Abstract
The most popular current hypothesis is that Alzheimer’s disease (AD) is caused by aggregates of the amyloid peptide (Aβ), which is generated by cleavage of the Aβ protein precursor (APP) by β-secretase (BACE-1) followed by γ-secretase. BACE-1 cleavage is limiting for the production of Aβ, making it a particularly good drug target for the generation of inhibitors that lower Aβ. A landmark discovery in AD was the identification of BACE-1 (a.k.a. Memapsin-2) as a novel class of type I transmembrane aspartic protease. Although BACE-2, a homologue of BACE-1, was quickly identified, follow up studies using knockout mice demonstrated that BACE-1 was necessary and sufficient for most neuronal Aβ generation. Despite the importance of BACE-1 as a drug target, development has been slow due to the incomplete understanding of its function and regulation and the difficulties in developing a brain penetrant drug that can specifically block its large catalytic pocket. This review summarizes the biological properties of BACE-1 and attempts to use phylogenetic perspectives to understand its function. The article also addresses the challenges in discovering a selective drug-like molecule targeting novel mechanisms of BACE-1 regulation.
Keywords: BACE-1, secretase, Memapsin, Alzheimer, Amyloid, aspartyl protease
Significance of BACE-1
AD is a chronic and progressive neurodegenerative disorder and is the most common cause of dementia in the elderly. The classic clinical symptoms of the disease are an amnesic type of memory impairment [1], visuospatial deficits [2] and deterioration of language [3]. Motor and sensory abnormalities, seizures and gait disturbances are uncommon until late stages of the disease [4]. There is a progression from loss of complex day-to-day activities such as using the public transport, to abnormalities in basic activities of daily living [5]. The incidence of AD increases exponentially with age and is thought to affect 4–8% of the population over 65 years. The current US estimate on the number of AD patients range from 3–5 million with an annual estimated cost of approximately $100 billion. By 2050, it is projected that the number of patients could be as high as 25 million [6]. The pathological hallmarks of AD include the presence of extracellular senile plaques and intracellular neurofibrillary tangles (NFT), which are aggregates of a microtubule associated protein called tau [7]. A major landmark in the study of AD is the identification of the major component of the senile plaque as the 42-residue amyloid peptide — Aβ42 [8, 9]. This finding was followed by the identification of APP and its further characterization as a large type-I integral membrane protein (695–770 aa) with the Aβ42 sequence spanning portions of the extracellular and transmembrane domains [10, 11]. The proteolytic processing pathways that release Aβ42 from the intact APP protein was quickly identified (Figure 1), which led to a large expansion in the study of membrane protein processing and turnover. Early studies discovered that APP was rapidly cleaved and secreted at high levels and this cleavage (by α-secretase) was mapped to residues K16 and L17 of Aβ and generated a secreted derivative, sAPPα, and a membrane-bound 83-residue fragment—CTFα [12–15]. Although initial investigation defined α-secretase as integral to the normal processing pathway, while the Aβ-production was considered pathological, subsequent studies discovered that Aβ, mostly of 40 aa (Aβ40) but approximately 5–10% of 42 aa (Aβ42), is indeed found normally in biological fluids [16]. The major processing pathway leading to Aβ was subsequently discovered by the presence of sAPPβ and CTFβ, the membrane bound 99-residue fragment. The unidentified enzyme responsible for CTFβ production was termed β-secretase [17, 18]. Intramembrane processing of CTFβ and CTFα by γ-secretase yielded Aβ and a smaller 3 kDa fragment named P3, respectively. Most Aβ and P3 terminate at residue 40 (starting at 1 of Aβ) but a smaller fraction (5–10%) ends at residue 42. However, Aβ42 is a more significant player in AD as most familial AD (FAD) mutations increase the ratio of Aβ42/Aβ40. Some of these mutations map to the C-terminal side of the Aβ sequence on APP but most are linked to presenilins 1 and 2 (PS1, PS2)—subunits of γ-secretase [19]. While most of the studies focused on Aβ, the intracellular fragment generated by γ-secretase processing of CTFα and CTFβ – CTFγ (a.k.a. CTFε or AICD) – was not detected in lysates for a long time until small amounts were detected in the brain by direct extraction and larger quantities were consistently detected in an in vitro γ-secretase assay [20, 21]. Most CTFγ consists of the cytoplasmic domain and 3 aa from the transmembrane domain of APP, leaving behind Aβ of 49 aa for further processing to Aβ40 and Aβ42 [22–24]. It is reported that the APP-derived CTFγ regulates transcription like a similar fragment – NICD — derived from Notch, thereby transducing signal from the cell surface to the nucleus [22–24].
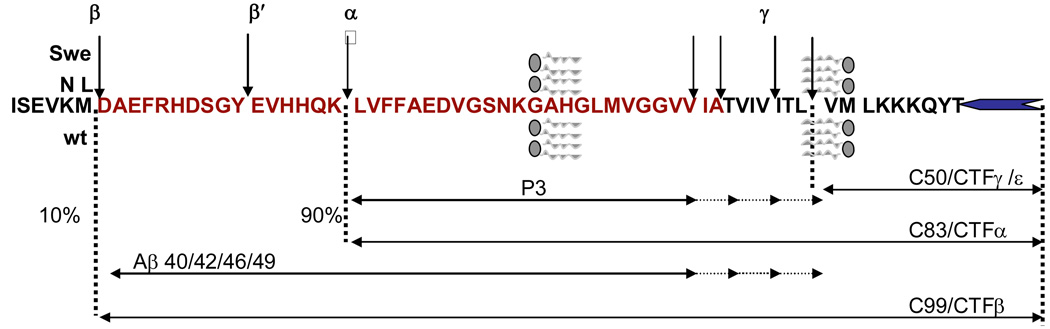
Figure 1. Schematic showing the cleavage sites and products of the APP processing pathway.
Full length APP is a type 1 integral-membrane protein. It has two processing pathways: Minor amyloidogenic (10%) and major nonamyloidogenic (90%). In the amyloidogenic pathway, β-secretase (BACE-1) cleaves the luminal domain of APP at the start of Aβ, releasing sAPPβ and CTFβ. CTFβ of 99 residues is cleaved by γ secretase to release Aβ40/42 and CTFγ of 50 residues. In the nonamyloidogenic pathway, α-secretase cleaves APP to release sAPPα and CTFα of 83 residues. CTFα is further cleaved by γ secretase to release P3 of 24/26 residues and CTFγ of 50 residues. The Swedish (Swe) APP670NL mutant of APP substitutes “KM” from wild type (wt) on the N-terminal side of Aβ with “NL” and is a better substrate for BACE-1 and yields higher levels of sAPPβ, CTFβ and both Aβ42 and Aβ40.
Evidence that Aβ plays a role in AD pathogenesis primarily comes from the study of mutations in the APP, PS1 and PS2 genes that are linked to early onset of FAD [25–31]. The APP mutations linked to FAD frame the Aβ sequence at the N and C terminal ends, and increase production of Aβ42 or total Aβ. The Swedish APP670NL mutant of APP substitutes “KM” on the N-terminal side of Aβ with “NL” and is a better substrate for β-secretase and yields higher levels of sAPPβ, CTFβ and both Aβ42 and Aβ40 [32, 33]. Since Aβ oligomers (a.k.a. ADDLs) are neurotoxic in vitro and in vivo, the currently popular hypothesis is that they are the cause of the cascade of events that lead to AD [31, 34]. As the limiting first step in generating Aβ, β-secretase is the key enzyme that mediates production of this potentially toxic peptide and is therefore an important drug target.
Discovery of BACE-1
As introduced above, detailed studies established that APP is processed by β-secretase to produce Aβ. A number of indirect studies were carried out on this enzyme, as it was the Holy Grail in AD research from its discovery in 1992 to its definitive identification in 1999. Several attempts were made to identify β-secretase including the suggestion that it was actually cathepsin-D, which cleaves APP-derived peptide substrates with specificity similar to β-secretase [35, 36]. However, knockout mice lacking cathepsin-D demonstrated that it was not the major β-secretase [37]. Additional studies suggested that β-secretase cleaves wild type APP in an intracellular compartment after endocytosis, but FAD mutant APP in the secretory pathway (probably in the same compartment as α-secretase), and also that 4-(2-Aminoethyl) benzenesulfonyl fluoride, a serine protease inhibitor, reduces the yield of β-secretase-cleavage products [38, 39]. Moreover, substrate mutation studies demonstrated that unlike α-secretase, β-secretase is sequence specific [40]. Additionally, most sAPPβ is secreted in the basolateral membranes of polarized MDCK cells expressing APPwt, but sAPPβ from the APP670NL mutant is shifted by ~20% to the apical surface [41, 42]. Despite their importance in understanding the cleavage process, none of these studies on the cell biology could aid in the identification of the elusive β-secretase.
