch dots. (b) 400-nm diameter and 800-nm pitch dots. (c) 600-nm diameter and 1200-nm pitch dots. (d) 150-nm width and 200-nm pitch lines. (e) 400-nm width and 800-nm pitch lines. (f) 600-nm width and 1200-nm pitch lines.
In the experiments, two types of nanopatterns, varying in size from 150-nm to 600-nm was used: dots (150-, 400-, 600-nm in diameter) and lines (150-, 400-, 600-nm in width). For consistency, the pattern height was fixed at a constant height of 500-nm, as verified by atomic force microscopy (AFM) measurements (not shown). As shown in Figure 2, the patterns were prepared over a large area (4 cm by 5 cm) with good physical integrity, allowing for large-area investigation of hMSCs differentiation into osteoblasts. It is noted in this regard that for cell biological applications, it would be beneficial to have large area patterns, not smaller than 1×1 cm2, for various cell density and conventional biochemical assays (e.g. western blotting, RT-PCR, etc).
Expression of ALP Activity in hMSCs
To investigate the role of surface nanotopography, hMSCs were cultured for 8 days at the initial cell density of 3000 cells/cm2 on the nanopatterns as well as on the flat PUA surface (control) in GM and OM, respectively. The initial plating density of hMSCs was fixed at 3000 cells/cm2 because the lineage-specific differentiation of hMSCs is significantly affected by initial cell density. 55 As surface roughness is a defining feature of the bone tissue, we hypothesized that osteogenic differentiation of hMSCs might be enhanced on nano-topographically defined surfaces. To determine whether surface nanotopography could enhance hMSCs differentiation into osteoblasts, we characterized the ALP activity level of hMSCs cultured on various nanostructured substrates. ALP is widely used as a marker of the osteogenic phenotypes, because cell membrane of osteoblasts is composed of glycoprotein, an isoform of ALP. 56 The ALP activity of hMSCs was measured using cell samples collected on day 8. As shown in Figure 3a-b, the ALP activity level of hMSCs was increased for the patterned substrates both in OM and GM. In particular, the hMSCs that were cultured on 150-nm line, 150-nm dot, and 400-nm dot patterns in OM showed significantly higher level of ALP activity compared to the other nanopatterns and the control group (p < 0.01). Among the patterns tested, the hMSCs on 400-nm dot patterns showed the highest level of ALP activity, with respect to which all the data were normalized both in OM and GM. The osteogenic differentiation is well represented by the characteristic spindle shapes of cells, as shown in Figure 3c. Also, a fluorescent image in Figure 3d demonstrates that the cells express a higher intensity of ALP when cultured on a nanopatterned substrate in OM (400-nm dot patterns used here).
Figure 3.
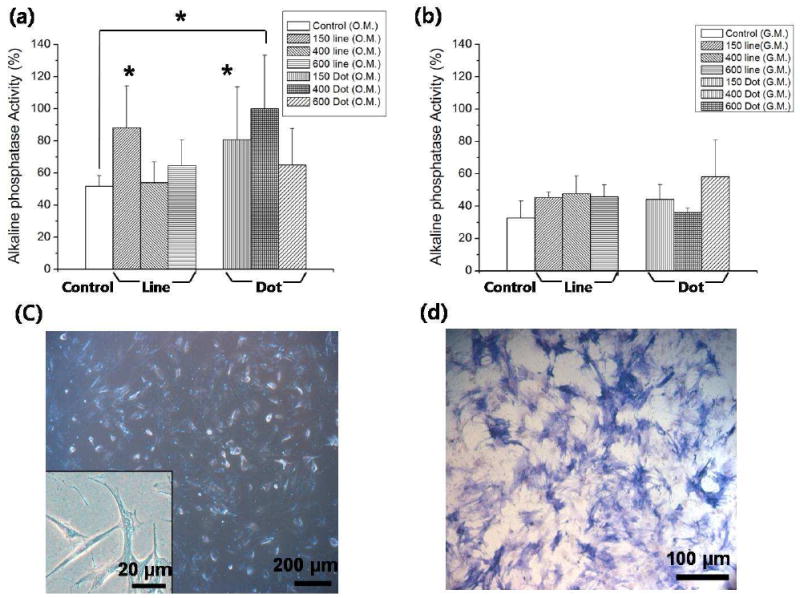
ALP expression level of the hMSCs plated at 3000 cells/cm2, cultured for 8 days in (a) OM and (b) GM. The data were normalized with respect to the highest level of ALP activity for 400-nm dot pattern both in OM and GM. As shown, the ALP activity was increased for the patterned substrates both in OM and GM compared to the control group. Especially, the hMSCs cultured on 150-nm line, 150-nm dot, and 400-nm dot patterns in OM showed significantly high ALP activity level (p < 0.01). *: p < 0.01. (c) Bright-field images of hMSCs cultured on 400-nm dot pattern in OM, plated at 3000 cells/cm2, cultured for 8 days. Bar = 200 μm. Inset shows a magnified image of osteogenic hMSCs exhibiting a spindle shape. Bar = 20 μm. (d) Fluorescent image of hMSCs cultured on 400-nm dot pattern in OM stained for ALP (osteogenic marker). Bar = 100 μm.