In 1999, five years after the discovery of β-secretase cleavage, five groups simultaneously reported the discovery of β-secretase as a novel integral membrane aspartyl protease, the first of its kind reported in vertebrates [43–47]. Three of these groups used the evidence that one of the secretases is an aspartyl protease to identify novel mammalian aspartyl proteases from the human genome databases. Two groups called the enzyme Asp-2 to denote the second novel aspartyl protease detected in their bioinformatics screens [44, 45]. A third group termed the enzyme memapsin-2 for “membrane-anchored protease” as per the convention for aspartyl proteases to end with “in” as in cathepsin, pepsin, gastricin, renin and napsin [43]. The fourth group isolated β-secretase cDNA in an expression screen for cDNAs that increase Aβ and termed the enzyme BACE for Beta Site APP-Cleaving Enzyme [46], which has been adopted by most scientists in the field. However, the immediate recognition of the presence of a homologue of BACE, named BACE-2, led to the former being named BACE-1. The fifth group used conventional biochemistry to isolate and purify the active enzyme from brain membranes and preferred to continue calling it β-secretase to avoid the confusion generated by changing nomenclature [47]. These findings generated a lot of excitement and have initiated a large body of studies aimed at understanding BACE-1 structure, function, localization, regulation and changes in AD. The availability of the pure enzyme has also allowed the discovery of specific inhibitors that lower Aβ and might prevent or treat AD.
Characterization of BACE-1
BACE-1 cleaves APP at the expected sites on its extracellular domains at positions D1 and E11 as indicated by arrows on the sequence shown below (Fig. 1). In addition, the APP670NL mutation that replaces KM on the N-terminal side of D1 is readily cleaved in vivo to generate higher levels of CTFβ and Aβ and is also a much better BACE-1 substrate in vitro as shown in Figure 1 [46, 48]. Complementarily, mutation of “KM” to “KV” markedly reduced processing of APP by BACE-1 and BACE-1 antisense RNA reduces production of Aβ cleaved at either position 1 or position 11 without affecting adventitious variants cleaved at V-3 or I-6 [46]. Consistent with the need for an acid compartment in vivo, BACE-1 cleavage displays an acid pH optimum. While it does cleave smaller peptide based substrates, it strongly prefers longer substrates and shows a higher degree of specificity with such substrates [47].
A close homolog of BACE-1, BACE-2, is 68% similar to BACE-1 and was also identified from the genome database [49]. Despite their homology and common structural organization, the two enzymes differ in their tissue distribution [49–51], subcellular localization [44, 50] and substrate specificity [51–53]. BACE-2 exhibits secretase activity with cleavage in the middle of the Aβ domain at amino acids 19 and 20 generating a CTF of 79 residues, and does not contribute to the process of Aβ generation [51, 53]. BACE-2 can cleave at the β-site, but consistent with its different substrate specificity, does not cleave a preferred peptide-based BACE-1 substrate [54].
The ultimate evidence for the role of BACE-1 in APP processing came from knockout mouse studies showing that both wild type and the Swedish APP670NL mutant forms of APP fail to produce Aβ in the absence of BACE-1 [33]. These initial studies also reported that BACE-1-knockout mice displayed no biological deficiency, suggesting that specific inhibition of its activity is likely to be well tolerated [33]. However, more recent studies found that neurons from BACE-1 knockout mice fail to generate Aβ, but that glial cells continue to generate Aβ. This production was lost in BACE-1/BACE-2 double knockout mice suggesting that BACE-2 plays a role in glial Aβ generation. In addition, BACE-1 knockout mice displayed significant neonatal lethality, which was increased in BACE-2 knockout mice suggesting that these enzymes do serve an essential function in mice, which may be partially fulfilled by other cellular proteases [55].
BACE-1 substrate specificity
When cells were transfected with APP lacking the transmembrane domain, they were not cleaved by BACE-1 [56], suggesting that the enzyme is a membrane bound protease, and only cleaves APP when membrane bound. However, it is important to note that the enzyme can cleave soluble substrates in vitro in a sequence specific manner. Instead, the failure to observe in vivo cleavage is likely related to the relatively high Km of the enzyme, which may be compensated by the high local concentration of enzyme and substrate as a single molecular layer in cellular membranes. This concept needs to be further elaborated for several membrane-bound enzymes.
BACE-1 accepts a wide variety of substrates, preferring acidic or polar residues in contrast to other known aspartyl proteases. Compared to other mammalian aspartyl proteases, BACE-1 is more like cathepsin D than renin in terms of substrate specificity [57]. Radiosequencing studies suggests that most Aβ secreted from APP670NL starts at Asp +1 but wild type APP is more ragged and can also begin at Val-3, Ile-6 and Glu+11 [58]. By using inhibitors, it was proved that only cleavages at Asp +1 and Glu+11 were generated by BACE-1 and unidentified alternative pathways were responsible for generating Val-3 and Ile-6 [46]. More careful peptide based analysis of target sequence specificity using purified BACE-1 has identified several specific rules for processing by this enzyme [54, 59]. Based on the nomenclature of Schlechter and Berger [60], four residues on either side of the cleavage site (P4, P3, P2, P1 ⇓ P1’, P2’, P3’, P4’) of APP and other substrates were analyzed and characterized (Table 1). The studies found that the requirements on the N-terminal side were more stringent than the C-terminal (Table 1). Longer sequences on the N-terminal side beyond P4 were also found to influence cleavage although these requirements were not systematically examined. Based on the sequence specificity, the consensus sequence identified — EIDLMVLDWHDR —had a kcat/Km almost 14-fold higher than the APP670NL peptide and resulted in the development of an even more potent inhibitor of the enzyme (OM00-3; E-L-D-L(hydroxyethylene isostere)-A-V-E-F) [59, 61]. However, the longer peptide substrate discovered in the screen (GLTNIKTEEISEISY-⇓ -EVEFRWKK) displayed an almost 50-fold greater kcat/km, making it an even more effective substrate and suggested the presence of an even larger catalytic pocket in BACE-1 for substrate interaction, consistent with the higher affinity for longer APP substrates [54]. Replacement of the natural cleavage site of APP with ISYEV results in a very large increase in the levels of CTFβ and Aβ at the expense of CTFα [54].
Table-1.
Substrate Specificity of BACE-1.
| P4 | N-term | E > Q > D > N > M > G > L = T = H = P > R > V = W > F > A = S > I > Y <K> |
| P3: | I > V > L > E > H > M = A = T = P > K > S > F > D > Q > G = Y <W N R> | |
| P2: | D > N > M > F > Y = L > S = E > A > Q > K > G <W I V T H R P> | |
| P1: | L > F > M > Y >>> T > S > D > G > N > H > A <W Q I V E R K P> | |
| Cleavage site | ||
| P1’: | C-term | M > E > Q > A > D > S >>> Y > L > T > V > I > F > R > K > G > N > W <P H> |
| P2’: | V > I > A > E > F > L > T > M > Y > S >> G > Q > N = D = W = K <H R P> | |
| P3’: | L > V > W > I > T > D > E > F > Y > M > R > K > A > G > S > Q > N > H > P | |
| P4’: | D > E > W > F = Y = M > V > L > I > T > A > Q > G = S >> R > H > K > N <P> | |
P4, P3, P2 and P1 and P1', P2', P3', P4' refer to the residues on the N-terminal side and the C-terminal side of the BACE-1 cleavage site, respectively. The substrates are listed in the order of preference from highest activity to no activity. Schlechter and Berger established this nomenclature for substrate specificity analysis of endoproteases [60]. Note that the residues on the N-terminal side are more critical for activity. Moreover, additional residues on the N-terminus are required for optimal activity of BACE-1 due to its large catalytic pocket (Not shown)
Structure of BACE-1
BACE-1 is a type-I integral membrane glycoprotein with a 21-residue cleavable signal sequence, a large ectodomain of ~434 aa, a single transmembrane domain of ~22 aa and a short cytoplasmic tail of 24 residues based on predictions using PSORT (www.psort.org) and Sig-Phred (www.bioinformatics.leeds.ac.uk/prot_analysis/Signal.html) (Figure 2). However, the prediction of the transmembrane domains from the primary sequence analysis is not very precise as palmitate residues, which also bind the sequence to the membrane, modify C-terminal cysteine residues in the cytoplasmic domain and can act as additional membrane anchors. A signal for intracellular transport motif, mapped to the C-terminal region of BACE-1 is the DDISLL sequence, also termed the acid cluster dileucine (ACDL) sequence, was found to interact with GGA proteins and facilitate intracellular transport and recycling [62, 63]. BACE-1 is a compact globular protein, which is formed by two domains: 1) residues 47–146; and 2) residues 146–385 [64]. The active site contains the two conserved aspartic acid residues, Asp32 and Asp228 within conserved motifs of eukaryotic aspartic proteinases. The residues responsible for catalytic activity are completely encoded within the ectodomain, and the molecule resembles cathepsin D with a membrane anchor [64]. In addition to the substrate-binding site, BACE-1 also contains an exosite that was mapped using bacteriophage display libraries [65]. According to crystal structures of BACE-1, the catalytic region is loctated between the N- and C-terminal lobes, within the substrate binding site in the cleft [64, 66, 67]. Crystal structures of BACE-1/inhibitor complexes have served to further elucidate subsite positions in the protease. When comparing the structures of other mammalian aspartyl proteases, BACE-1 seems to have an extra loop, which could facilitate the addition of more subsites and increase the size of the target recognition site. There are currently eleven such sites on BACE-1 that recognize the sequence from P7 through P4’ as discussed further above (BACE-1 substrate specificity subsection) [59]. Using the OM99-2 inhibitor, the structure of the BACE-1 catalytic unit was revealed along with the features of the active site, including eight subsites that bind the eight substrate-like residues on OM99-2 (P4 to P4’ as described under substrate specificity) [67]. Subsites S5 through S7 that bound the substrate were then mapped using a larger P10-P4’StatVal inhibitor [47], which extends the structure of OM99-2. These three new subsites, P7 through P5 [68] are found in the active site cleft in a region that extends from the previous eight residues (aa 158–167) in an insertion helix [64]. This region is thought to be unique for BACE-1, as it is not seen in the structure of other aspartyl proteases and creates the extendend substrate requirement for BACE-1. The side chains for P7 through P5 have a preference for hydrophobic residues, particularly tryptophan [68]. The impact on cellular activity of these subsites has not been fully characterized by mutagenesis of the enzyme and the substrate.
Figure 2. Protein sequence of BACE-1 illustrates some of its key structural elements.
The italicized leader sequences represent the cleaved signal peptide followed by the furin-cleaved pro-domain. The bold, underlined fragments in hexagonal boxes indicate the 2 active sites of BACE-1 conserved in all aspartyl proteases. The limits of the transmembrane domain are shown as bold, italicized, and underlined sequences near the C-terminal end and the core 17 residues are shaded and boxed. The cytoplasmic tail that follows are shaded and the boxed DDISLL sequence is the ACDL motif that is important in endocytosis of BACE-1. The Cys residues in the cytoplasmic domain are marked with an “*” to indicate that they are palmitoylated.
Processing of BACE-1
The BACE-1 cDNA encodes a longer pre-pro-enzyme with an open reading frame of 501 amino acids. The BACE-1 protein has a co-translationally cleaved signal peptide of 21 amino acids followed by a short pro-domain (22–45 aa), a large luminal catalytic domain (residues 46–451) a single transmembrane domain (residues 452–483) and a short cytoplasmic domain (residues 484–501) [47]. ProBACE-1 does not spontaneously convert to mature BACE-1 like some aspartic protease zymogens, but requires other proteases to eliminate the pro-peptide. Contrary to expectation, prevention of proBACE maturation does not inhibit its activity in vivo [69]. However, in vitro studies reveal that the recombinant proBACE-1 activity increased when a portion of its prodomain is removed by clostripain cleavage, and there was a three-fold increase in BACE-1 activity upon complete prodomain removal after cleavage at E46, indicating that maturation may regulate its optimal activity [70]. Thus, the enzymes that process proBACE-1 may be useful therapeutic targets and aid in understanding how BACE-1 activity is regulated through maturation. The cleavage site shows a typical proprotein convertase substrate motif, RLPR. There are seven known proprotein convertases — furin, PC1/3, PC2, PACE4, PC4, PC5/6 and PC7 — of which furin, PACE4, PC5/6 and PC7 are most widely expressed in the constitutive secretory pathway [71, 72]. Our studies with RPE40, a furin-deficient cell line, showed that there was reduced maturation of proBACE-1; thus, any of the other proprotein convertase activities in the constitutive secretory pathway can effect BACE-1 maturation providing redundancy in this pathway [73]. More recent studies reveal that PS1 binds proBACE and may modulate its maturation process [74, 75].
Post-translational modifications
Several post-translational modifications have been reported on BACE-1, including phosphorylation of a serine in the cytoplasmic domain [76], palmitoylation of three cysteines in the transmembrane/cytoplasmic domain, sulfation of the N-linked oligosaccharides [77] and inclusion of four glycosylation sites in the catalytic domain [78].
Glycosylation is a well-known player in protein trafficking, folding and stabilization. Our unpublished studies have shown that lack of N-glycosylation can reduce Aβ and sAPPβ. BACE-1 has been found to have four potential glycosylation sites: N153, N172, N223, and N354 [78, 79]. Three of these sites are modified in the endoplasmic reticulum and subsequently processed in the Golgi stacks, thus leading to the formation of mature BACE-1 [78–80]. The significance of these four sites and their proximity to the active site is of great interest [78], especially with the knowledge that these types of modifications can alter protein folding and possibly prevent proteolytic attack [81]. These residues impact the proteolytic activity of BACE-1 in CHO cell lines [82], are essential to the maintenance of an active conformation of this protein, and therefore ensure its optimal activity and substrate interactions [82]. However, unmodified recombinant BACE-1 was found to be fully active and substrate specific, indicating that these modifications are not needed for the enzyme to function [43, 59, 70] although the activity may not be optimum.
BACE-1 undergoes homodimerization in vivo and the purified native BACE-1 dimer shows a higher affinity towards APP and a higher catalytic turnover rate than monomeric soluble BACE-1. Hence, it is thought that dimerization of BACE-1 facilitates binding and cleavage of physiological substrates in vivo [83].
Palmitoylation of BACE was shown to regulate its recruitment into the cholesterol-rich rafts [77]. However, studies with cerulenin, an inhibitor of fatty acid synthase that inhibits palmitoylation, showed that the association of BACE-1 with APP was not hindered [84]. However, inhibiting palmitoylation by cerlenin and farnesylation by the Cys-Val-Phe-Met peptide prevented the dimerization of BACE-1 signifying the complexity of the process and its relationship to lipid modification [84, 85]. It was also shown that ectodomain shedding of BACE-1 could be facilitated by loss of palmitoylation [77]. It was suggested that the shedding might be important in the turnover of BACE-1 and therefore might modulate the production of Aβ; direct evidence for this type of regulation remains to be presented.
Isoprenoids such as geranylgeranyl pyrophosphate (GGPP) and farnesyl pyrophosphate can be post-translationally linked to proteins and has been shown to affect Aβ production [86, 87]. Previous studies have shown that isoprenoids regulate BACE-1, which may explain the Aβ-lowering effects of statins [86]. However, we have shown that isoprenoids such as GGPP stimulate gamma secretase activity, which may also explain at least part of their Aβ-lowering properties [87]. Although we were unable to detect prenylation sequence motifs on BACE-1, BACE-1-associated proteins such as BRI3 are known to be farsenylated. Several studies have shown that cholesterol increases Aβ by increasing BACE-1 cleavage, but the mechanisms involved, including the role of isoprenoids is not understood [88, 89].
Another process involved in the maturation of BACE-1 involves disulphide bond formation within the catalytic domain between Cys 216/Cys420, Cys278/Cys443, Cys330/Cys380. When these disulphide bonds were abolished by replacement of Cys residues by mutagenesis, BACE-1 maturation was impaired. Moreover, the Cys330/Cys380 residues that were conserved amongst the family members were found to be crucial for the activity of the enzyme [90].
Regulation of BACE-1
BACE-1 has a tissue-specific expression pattern with mRNA preferentially detected in neuronal cells and the pancreas [45]. Mutations in the BACE-1 gene have not been identified in FAD cases, but increased BACE-1 activity has been reported in some patients with FAD [91] and some increase in BACE-1 mRNA levels was also reported in the cortex of sporadic AD patients [92, 93]. It is possible that BACE-1 gene expression is regulated at the level of transcription or translation, which could contribute to increased Aβ levels in sporadic AD cases. However, the evidence for increased Aβ production in sporadic AD in the cerebral spinal fluid is weak, although one cannot rule out possible increases in production in specific regions. We have previously cloned and characterized a 4129 bp BACE-1 promoter, including the majority of the 5’UTR [94–96].
The proximal BACE-1 promoter region does not include TATA or CATA boxes and has a high GC content unlike the typical eukaryotic promoter. The BACE-1 5’UTR consists of 457 bp and is reported to contain negative regulatory elements to attenuate promoter activity. In addition, there are three upstream initiation codons in the 5’ UTR region with the fourth codon being responsible for translation. The rest of the AUGs are skipped due to leaky ribosomal scanning. Hence, it is thought that leaky scanning in the 5’UTR and reinitiation at the 4th AUG were responsible for the low level of expression of BACE-1 under normal conditions [34].
For this report, we carried out a fresh analysis of the BACE-1 promoter using MatInspector (www.genomatix.de). Sequence analysis of the BACE-1 promoter region and 5’ UTR revealed the presence of multiple potential transcription factor binding sites as reported previously [94–96]. This review focuses on only two regions: the 5’ UTR and the 5’ flanking promoter to residue −1053. This region includes domains that were previously described as the neutral region IV, and the active regions V and VI, and includes a number of transcription factors [94–96] (Table 2). The most important factors for expression that have been identified are SP1 and YY1. Gel shift assay revealed the presence of a SP1 response element with the binding sequence centered at −908. It was also shown that overexpression of SP1 facilitates BACE-1 gene expression leading to an increase in Aβ biogenesis [97]. SP1 was found to play a central role in the control of BACE-1 gene expression, both in neuronal and nonneuronal cells (Table 2). Furthermore, it was also reported that the transcription factor YY1 could activate the BACE-1 promoter. However, YY1 is not expressed in most neurons and may be involved in promoting BACE-1 expression in reactive astrocytes [98]. Studies suggest that NF-kB acts as a repressor for BACE-1 transcription in differentiated neuronal cultures and nonactivated glial cultures, but as an activator for BACE-1 in activated astrocytic and Aβ-exposed neuronal cultures [95, 99].
Table-2.