As mentioned above, the hMSCs in OM showed higher up-regulation of osteogenic marker (Figure 3a). Interestingly, the hMSCs in GM also showed increasing ALP activity for osteogenesis as compared to control group (Figure 3b). The increased level, however, appears to be less significant than that for OM and the variations among different patterns were observed to be small. These findings suggest that mechanical topographic cues induce the osteogenic differentiation of hMSCs, the level of which can further be enhanced when combined with a suitable chemical definition of the cell medium. In addition, a specific topography (e.g, 400-nm dot patterns) could more synergistically affect osteogenic differentiation in the presence of OM. In fact, in clinical uses of dental implants, rough surfaces in bone-implant contact have already shown a better osseous fixation than smooth surface. 57-58
Expressions of Cbfa1, ALP, OC, OP, PPARγ2, and Sox 9
As reported above, we observed that the ALP activity level of hMSCs was increased on all the nanopatterns tested as compared to the control group. To explore the differentiation outcomes further, we conducted semi-quantitative RT-PCR on day 8 to detect molecular markers of adipocyte, chondrogenic, and osteoblast lineage. For this purpose, hMSCs were cultured in OM and GM for 8 days at the same density of 3000 cells/cm2 on the same surfaces as described in the previous section.
As shown in Figure 4, adipocyte marker (peroxisome proliferator activator receptor γ2, PPARγ2), osteoblast markers (ALP, Cbfa1: early osteogenic marker, OC, OP: late osteogenic marker), and chondrogenic marker (Sox9) displayed different expressions. In OM, ALP level was expressed for various nanopatterns except for 600-nm lines and dots. ALP level in GM was also expressed for various nanopatterns except for 600-nm lines and dots. The reason for the absence of ALP activity in the case 600-nm patterns is not clear at this stage, which appears to be associated with different cell attachment and penetration into the spaces of nanopatterns. 59
Figure 4.
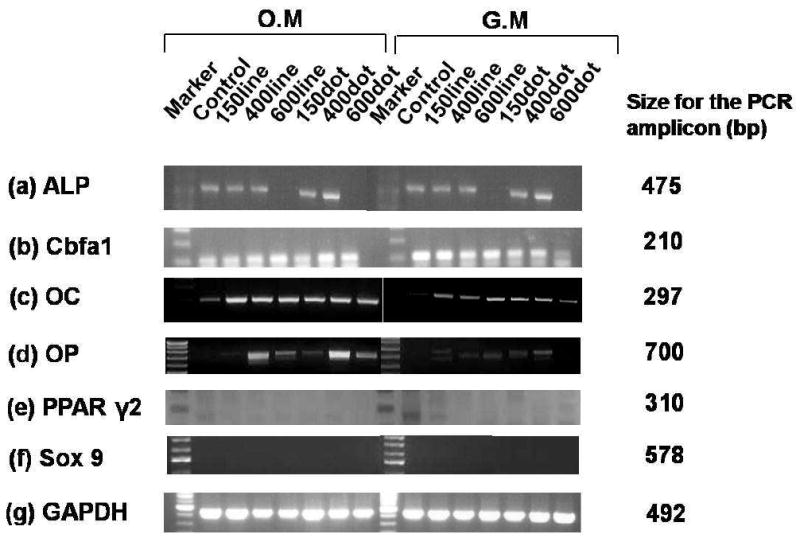
RT-PCR analysis: (a) ALP, (b) Cbfa1, (c) OC, (d) OP, (e) PPAR γ2, (f) Sox9, and (g) glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All samples were collected after 8 days in culture.