Selected Transcription factor binding sites on the BACE-1 promoter (Range −1053 to 493).
| Factor | Description | Position (−1053 to 493) |
|---|---|---|
| ALS | Acid-Labile Subunit | 64 |
| AP–2 | AP2 and related proteins | −58, 94, 173, 305 |
| AP-3 | AP3 and related proteins | −858 |
| AP–4 | AP4 and related proteins | 33 |
| ApoAI RP–1 | Apolipoprotein AI | −774, 86 |
| bHLH | Basic helix–loop–helix | −202, 247 |
| CLS | Cardiolipin synthase | −433 |
| GATA | GATA binding factors | −867, −847, −846, −644, −713, − 277 |
| GC box | GC box binding protein | 98, 155, 161, 194 |
| HSF–1/2 | Heat Shock Factor 1 and 2 | −919, −918, −404 |
| IL-6 REBP | Interleukin 6 | −1043, −852, −504, −395 |
| IRF-1 | Interferon regulatory factors | −319 |
| IRF-3 | Interferon 3 regulatory factors | −913 |
| MZF1 | Myeloid zinc finger 1 factors | −2 |
| NF-1 | Nuclear factor 1 | −857, −687, −686, −598, −544, 177, 57 |
| NF-κB | Nuclear factor kappa B/c-rel | −921, −342 |
| ONSF | ON sheath fenestration | 13 |
| SF1 | Vertebrate steroidogenic factor | −634 |
| Smad3 | Vertebrate SMAD family of transcription factors | −986, −686, −204, −176, −118 |
| SP1 | GC-Box factors SP1/GC | −169. −152, 4, 8, 106, 138, 147, 157, 162, 229 |
| STAT6 | Signal transducer and activator of transcription | −931 |
| YY1 | Yin Yang | −447 |
| Zeste | Drosophila sequence-specific DNA-binding protein |
−968, −733, −436, −407 |
There is evidence to support the idea that non-steroidal anti-inflammatory drugs (NSAIDs) decrease the risk of AD. Peroxisome proliferator–activated receptor-gamma (PPARγ), a nuclear transcriptional regulator, can be activated by certain NSAIDs. It was shown that depletion of PPARγ resulted in an increase in the levels of the BACE-1 promoter, wherein mutation at PPRE increased the promoter activity [100]. Peptidylprolyl isomerase (cyclophilin) - like 2 (PPIL2), a U box-containing gene is a member of the cyclophilin family with poorly defined function. It was demonstrated that BACE-1 mRNA levels were affected by the expression of PPIL2 (i.e., knocking down PPIL2 knocked down BACE-1 mRNA) and over expression of PPIL2 increased BACE-1 mRNA synthesis. It was also found that PPIL2 co-immunoprecipitates with CD147 and is thought to act as a chaperone for proteins expressed at the cell surface [101]. It was shown that CD147 is a component of the γ secretase complex, and hence the complex may modulate BACE-1 expression [102].
Oxidative stress induced by hydrogen peroxide appears to increase the promoter activity of BACE-1, which can readily predict a higher level of BACE-1 activity in the brain of the elderly [103]. It was also observed that hydrogen peroxide treatment facilitated BACE-1 activity and Aβ generation [103]. Palmitic acid caused an up-regulation of BACE-1 expression in cortical neurons [104]. 4-Hydroxynonenal, an aldehyde product of lipid peroxidation was found to up-regulate BACE-1 expression as well [105]. BACE-1 expression was increased in cells stimulated with the M1/M3-selective muscarinic ACh receptor agonist talsaclidine. Protein kinase C activation by phorbol esters led to up regulation of BACE-1 [106]. The lipid second messenger ceramide was found to regulate several biochemical events as well. Ceramide levels were found to be highly elevated in the AD brain [107]. Pulse chase and time course degradation experiments showed that ceramide post-translationally stabilizes BACE-1, thus increasing the half-life of BACE-1 and thereby promoting Aβ biogenesis [107].
Subcellular localization of BACE-1
There is evidence suggesting that BACE-1 is found in the endoplasmic reticulum, Golgi network, cell surfaces, and endosomes [79, 80]. BACE-1 is enriched in neuronal Golgi membranes [108] and the phosphorylation of a serine residue at the BACE-1 C-terminal region may regulate its intracellular trafficking [76]. After synthesis, BACE-1 is transported to the cell surface via the endoplasmic reticulum and Golgi. When it is translocated into the endoplasmic reticulum, four Asn residues are N-glycosylated and disulphide bonds are formed between the Cys residues [78]. As the protein gets transported through the secretory pathway, it is further glycosylated in the Golgi to the mature endoglycosidase-H-resistant form [80]. The enzyme is then transported to the Golgi apparatus; by furin or furin-like proteases remove the prodomain [77, 109].
The mature enzyme is trafficked to the cell surface where it is internalized into early endosomal compartments which can either recycle directly back to the cell surface, fuse with the trans Golgi network or move to late endosomal/lysosomal compartments [76, 79, 108]. There are two distinct signals within the cytoplasmic tail of BACE-1 that regulate its subcellular trafficking. First, a dileucine motif (499/500) present in the intracellular domain is thought to regulate trafficking as well as endocytosis [110]. BACE-1-LL/AA mutations therefore also increase the protein levels of mature BACE-1 [110]. Second, the serine residue (S498) modulates the recycling from the early endosome via the late endosome and/or trans Golgi network to cell surface, but does not significantly alter the endocytic pathway per se [76]. BACE-1 appears to be localized at the Golgi when S498 is phosphorylated, and at the endosome when it is not [76].
After synthesis, BACE-1 is transported to the cell surface via the endoplasmic reticulum and Golgi. Membrane-associated APP and BACE-1 are both endocytosed where the BACE-1 cleavage of APP occurs. As the optimum pH of BACE-1 is 3.5–4.4, it is unlikely that the enzyme is fully active in Endoplasmic Reticulum/Golgi where the pH is around 6–7. It is possible that the majority of cleavage occurs in endosomes based on the observation of bafilomycin A1 inhibition of BACE-1. Lysosomal and endosomal aberrations have been reported in AD, and although subtle, these changes may result in large changes in BACE-1 processing of APP leading to increases in amyloid overproduction and deposition [110]. Nevertheless, recent studies using dominant negative dynamin to inhibit endocytosis where Aβ production is increased, suggests that some BACE-1 cleavage occurs at the cell surface [111]. However, this study cannot rule out the possibility that CTFβ is generated intracellularly and cleaved by γ-secretase to Aβ when the CTFβ generated intracellularly is retained on the surface.
It is important to note that despite having mutually exclusive cleavage sites, α-secretase and β-secretase normally do not compete with each other, as shown by the lack of increase in Aβ upon inhibition of α-secretase. However, increase in APP expression results in increased Aβ yields in most cultured cells indicating that BACE-1 activity is not limiting. Thus, BACE-1 must be highly concentrated in an active form in a specialized subcellular compartment, which has not yet been identified [112].
Even before the discovery of BACE-1, we found that glycosylphosphatidylinositol (GPI)-anchored proteins facilitate wild type APP processing by β-secretase [113]. We proposed that GPI-anchored proteins comprise a small group of membrane proteins that play an important role in β-secretase activity and Aβ secretion, but not in α-secretase activity [113]. Removal of these proteins by phosphatidylinositol-specific phospholipase C (PI-PLC) treatment lowered Aβ and sAPPβ, without affecting sAPPα levels [114]. Analysis of BACE-1 shows that it is not GPI anchored, so GPI-anchored proteins are likely to be regulators of the pathway rather than its effectors. One of the possible explanations is that BACE-1 is localized in lipid rafts, which are enriched in GPI-anchored proteins [114]. Lipid rafts are cholesterol-enriched membrane microdomains implicated in signal transduction, protein trafficking and proteolytic processing and may lead to regulation of activity by several mechanisms. The GPI-anchored proteins may drive the localization of BACE-1 to lipid rafts and also facilitate its binding to the APP substrate. Consistent with this hypothesis, the recombinant BACE-1 ectodomain is rapidly taken up from the medium by using APP as a receptor. This uptake is dependent on the presence of GPI-anchored proteins [115].