As for other osteogenic markers, Cbfa1 was expressed for various nanopatterns both in GM and OM. In OM, the OC level was more intensively expressed for various nanopatterns than that in GM. The OP level also showed higher expression with nanopatterns and especially the highest intensity for 400-nm dots in OM. In contrast, the intensities of OC and OP were relatively lower for the nanopatterns in GM. The fact that the Cbfa1 is similarly expressed both in GM and OM indicates that early differentiation occurs in both media. Moreover, the cell density was properly controlled in the experiment, which can further facilitate osteogenic differentiation even in GM sample. However, late stage differentiation, as evidenced from staining results from OP and OC, was only observed in OM, supporting the existence of synergistic role of nanotopography and osteogenic induction media.
Figure 4 also demonstrates differentiation into other lineage using molecular makers of PPARγ2 and Sox 9, in which the markers were not detected for all the nanopatterns in OM and GM (except for expressing PPARγ2 marker in GM control). On the basis of these results, osteogenesis appears to predominate over adipogenesis and chondrogenesis for 8 days in the presence of the induction media substance. It is noted that the cell proliferation was low, so that the cell density was almost maintained at ∼3000 cells/cm2 for 2∼3 weeks, in agreement with the previous study. 31
Real time PCR
We also conducted real time quantitative RT-PCR using hMSCs on the 400-nm dot pattern and control on day 8. As shown in Figure 5a-c, the expression levels of Cbfa1, OC, and OP on the 400-nm dot pattern (GM and OM) increased significantly in comparison to the control group (p < 0.05), further confirming strong influence of the surface nanotopography on osteogenic differentiation. Here, ALP and Cbfa1 are expected to increase early in osteogenesis and decrease with mineralization. Also, OC and OP are the main components of mineralized ECM for osteoblasts. As can be seen from the figures, ALP and Cbfa1 were expressed both in GM and OM, with significantly lower levels of OC and OP in GM samples. This suggests that early osteogenic differentiation can be initiated regardless of the choice of media with the help of a proper cell density; the late stage osteogenesis (mineralization), however, is hardly observed in GM-control, supporting the synergistic role of nanotopography and differentiation induction media. As with other staining and PCR results, the 400-nm dot pattern showed the higher mineralization with some local distribution of the osteogenic differentiation.
Figure 5.
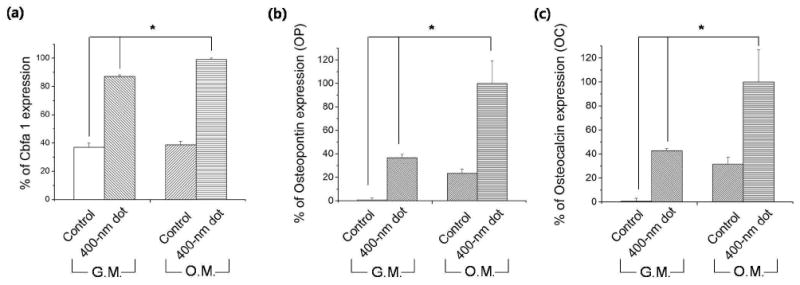
Quantification of osteogenic-specific marker expression by real-time PCR at day 8 (* and * p < 0.05): (a) Cbfa1, (b) OP, (c) OC.
Conclusions
In this study, we have presented that nanotopographic surface definition could synergistically induce a significant up-regulation of osteogenic markers such as ALP, Cbfa1, OC, and OP with osteogenic induction media (OM), demonstrating directed differentiation into osteogenic lineage of hMSCs. We have used UV-assisted capillary lithographic technique with a polymeric biomaterial of PUA for simple and scalable (4 cm × 5 cm) fabrication of surface nanotopography. Based on alkaline phosphatase (ALP) assay, RT-PCR, and real-time quantitative RT-PCR analysis, we found that the hMSCs responded to the underlying surface nanotopography by up-regulation of mRNA osteogenic level, and the 400-nm dot pattern appeared more effective in facilitating osteogenic differentiation synergistically with osteogenesis induction media (OM). In normal culture media (GM), the hMSCs also showed increased level of ALP, OP, and OC activity for osteogenesis but the increased level was less significant than that for OM and the variations among different patterns were observed to be small.