BACE-1 binding partners
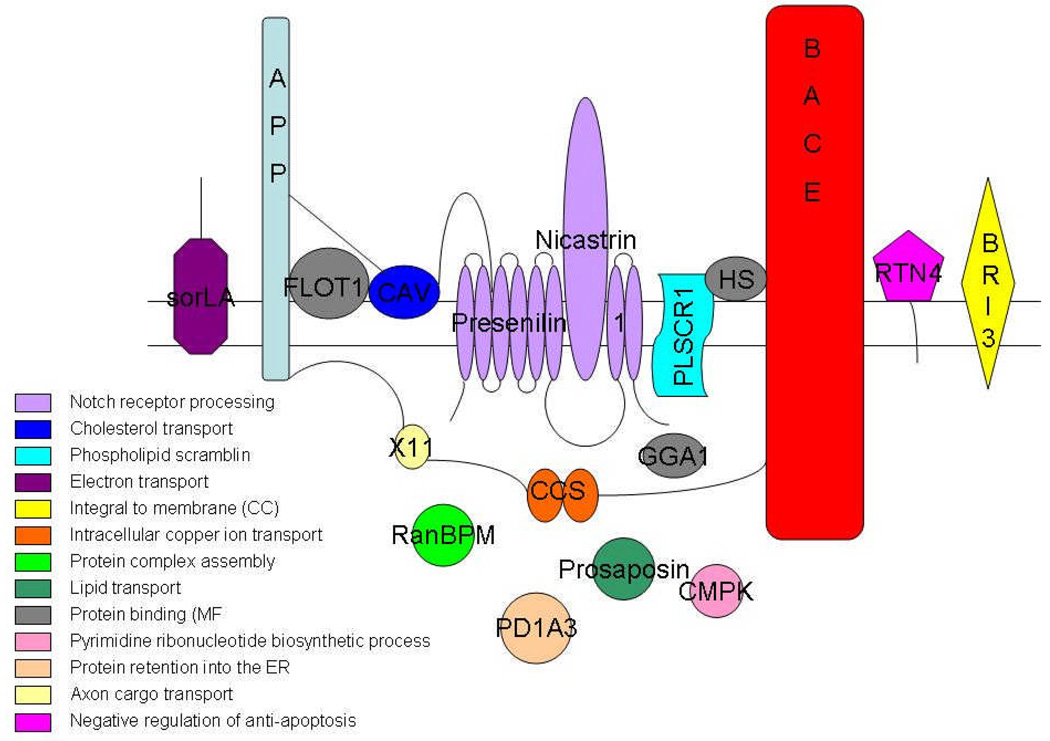
A number of properties of BACE-1 are very enigmatic, including its potential localization in lipid rafts and its high in vivo activity and compartmentalization [112, 114, 116]. These enigmatic properties are best explained by the binding of BACE-1 to other partner proteins that modulate its localization and activity. A representative group of these proteins is listed below (see Figure 4), but do not fully explain the phenomena, suggesting that other proteins may also be involved. However, at least four proteins in lipid rafts were reported to interact with BACE-1 – prion protein (PrP), reticulon (RTN), caveolin and flotillin. In addition, extensive screens have identified other BACE-1-binding proteins although their relevance and functions remain to be determined. Two significant proteins worth mentioning are Sortillin related receptor (SORL1) and BRI due to their linkage to amyloidosis and AD [117, 118]. The roles of these proteins are enumerated below.
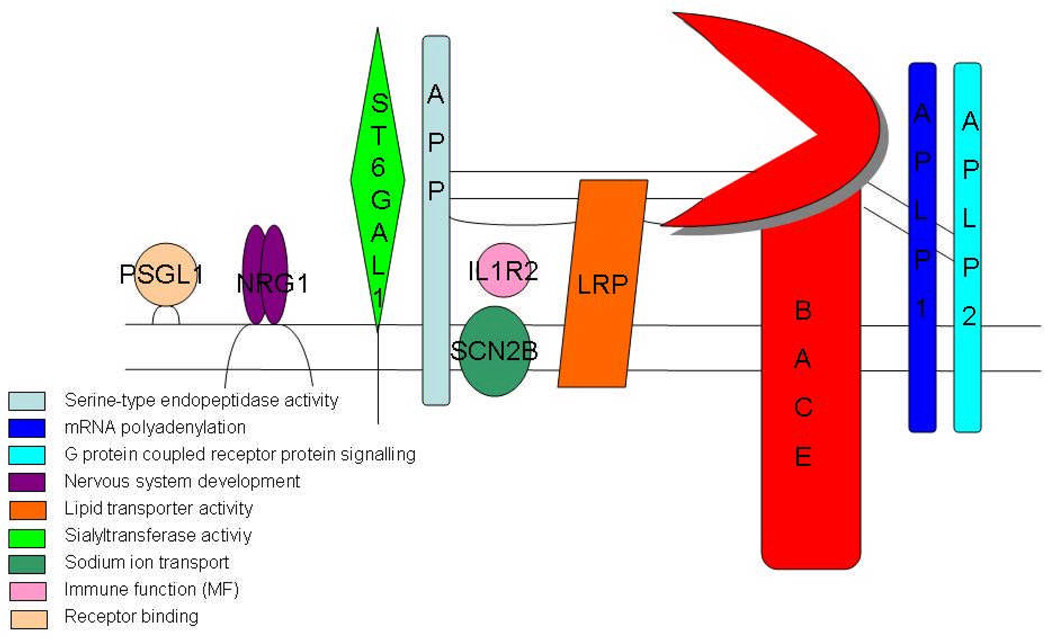
Figure 4.
Representation of the substrates of BACE-1 and their most significant gene ontology molecular function terms: These terms were deciphered using Bioconductor and the R Project (http://r-project.org).
Prion Protein (PrP): The prion protein is a causative agent of the transmissible spongiform encephalopathies that include Creutzfeldt–Jakob disease, Gerstmann-Scheinker-Straussler disease, bovine spongiform encephalopathy in cattle, and scrapie in sheep [119]. The role of PrP is not yet fully known, though evidence has shown its involvement in metal ion homeostasis, neuroprotection, and oxidative stress [120, 121]. There has long been a debate about the implication of PrP in AD and a Val/Met-129 polymorphism has been found to be a risk factor for typical late onset AD [122, 123]. Interestingly, as a GPI-anchored protein, PrP is localized to lipid rafts. PrP was found to inhibit β-secretase processing of APP, possibly through lipid raft localization of all three components. However, the mechanisms of inhibition remain to be revealed, as PrP does not appear to directly inhibit the enzyme activity of BACE-1 [124].
Flotillin-1 and Caveolin-1: Lipid rafts have been known to play a crucial role in Aβ biogenesis. Cholesterol is a main component of lipid rafts that has been implicated in the regulation of APP processing, so the finding of APP, Aβ and the Presenilins in lipid rafts was significant. Lipid raft proteins such as Caveolin (CAV) and Flotillin (FLOT-1) have also been implicated in APP processing. FLOT-1 was found to bind BACE-1, and the overexpression of CAV-1 or FLOT-1 resulted in the recruitment of BACE-1 to lipid rafts [125].
Reticulon 4 (Nogo; RTN4): The reticulon family of proteins has four members: RTN1, RTN2, RTN3 and RTN4 (also known as Nogo). RTN4 is well known for its role in inhibiting neuritic outgrowth after injury. He and colleagues showed that the reticulon proteins are binding partners of BACE-1. They also suggested a role for them as negative regulators of BACE-1, probably by blocking the access of BACE-1 to APP [126].
SORL1: SORL1 or is member of the vacuolar protein sorting 10 protein (Vps10p) receptors, which interacts with BACE-1 in the Golgi, directly inhibiting the BACE–APP complex and activity [127]. Mice lacking SORL1 show accelerated amyloidosis highlighting its importance in the disease [128]. Most prominently, SORL1 levels are reduced in the AD brain, which may facilitate amyloid deposition; polymorphisms in the gene are linked to AD [117, 129]. Thus SORL1 appears to play a role in AD pathogenesis by modulating BACE-1 activity.
Brain specific type II membrane bound protein (BRI3), Ran-binding protein (RanBPM), protein disulfide isomerase family A, member 3 (Erp60, PDIA3), KH–type splicing regulatory protein, Na/K ATPase Beta subunit (UMP-CMP kinase), Prosaposin (PSAP): A yeast two-hybrid screen with a human cDNA library and cytosolic tail of BACE-1 as bait has identified several proteins such as BRI3, (RanBPM, KH–type splicing regulatory protein, Na/K ATPase Beta subunit (UMP-CMP kinase), ERp60 (PD1A3) and Prosaposin (PSAP) [130] as possible partners. Co-localization and co-immunoprecipitation studies confirmed that BACE-1 and BR13 co-localized and interacted with each other via the cytoplasmic tail of BACE-1. BRI3 assumes specific significance as mutations in this protein lead to deposition of familial British dementia amyloid – ABRI [118]. ABRI is a small peptide synthesized as an abberant extension from a faulty stop codon and released by furin cleavage that forms an extracellular amyloid deposit similar to Alzheimer’s Aβ and also induces deposition of tau as neurofibrillary tangles [131].
ADP ribosylation factor (GGA): GGA proteins are thought to be important sorting adaptors. GGA1, GGA2 and GGA3 are the known GGA proteins in humans. The GGA binds to the ACDL motif containing membrane proteins and recruits it to coated vesicles on the Golgi membrane [132]. BACE-1 can undergo phosphorylation at S498 in cell culture, but the physiological significance is unclear [76]. However, BACE-1 interaction was found to be sensitive to BACE-1 phosphorylation and occurs in the trans Golgi network [133, 134]. Moreover, depletion of GGA proteins with RNA interference or mutation at S498 to prevent phosphorylation results in accumulation of BACE-1 in early endosomes. These observations suggest that GGA1 binds phosphorylated BACE-1 and regulates its recycling to the cell surface via the trans Golgi network [62].
Nicastrin and Presenilins: PS1, PS2, and Nicastrin are important γ secretase components, which cleave the APP-CTFs inside the membrane. Nicastrin binds BACE-1 and increases its activity as demonstrated by an increase in levels of sAPPβ in transfected cells [135]. In addition, PS1 binds the immature proBACE-1 as demonstrated by co-immunoprecipitation, suggesting that PS1 may regulate its maturation [136]. The role of Nicastrin in the PS1-BACE interactions remains unclear, but unlike Nicastrin, PS1 binds proBACE rather than mature BACE [137]. The complex regulation of BACE-1 by γ-secretase components and its implications in coordinating the two activities need to be examined in greater detail.