In conjunction with our findings, a previous study by Dalby et al. 22 reported that the differentiation of hMSCs could be enhanced when cultured on dot patterns. They found that when hMSCs were cultured on dot patterns and displaced randomly by up to 50-nm on both axes from their initial position, the cells showed more osteogenic differentiation. This observation, together with our findings, suggests that the hMSCs can sense the underlying surface nanotopography and differentiate into different cell lineage. Furthermore the dot patterns might be more effective in up-regulating osteogenic differentiation presumably due to its biomimetic character with respect to the actual bone surface, stimulating Runx 2 gene of hMSCs. A further study would be required to elucidate the biomechanical or physical role of nanopatterns on the differentiation of hMSCs. Our study further demonstrated that tunable nanotopographic features may have synergistic effect with the chemical medium composition on directed stem cell differentiation into osteogenic lineage.
Acknowledgments
This research was supported by the WCU (World Class University) program (R31-2008-000-10083-0), the Institute of Advanced Machinery and Design, and the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (Grant KRF-J03003). This work was also supported in part by National Institute of Health (R21-EB008562).
References
- 1.Chen CS, Tan J, Tien J. Mechanotransduction at cell-matrix and cell-cell contacts. Annual Review of Biomedical Engineering. 2004;6(1):275–302. doi: 10.1146/annurev.bioeng.6.040803.140040. [DOI] [PubMed] [Google Scholar]
- 2.Kim DH, Wong P, Park JY, Levchenko A, Sun Y. Microengineered platforms for cell mechanobiology. Annual Review of Biomedical Engineering. 2009;11:203–233. doi: 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- 3.Zaari N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Photopolymerization in Microfluidic Gradient Generators: Microscale Control of Substrate Compliance to Manipulate Cell Response. Advanced Materials. 2004;16(23-24):2133–2137. [Google Scholar]
- 4.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 5.Kunzler TP, Drobek T, Schuler M, Spencer ND. Systematic study of osteoblast and fibroblast response to roughness by means of surface-morphology gradients. Biomaterials. 2007;28(13):2175–2182. doi: 10.1016/j.biomaterials.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JM, Kushwaha M, Tambralli A, Bellis SL, Camata RP, Jun HW. Osteogenic Differentiation of Human Mesenchymal Stem Cells Directed by Extracellular Matrix-Mimicking Ligands in a Biomimetic Self-Assembled Peptide Amphiphile Nanomatrix. Biomacromolecules. 2009;10(10):2935–2944. doi: 10.1021/bm9007452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park J, Bauer S, von der Mark K, Schmuki P. Nanosize and Vitality: TiO2 Nanotube Diameter Directs Cell Fate. Nano Letter. 2007;7(6):1686–1691. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 8.Jiang W, KimBetty YS, Rutka JT, ChanWarren CW. Nanoparticle-mediated cellular response is size-dependent. Nature Nanotechnology. 2008;3(3):145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 9.Wójciak-Stothard B, Madeja Z, Korohoda W, Curtis A, Wilkinson C. Activation of macrophage-like cells by multiple grooved substrata. Topographical control of cell behaviour. Cell Biology International. 1995;19(6):485–490. doi: 10.1006/cbir.1995.1092. [DOI] [PubMed] [Google Scholar]