Copper chaperone for superoxide dismutase-1 (CCS): Copper metabolism has been implicated in AD, as both APP and Aβ bind copper [138, 139]. Interestingly, BACE-1 interacts with CCS and may therefore modulate the Aβ-Cu interaction [140]. It was also shown that BACE-1 and soluble CCS are co-transported through axons in primary cortical neurons. Copper chaperones have been found in yeast, nematode, bacterial and human genomes [141, 142]. Since BACE-1 is not found in these primitive organisms, the physiological relevance of this protein is unclear. Moreover, the effects of this interaction on BACE-1 activity need to be clarified.
Phospholipid scramblase 1 (PLSCR1): With a yeast two-hybrid screening method, it was found that the cytoplasmic domain of PLSCR1, a type II integral membrane protein, interacts with the cytoplasmic domain of BACE-1 [143]. It was reported that PLSCR1 was a component of the lipid microdomain, along with BACE-1 and APP in SH-SY5Y cells [143]. It was thought that PLSCR1 might alter the local composition or topology of plasma membrane phospholipids, which could influence the process of endocytosis of BACE-1.
Heparan sulphate: Glycosaminoglycan chains attached to core proteins, forming heparin sulphate proteoglycans, and heparin, are known to regulate the activity of many proteases [144, 145]. It was shown that heparin promoted the BACE-1 cleavage of APP in cell culture [146]. Low concentrations of heparin promoted BACE-1 activity, whereas it was inhibited by high concentrations by mechanisms that have not yet been characterized [147].
Adaptor protein - X11: The X11 adaptor proteins α and β are well-known modulators of APP processing and accumulation of Aβ, making them potential therapeutic targets for Alzheimer's disease [148]. The PDZ2 domain of X11α binds to CCS, the copper chaperone for SOD1 and leads to the suppression of SOD1 activity, possibly due to failure of CCS to deliver the copper required for SOD1 activity. The amino terminus of CCS has been demonstrated to bind the cytoplasmic domain of BACE 1, thus making X11α a modulator of BACE-1 activity [140, 148].
Prostate apoptosis response-4 (PAR-4): PAR-4 is a leucine zipper protein that was previously identified to be involved in neuronal degeneration and abnormal Aβ production in models of AD. It was reported that the cytoplasmic domain of PAR-4 formed a complex with the cytosolic tail of BACE-1 using co-immunoprecipitation assays and in vitro pull-down experiments [149]. It was also shown that the BACE-1 cleavage of APP was increased by over expression and decreased by inhibition of PAR-4.
BACE-1 targets
APP and its homologues — APLP1 and APLP2: The biological function of BACE-1 is not fully known and based on its numerous partners, its regulation remains largely unexplored. Weak affinity for wild-type APP and low yield of BACE-1 cleavage products suggest that APP is not the primary endogenous substrate for BACE-1 [150]. Even the mammalian homologs of APP, APLP1 and APLP2 do not appear to be the primary essential substrates as a study using the MDCK cell line (a system used to study the polarized sorting mechanisms of proteins) demonstrated that the majority of BACE-1 is sorted to the apical surface in contrast to the basolateral sorting of APP [41, 42, 151]. Studies of APP and its homologs in C. elegans and D. melanogaster, showed that it was essential for viability and important for metabolism, respectively [152, 153]. However, BACE-1 knockouts that fail to generate Aβ are viable as are APP-knockout mice, providing support to the notion that production of Aβ is not the primary function of BACE-1. In contrast mutations at the α-secretase cleavage site lead to progressive toxicity in mice demonstrating its fundamental importance in the metabolism and function of APP [154]. Additionally, lower invertebrates such as C. elegans and D. melanogaster process APP homologues by the α-secretase pathway, but do not display BACE-1-like processing of at least the human APP substrate, reinforcing the idea that BACE-1 function in evolution is related to other substrates [155]. Consistent with this idea, several BACE-1 substrates have been discovered, as discussed below (Figure 2).
P-selectin glycoprotein ligand-1 (PSGL-1): BACE-1 knockout mice are viable, suggesting that the relevant substrates could be processed by other activities in the cell. It is now clear that BACE-1 is involved in the processing of immunologically important proteins via PSGL-1 [156], which mediates leukocyte adhesion and trafficking. An evolutionary perspective suggests that PSGL-1 proteins are highly conserved in mammals and may therefore share function [157]. The role of BACE-1 cleavage in PSGL-1 functions needs to be examined in greater detail.
Sialyltransferase (ST6Gal1): Sialyltransferase ST6Gal1 [156, 158] is an enzyme that is active after cleavage and found to be involved in glycosylation regulation of the immune response. Involvement of BACE-1 in the cleavage of PSGL1 and ST6Gal1 suggests a possible regulatory role of the protease in glycoconjugate metabolism. While PSGL-1 is a type I integral membrane protein like APP and BACE-1, ST6Gal1 is a type II protein with a C-terminal ectodomain, suggesting that proteins with either topology can be readily processed by BACE-1. The functional consequence of this processing is not clear, but ST6Gal1 activity appears to be increased in vivo.
Low-density lipoprotein receptor (LDLR)-related protein (LRP): Another important BACE-1 substrate is LRP, a multifunctional endocytic and signaling receptor that binds apolipoprotein E –containing lipoproteins such as high density lipiprotein and very low density lipoprotein. Studies using a FRET-based assay of protein proximity, fluorescence lifetime imaging and co-immunoprecipitation techniques demonstrated that there is an interaction of LRP with BACE-1 and that LRP is a BACE-1 substrate [133]. It is important to remember that LRP ligand binding domains interact with APP that has the Kunitz protease inhibitor domain. This is an important finding because it could lead to the discovery of the function of peripheral APP, found specifically in platelets, that does harbor the Kunitz protease inhibitor domain [159]. LRP, which is enriched in lipid rafts, may act as a scaffold for APP and BACE-1; such interaction may enhance the BACE-1 cleavage of APP and increase Aβ generation. LRP and APP independently interact via their C-terminal domains and this interaction appears to affect the relative α-secretase and BACE-1 cleavage of APP and Aβ yield in addition to competition as substrates [159].
The family of low-density lipoprotein (LDL) receptors is an ancient group of multifunctional cell surface proteins that have been implicated in extracellular protein endocytosis, cross-membrane signal transduction, and modulation of synaptic function. These receptors have highly conserved proteins in many species including Aplysia, C. elegans and D. melanogaster [160]. In annelids, the hemoglobin linker chain shares a 39-residue cysteine rich module within the N-terminus that is similar to repeats in low-density lipoprotein class A receptors in metazoans, Xenopus [161] and C. elegans [162]. A similar module could have been incorporated. Due to sequence similarity, it is also likely that the annelid linkers would exhibit the same disulfide connectivity as the LDL receptor proteins [163]. Further up the evolutionary tree, we come to insects, which have lipophorins that are responsible for lipid transport. Phylogenetic analysis of these proteins revealed that they form a monophyletic group within the insects, with vertebrate receptors forming a sister group in the same tree. Functionally, in insects, this group of proteins showed an increase after immune challenge, indicating that lipid metabolism may have been involved in the immune response, as shown in Aedes aegypti [164], and could implicate this involvement in higher organisms as well, creating a more integrative vision of the role of BACE-1 activity in AD.
Sodium-gated channel – β subunit – (VGSCβ, SCN2B): Additional important substrates for BACE-1 include VGSCβ and SCN2B [165]. SCN2B is cleaved by BACE-1 in a similar manner as ST6Gal1 and PSGL-1, after a leucine residue, and is subsequently cleaved by γ secretase. Voltage-gated sodium channels are the most abundant ion channel type, and are responsible for the initiation and propagation of action potentials. The hypothesis that neural activity is able to regulate the production of Aβ through β- and γ-secretase and that Aβ depresses synaptic transmission and neuronal activity makes this relationship interesting [165]. It will be interesting to follow the cell biology of this processing in neurons, as these channels are preferentially located in the axonal hillock.
Neuregulin1 (NRG1): BACE-1 expression was detected at its highest levels in early postnatal stages, when myelination occurs. The levels of BACE-1 then declined in the second postnatal weeks, and reached low levels in later phases [166]. In addition, levels of unprocessed NRG1 were higher in BACE-1 −/− mice than in wild type mice. Thus, these studies showed that BACE-1 was required for myelination and correct bundling of neurons by Schwann cells probably through processing of type III NRG1 [166, 167]. However, a recent study showed that pharmacological inhibition of BACE-1 in adult mice led to a significant lowering of Aβ without any significant effect on brain NRG-1 processing [168]. Neuregulin is a member of the heregulins, a family of Epidermal growth factor (EGF)-like growth and differentiation factors. All of the isoforms of this family were produced by alternative splicing of a single gene [169]. A neuregulin homolog has been found in Xenopus, and has been shown to have very close homology to mammalian neuregulin [170]. The only other sequences that have been studied are the mammalian sequences, so very little is known about the evolution history beyond amphibians and invertebrates. It will be interesting to determine the co-evolution of BACE-1 and NRG1 to determine whether a functionally critical enzyme-substrate pair is maintained through phylogeny.