- 10.Ward BC, Webster TJ. The effect of nanotopography on calcium and phosphorus deposition on metallic materials in vitro. Biomaterials. 2006;27(16):3064–3074. doi: 10.1016/j.biomaterials.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 11.Mahdavi A, Ferreira L, Sundback C, Nichol JW, Chan EP, Carter DJD, Bettinger CJ, Patanavanich S, Chignozha L, Ben-Joseph E, Galakatos A, Pryor H, Pomerantseva I, Masiakos PT, Faquin W, Zumbuehl A, Hong S, Borenstein J, Vacanti J, Langer R, Karp JM. A biodegradable and biocompatible gecko-inspired tissue adhesive. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2307–2312. doi: 10.1073/pnas.0712117105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez E, Lagunas A, Mills C, Rodriguez-Segui S, Estevez M, Oberhansl S, Comelles J, Samitier J. Stem cell differentiation by functionalized micro- and nanostructured surfaces. Nanomedicine. 2009;4(1):65–82. doi: 10.2217/17435889.4.1.65. [DOI] [PubMed] [Google Scholar]
- 13.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sniadecki N, Desai RA, Ruiz SA, Chen CS. Nanotechnology for cell-substrate interactions. Annals of Biomedical Engineering. 2006;34(1):59–74. doi: 10.1007/s10439-005-9006-3. [DOI] [PubMed] [Google Scholar]
- 15.Suh KY, Park MC, Kim P. Capillary Force Lithography: A Versatile Tool for Structured Biomaterials Interface Towards Cell and Tissue Engineering. Advanced Functional Materials. 2009;19(17):2699–2712. [Google Scholar]
- 16.Patel AA, Thakar RG, Chown M, Ayala P, Desai TA, Kumar S. Biophysical mechanisms of single-cell interactions with microtopographical cues. Biomedical Microdevices. 2009;12(2):287–296. doi: 10.1007/s10544-009-9384-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westcott NP, Lou Y, Muth JF, Yousaf MN. Patterned Hybrid Nanohole Array Surfaces for Cell Adhesion and Migration. Langmuir. 2009;25(19):11236–11238. doi: 10.1021/la9023234. [DOI] [PubMed] [Google Scholar]
- 18.Hwang SY, Kwon KW, Jang KJ, Park MC, Lee JS, Suh KY. Adhesion Assays of Endothelial Cells on Nanopatterned Surfaces within a Microfluidic Channel. Analytical Chemistry. 2010 doi: 10.1021/ac100107z. [DOI] [PubMed] [Google Scholar]
- 19.Thapa A, Webster TJ, Haberstroh KM. Polymers with nano-dimensional surface features enhance bladder smooth muscle cell adhesion. Journal of Biomedical Materials Research Part A. 2003;67A(4):1374–1383. doi: 10.1002/jbm.a.20037. [DOI] [PubMed] [Google Scholar]
- 20.Kim P, Kim DH, Kim B, Choi SK, Lee SH, Khademhosseini A, Langer R, Suh KY. Fabrication of nanostructures of polyethylene glycol for applications to protein adsorption and cell adhesion. Nanotechnology. 2005;16(10):2420–2426. doi: 10.1088/0957-4484/16/10/072. [DOI] [PubMed] [Google Scholar]
- 21.Teixeira AI, McKie GA, Foley JD, Bertics PJ, Nealey PF, Murphy CJ. The effect of environmental factors on the response of human corneal epithelial cells to nanoscale substrate topography. Biomaterials. 2006;27(21):3945–3954. doi: 10.1016/j.biomaterials.2006.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nature Materials. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 23.Kim DH, Seo CH, Han K, Kwon KW, Levchenko A, Suh KY. Guided Cell Migration on Microtextured Substrates with Variable Local Density and Anisotropy. Advanced Functional Materials. 2009;19(10):1579–1586. doi: 10.1002/adfm.200990041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh S, Brammer KS, Li YSJ, Teng D, Engler AJ, Chien S, Jin S. Stem cell fate dictated solely by altered nanotube dimension. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(7):2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng L, Barczak AJ, Barbeau RA, Xiao Y, LaTempa TJ, Grimes CA, Desai TA. Whole Genome Expression Analysis Reveals Differential Effects of TiO2 Nanotubes on Vascular Cells. Nano Letters. 2009;10(1):143–148. doi: 10.1021/nl903043z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira L, Karp JM, Nobre L, Langer R. New opportunities: The use of Nanotechnologies to manipulate and track stem cells. Cell Stem Cell. 2008;3(2):136–146. doi: 10.1016/j.stem.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 27.Shin YM, Kim KS, Lim YM, Nho YC, Shin H. Modulation of Spreading, Proliferation, and Differentiation of Human Mesenchymal Stem Cells on Gelatin-Immobilized Poly(l-lactide-co-ε-caprolactone) Substrates. Biomacromolecules. 2008;9(7):1772–1781. doi: 10.1021/bm701410g. [DOI] [PubMed] [Google Scholar]