Interleukin-1 receptor II (IL-1R2): IL-1R2 functions as a decoy receptor, which limits the effects of IL-1 in the brain, most of which can be detrimental possibly by stimulation of inflammatory cascades. It has been demonstrated that increased secretion of IL-1R2 is associated with the pathogenesis of AD, and that it can be cleaved by all three secretases of the APP processing pathway [171, 172]. There is still some controversy about the validity of IL-1R2 as a substrate due to the similarity in the CTFs produced by α and β secretases. There are over 80 Il-1 family members from various species. Within this family is the Il-1R/Toll like (TLR) superfamily, which comprises a group of proteins vital to immunity in many aspects, including host defense, allergic and non-allergic inflammation, injury and stress [173]. The members of this group contain an ancient domain similar to that of Toll in D. melanogaster (TIR domains) that transduces signals via similar pathways in invertebrates and vertebrates [174]. TIRs have been found in species from plants to mammals; these have differentiated functions, in that in invertebrates, function is centered around development and immunity, while the function in vertebrates is for defense and inflammation [173]. This divergence in function may be a clue to the separation of plant and animal kingdoms by the regulation of immune response, and thus could lead to further elucidation of the regulation of this response in disease as well [173]. A detailed analysis of this relationship is particularly warranted by the inflammatory nature of neurodegeneration in AD, which may be mediated by cytokines such as IL-1, but reflected in the deposition of Aβ.
BACE-1 Inhibitors
Upon finding that BACE-1 knockouts appeared healthy and that the inhibition of BACE-1 reduced the levels of Aβ, searching for suitable BACE inhibitors started. Interestingly, BACE-1 is resistant to a broad-spectrum aspartic protease inhibitor, pepstatin A [47]. In addition, the HIV inhibitors as well as renin inhibitors failed to inhibit the enzyme [57]. This led to the conclusion that the active site of the enzyme was different from other aspartyl proteases. From the elucidation of the protein structure and activity assays, it is now possible to know more about the catalytic as well as inhibitory mechanisms. Many different types of assays were invented to tackle this problem; one of the more successful ones was a fluorogenic assay where inhibitors for both BACE-1 and BACE-2 have been described [175]. Peptidomimetic statin inhibitors were initially developed based on the sequence of the Swedish mutation of APP. However, other inhibitors were developed based on the substrate specificity of this enzyme, including its peripheral sites to probe its structure as described earlier [65]. Some of these inhibitors are very effective in vitro and reduce secretion of Aβ in cultured cells [176]. However, drug development has still been hindered by the lack of inhibitors that can be successfully delivered to the brain. Nonetheless, , there is a very large effort from pharmaceutical companies and academia to overcome this obstacle. The development of BACE-1 inhibitors is indeed a very large topic, placing it beyond the scope of this review (see [176]).
BACE-1 Function
Since BACE-1 is enriched in neurons and transported to axons [177], the potential role of BACE-1 in axonal growth and brain development was investigated by examining BACE-1-null mice. Significant reduction in myelin sheath thickness of both central and peripheral nerves was found, and was correlated with a reduction in myelin proteins [167]. Further, mice deficient in BACE-1 have altered hippocampal synaptic plasticity, decreased cognitive performance [178] and reduced lifespan [55]. It has been suggested that high expression of BACE-1 in neurons at birth could be linked to the onset of myelination by Schwann cells, and depends on signaling from the accompanying axons. In particular, the type III isoform of the epidermal growth factor-like protein, NRG1, is important during Schwann cell development and myelination [179, 180]. Interestingly, this function must be very tightly regulated and independent of Aβ as overexpression of BACE-1 also causes neurodegeneration despite reducing levels of Aβ [181].
Immunohistochemical analyses of brain sections revealed significant reductions in myelin in BACE-1-null mice relative to wild type controls. Hypomyelination was easily discernible in the hippocampus and cerebral cortex of BACE-1-null mice, whereas axonal development appeared normal [167]. BACE-1 seemed to affect myelination from early stages of development, hypomyelination being seen at postnatal day 15 and 30 in BACE-1-null mice [167]. Thus, myelin sheaths are significantly thinner in the optic nerves of BACE-1-null mice compared with littermate wild type controls. Sheath thickness was lower for axons of all diameters, and a higher percentage of small myelinated axons were present in BACE-1-null mice. These axonal changes were consistent with observations made for other hypomyelinated mouse models [182]. Abnormal myelination may have neurological consequences, including impaired motor and sensory functions. BACE-1-null mice show decreased grip strength and have increased pain sensitivity as compared to controls [167].
In addition to the role of BACE-1 in myelination, there is also evidence regarding its expression in platelets, where it might play a role in inflammation. BACE-1 expression has indeed been found, along with APP in platelets, though its function in the periphery is not clear [183].
BACE-1: An evolutionary perspective
BACE-1 belongs to the class of eukaryotic aspartyl proteases. Phylogenetic relationships among aspartyl proteases were used for the discovery of BACE-1 homologues as discussed earlier. In this section, we attempt to understand the phylogenetic structure-function relationship of these proteases. Aspartyl proteases are a large family of proteolytic enzymes [184] that are known in vertebrates (pepsins, cathepsins, etc), fungi, plants, and retroviruses. Eukaryotic aspartyl proteases are monomeric enzymes that consist of two domains containing an active site on a catalytic aspartyl residue. The N-terminal active site of most eukaryotic aspartyl proteases is characterized by the consensus motif DTGSSNLW. These enzymes are synthesized as zymogens that are subsequently proteolytically processed [185]. It is thought that eukaryotic aspartyl proteases come from a common ancestor by gene duplication and fusion. This hypothesis is supported by the structure of the enzymes, which have two homologous lobes [186] and by the organization of vertebrate genes for aspartyl proteases, which have two clusters of four exons [187]. The ancestral proteinases may have been homodimers like those found in retroviruses [188].
The origin of metazoan genome complexity is thought to be correlated to gene duplication events [189]. There is evidence to indicate that gene duplication is followed by a period of positive selection [189]. According to the data shown, the evolution of aspartyl proteases includes numerous gene duplication events, leading to the segregation of various groups of paralogs. One hypothesis is that duplicated genes remain in the genome only if mutations create new and critical protein function [190]. Increasing evidence indicates that the persistence of duplicated genes can be found by sub-functionalization, a mechanism by which each duplicate retains some ancestral function [191]. One way this can occur is by the division of gene expression after gene duplication in the tissue-specific expression of the daughter genes produced [189]. Comparisons of genes encoding aspartyl proteases from organisms including yeast, fungi, nematodes, protozoa, plants and vertebrates shows that though intron position varies throughout the protein family, the genes encoding the enzymes share structural similarity [192]. The only group with no introns in the aspartyl protease genes is yeast, which have small genomes and thus the aspartyl protease genes may have developed by losing introns during evolution [193]. Unlike vertebrate aspartyl protease genes with a 9-exon structure and conservation of the exon-intron boundaries, aspartyl protease genes from other organisms have a variable number of exons suggesting that the introns can be mobile and vulnerable to rapid rearrangements or deletions [194].
Analysis of the homologs of aspartyl proteases revealed that all six C. elegans aspartyl proteases contain two pairs of conserved cysteine residues that may form intra-strand disulfide bonds similar to those in mammalian cathepsins [195]. C. elegans aspartyl proteases show no closer relationship to each other than to aspartyl proteases from other organisms showing that this group of proteases diverged early in its evolution. The highest sequence similarities were found in three invertebrate aspartyl proteases: a lysosomal enzyme from Aedes aegpytii [196], Ancylostoma caninum anticoagulation protein [197] and Schistosoma japonicum aspartyl protease [198]. Syntichaki and coworkers reported that two C. elegans aspartyl proteases, asp3 and 4, nematode orthologues of Drosophila melanogaster aspartyl proteases (DASPs), mediate neuronal necrotic cell death in C. elegans [199]. There are also two D. melanogaster aspartyl proteases that show a 24 % homology to BACE-1: DASP1 and 2 [200]
Some exon-intron boundaries are shared between lineages. Specifically, correlations have been shown between vertebrates and non-parasitic nematodes and between apicomplexans and parasitic nematodes [194]. There have been reports of ancient intron positions in the plant and vertebrate genes [201] and in the duplicated glyceraldehyde-3-phosphate dehydrogenase genes [202]. Finally, the divergent exon-intron structure in aspartyl proteases from different groups of organisms suggests a process of gene evolution involving intron insertion [203], intron loss [204] and intron drift [205]. Though the proportion of these events is unknown, the existence of identified ancient intron positions does not support an extreme model that the origin and evolution of all aspartyl protease introns is relatively recent [194]. The finding of introns conserved between aspartyl proteases from nematodes and apicomplexans extends the idea of similarly positioned introns in more divergent organisms, suggesting an extremely early divergence time [206].
Evolution and function
As one of the crucial aspartyl proteases involved in AD, much attention has focused on the molecular function and pathological significance of BACE-1. Mice lacking BACE-1 display developmental defects that lead to hypomyelination [166, 167]. Further, it has been proposed that BACE-1 is needed for neuregulin processing and therefore its loss leads to hypomyelination [180]. However, recent studies suggest that BACE-1 inhibitors do not block neuregulin maturation in the brain [168]. Understanding the phylogenetic history of BACE-1 and neuregulin and other BACE-1 substrates can shed light on their functional conservation. Although the evolution of aspartyl proteases has been studied in detail in many disparate species and the conserved sequences within this family of proteases were used to identify BACE-1, the phylogenetic relationships of BACE-1 remain poorly described.