- 28.Andersson AS, Backhed F, von Euler A, Richter-Dahlfors A, Sutherland D, Kasemo B. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials. 2003;24(20):3427–36. doi: 10.1016/s0142-9612(03)00208-4. [DOI] [PubMed] [Google Scholar]
- 29.Miller DC, Thapa A, Haberstroh KM, Webster TJ. Endothelial and vascular smooth muscle cell function on poly(lactic-co-glycolic acid) with nano-structured surface features. Biomaterials. 2004;25(1):53–61. doi: 10.1016/s0142-9612(03)00471-x. [DOI] [PubMed] [Google Scholar]
- 30.Yim EK, Pang SW, Leong KW. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Experimental Cell Research. 2007;313(9):1820–9. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo W, Jones SR, Yousaf MN. Geometric Control of Stem Cell Differentiation Rate on Surfaces. Langmuir. 2008;24(21):12129–12133. doi: 10.1021/la802836g. [DOI] [PubMed] [Google Scholar]
- 32.Koepsel JT, Murphy WL. Patterning Discrete Stem Cell Culture Environments via Localized Self-Assembled Monolayer Replacement. Langmuir. 2009;25(21):12825–12834. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins JM, Ayala P, Desai TA, Russell B. Three-Dimensional Culture with Stiff Microstructures Increases Proliferation and Slows Osteogenic Differentiation of Human Mesenchymal Stem Cells. Small. 2010;6(3):355–360. doi: 10.1002/smll.200901757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khor HL, Kuan Y, Kukula H, Tamada K, Knoll W, Moeller M, Hutmacher DW. Response of cells on surface-induced nanopatterns: fibroblasts and mesenchymal progenitor cells. Biomacromolecules. 2007;8(5):1530–40. doi: 10.1021/bm0611533. [DOI] [PubMed] [Google Scholar]
- 35.Costa-Pinto AR, Correlo VM, Sol PC, Bhattacharya M, Charbord P, Delorme B, Reis RL, Neves NM. Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells Seeded on Melt Based Chitosan Scaffolds for Bone Tissue Engineering Applications. Biomacromolecules. 2009;10(8):2067–2073. doi: 10.1021/bm9000102. [DOI] [PubMed] [Google Scholar]
- 36.Heinemann C, Heinemann S, Lode A, Bernhardt A, Worch H, Hanke T. In Vitro Evaluation of Textile Chitosan Scaffolds for Tissue Engineering using Human Bone Marrow Stromal Cells. Biomacromolecules. 2009;10(5):1305–1310. doi: 10.1021/bm900172m. [DOI] [PubMed] [Google Scholar]
- 37.Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276(5309):71–4. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 38.Pittenger MF, Mosca JD, McIntosh KR. Human mesenchymal stem cells: progenitor cells for cartilage, bone, fat and stroma. Current Topics in Microbiology and Immunology. 2000;251:3–11. doi: 10.1007/978-3-642-57276-0_1. [DOI] [PubMed] [Google Scholar]
- 39.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Engineering. 2001;7(2):211–28. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 40.Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Runx2 Regulates G Protein-coupled Signaling Pathways to Control Growth of Osteoblast Progenitors. Journal of Biological Chemistry. 2008;283(41):27585–27597. doi: 10.1074/jbc.M802453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HJ, Kim JH, Bae SC, Choi JY, Ryoo HM. The Protein Kinase C Pathway Plays a Central Role in the Fibroblast Growth Factor-stimulated Expression and Transactivation Activity of Runx2. Journal of Biological Chemistry. 2003;278(1):319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- 42.Marie PJ. Fibroblast growth factor signaling controlling osteoblast differentiation. Gene. 2003;316:23–32. doi: 10.1016/s0378-1119(03)00748-0. [DOI] [PubMed] [Google Scholar]
- 43.Kim DH, Kim P, Song I, Cha JM, Lee SH, Kim B, Suh KY. Guided three-dimensional growth of functional cardiomyocytes on polyethylene glycol nanostructures. Langmuir. 2006;22(12):5419–5426. doi: 10.1021/la060283u. [DOI] [PubMed] [Google Scholar]
- 44.Suh KY, Kim YS, Lee HH. Capillary force lithography. Advanced Materials. 2001;13(18):1386–1389. [Google Scholar]
- 45.Suh KY, Jeong HE, Kim DH, Singh RA, Yoon ES. Capillarity-assisted fabrication of nanostructures using a less permeable mold for nanotribological applications. Journal of Applied Physics. 2006;100(3) [Google Scholar]
- 46.Yoon H, Kim TI, Choi SJ, Suh KY, Kim MJ, Lee HH. Capillary force lithography with impermeable molds. Applied Physics Letters. 2006;88(25) [Google Scholar]
- 47.Bang OY, Lee JS, Lee PH, Lee G. Autologous mesenchymal stem cell transplantation in stroke patients. Annals of Neurology. 2005;57(6):874–882. doi: 10.1002/ana.20501. [DOI] [PubMed] [Google Scholar]
- 48.You MH, Kim WJ, Shim W, Lee SR, Lee G, Choi S, Kim DY, Kim YM, Kim H, Han SU. Cytosine deaminase-producing human mesenchymal stem cells mediate an antitumor effect in a mouse xenograft model. Journal of Gastroenterology and Hepatology. 2009;24(8):1393–1400. doi: 10.1111/j.1440-1746.2009.05862.x. [DOI] [PubMed] [Google Scholar]
- 49.Citeau A, Guicheux J, Vinatier C, Layrolle P, Nguyen TP, Pilet P, Daculsi G. In vitro biological effects of titanium rough surface obtained by calcium phosphate grid blasting. Biomaterials. 2005;26(2):157–165. doi: 10.1016/j.biomaterials.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 50.Luthen F, Lange R, Becker P, Rychly J, Beck U, Nebe JGB. The influence of surface roughness of titanium on beta 1-and beta 3-integrin adhesion and the organization of fibronectin in human osteoblastic cells. Biomaterials. 2005;26(15):2423–2440. doi: 10.1016/j.biomaterials.2004.07.054. [DOI] [PubMed] [Google Scholar]
- 51.Wieland M, Textor M, Chehroudi B, Brunette DM. Synergistic interaction of topographic features in the production of bone-like nodules on Ti surfaces by rat osteoblasts. Biomaterials. 2005;26(10):1119–1130. doi: 10.1016/j.biomaterials.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 52.Park MC, Hur JY, Kwon KW, Park SH, Suh KY. Pumpless, selective docking of yeast cells inside a microfluidic channel induced by receding meniscus. Lab on a Chip. 2006;6:988–994. doi: 10.1039/b602961b. [DOI] [PubMed] [Google Scholar]
- 53.Kwon KW, Choi SS, Lee SH, Kim B, Lee SN, Park MC, Kim P, Hwang SY, Suh KY. Label-free, microfluidic separation and enrichment of human breast cancer cells by adhesion difference. Lab on a Chip. 2007;7:1461–1468. doi: 10.1039/b710054j. [DOI] [PubMed] [Google Scholar]
- 54.Kim DH, Han K, Gupta K, Kwon KW, Suh KY, Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials. 2009;30(29):5433–5444. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Developmental Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 56.Stigbrand T. Present status and future trends of human alkaline phosphatases. Progress in Clinical Biological Research. 1984;166:3–14. [PubMed] [Google Scholar]
- 57.Thomas KA, Cook SD. An evaluation of variables influencing implant fixation by direct bone apposition. Journal of Biomedical Materials Research. 1985;19(8):875–901. doi: 10.1002/jbm.820190802. [DOI] [PubMed] [Google Scholar]
- 58.Buser D, Nydegger T, Hirt HP, Cochran DL, Nolte LP. Removal torque values of titanium implants in the maxilla of miniature pigs. The International Journal of Oral & Maxillofacial Implants. 1998;13(5):611–9. [PubMed] [Google Scholar]
- 59.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proceedings of the National Academy of Sciences. 2010;107(2):565–570. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]