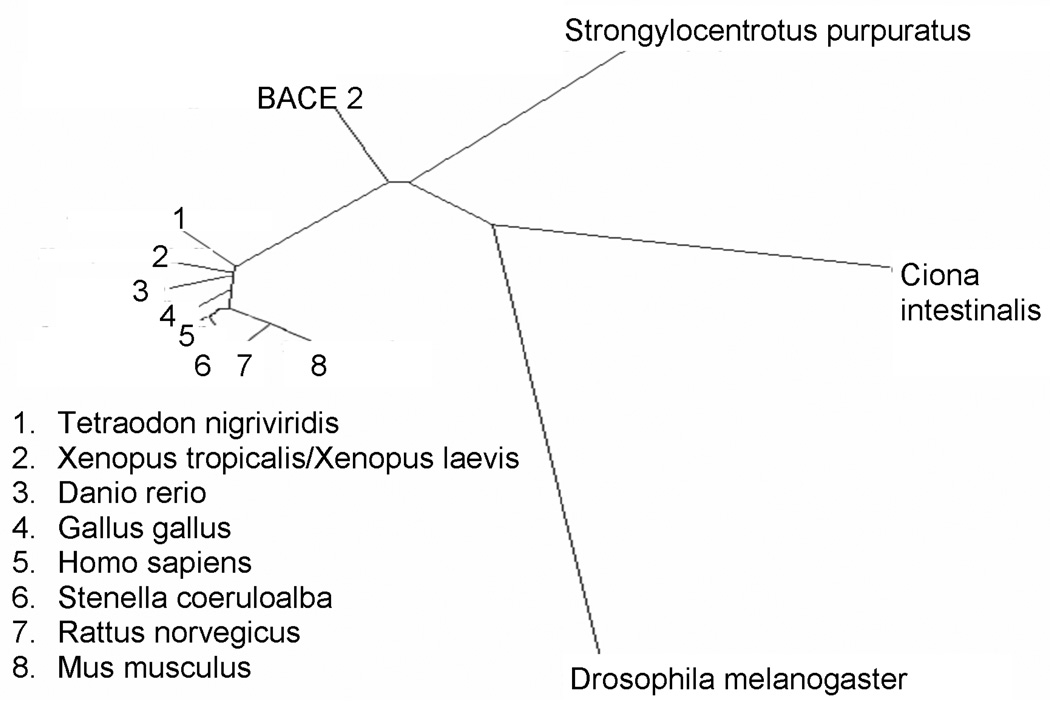
In our analysis of BACE-1 phylogeny (Figure 5), using an array of organisms including urochordates, insects, nematodes, fish, amphibians and mammals, we see that the sequences of the vertebrates are closely clustered to humans while D. melanogaster is the farthest removed. Even within the vertebrates, there is a considerable divergence in the BACE-1 sequence suggesting that it has limited functional specialization allowing it to rapidly evolve. For example, human APP is 87% similar to Xenopus APP whereas human BACE-1 is only 75% conserved. The Ciona intestinalis and Strongylocentrotus purpuratus BACE-1 homologues lie between insects and vertebrates, suggesting that they could be the ancestral sequences for mammals. Also, BACE-1 is clearly separate from BACE-2, which is likely the result of gene duplication events early in evolution. This phylogenetic pattern of BACE-1 fits the predicted evolutionary sequence of these organisms. Taking a representative substrate, we find that BACE-1 evolved prior to neuregulin, which is only seen in the vertebrate lineage along with other key myelin markers such as the proteolipid protein (PLP). Taking APP as a substrate, it has been reported that D. melanogaster cells are incapable of processing APP by the BACE-1 pathway despite the existence of structural homologues of the latter. This finding supports a conclusion that APP is not a functionally relevant substrate of BACE-1, at least early in the phylogenetic sequence. This hypothesis is supported by the observaton that BACE-1 is transported to the apical surface whereas APP is preferentially transported towards the basolateral plasma membrane in a polarized cell line [42].
Figure 5. Phylogenetic analysis of BACE-1.
The dendrogram shows the ortholog sequences found in an array of organisms dating back to early Deuterostomes. As apparent, these sequences are related to the mammalian form of BACE-1. BACE-2 separates early into its own separate branch.
Conclusion
BACE-1 is a novel integral membrane aspartyl protease responsible for the generation of Aβ in the brain and is therefore a key target for treatment of AD. While inhibitors against BACE-1 are being developed, the regulation and function of this enzyme needs to be thoroughly established. Some deficiencies in myelination have been discovered in the absence of BACE-1, which might be important in early development. However, a major question remains whether BACE-1 function remains essential in adults. We have presented a preliminary version of the BACE-1 phylogenetic tree and propose that a more thorough examination of the co-evolution of substrates and protein partners will provide important new insights into the critical functional aspects these molecules as they impact normal and pathophysiological states. An important observation is that both deletion of BACE-1 as well as BACE-1 overexpression leads to dysfunction in the nervous system. BACE-1 knockout results in loss of myelin and overexpression causes neurodegeneration. However, the increase in neurodegeneration is accompanied by a paradoxical reduction in Aβ levels. BACE-1 structure has been probed at the X-ray crystallography level, but the relationships of the BACE-1 complex with other partner proteins and the effects of post-translational modifications remain to be examined in detail. Providing a detailed evolutionary timeline for BACE-1 and its interacting partners and substrates could provide a clear picture of the original functions and mechanisms for these proteins and genes, and how they came to be involved in the pathogenesis and progression of AD. These studies will increase our detailed understanding of the role of BACE-1 in AD pathogenesis enabling discovery of drugs and other therapeutic strategies to regulate its activity for the benefit of future AD patients.
Figure 3.
Representation of the interaction partners of BACE-1 and their most significant gene ontology molecular function terms: These terms were deciphered using Bioconductor and the R Project (http://r-project.org).
Acknowledgements
The authors thank the National institutes on health (NIH) for grants AG022103 and AG023055 that supported this work.
Abbreviations
- Aβ40/42
40 or 42-residue amyloid peptide
- ACDL
acid cluster dileucine
- AD
Alzheimer’s disease
- AICD
Aβ protein precursor intracellular domain
- APP
Aβ protein precursor
- APLP
APP-like protein
- Asp
Aspartyl protease
- BACE-1
β-secretase
- BACE-2
BACE-1 homologue
- bp
base pair
- BRI
Brain specific type II membrane bound protein
- CAV
Caveolin
- CCS
copper chaperone for superoxide dismutase-1
- CD147
Cluster of differentiation 147
- CTF
Carboxy-terminal fragment
- DASP
Drosophila aspartyl protease
- EGF
Epidermal growth factor
- FAD
familial Alzheimer’s disease
- FLOT
flotillin
- FPP
Farnesyl pyrophosphate
- GGPP
Geranylgeranyl pyrophosphate
- GGA
ADP ribosylation factor
- GPI
glycosylphosphatidylinositol
- IL-1R2
interleukin-1 receptor II
- LDL
Low density lipoprotein
- LDLR
Low density lipoprotein receptor
- LRP
low-density lipoprotein receptor-related protein
- MDCK
Madin Darby Canine Kidney
- NF-kB
nuclear factor-kappa B
- NSAID
non-steroidal anti-inflammatory drugs
- P3
3 kDa protein generated by α- and γ-secretase processing of APP
- PAR-4
prostate apoptosis response-4
- PI-PLC
phosphatidylinositol-specific phospholipase C
- PLSCR1
phospholipid scramblase 1
- PPARγ
peroxisome proliferator–activated receptor-gamma
- PPIL2
peptidylprolyl isomerase (cyclophilin)-like 2
- PPRE
Peroxisome proliferator-activated receptor response element
- PrP
prion protein
- PS
presenilin
- PSAP
prosaposin
- PSGL
P-selectin glycoprotein ligand-1
- RanBPM
Ran-binding protein
- RTN
reticulon
- sAPP
secreted APP
- SOD1
superoxide dismutase 1
- SORL1
sortilin-related receptor
- SP1
specificity protein 1
- ST6Gal1
sialyltransferase
- YY1
Yin Yang1
Footnotes
The authors have no conflicts of interest.
References
- 1.Pillon B, Deweer B, Agid Y, Dubois B. Arch Neurol. 1993;50:374–379. doi: 10.1001/archneur.1993.00540040036010. [DOI] [PubMed] [Google Scholar]
- 2.Esteban-Santillan C, Praditsuwan R, Ueda H, Geldmacher DS. J Am Geriatr Soc. 1998;46:1266–1269. doi: 10.1111/j.1532-5415.1998.tb04543.x. [DOI] [PubMed] [Google Scholar]
- 3.Price BH, Gurvit H, Weintraub S, Geula C, Leimkuhler E, Mesulam M. Arch Neurol. 1993;50:931–937. doi: 10.1001/archneur.1993.00540090038008. [DOI] [PubMed] [Google Scholar]
- 4.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 5.Galasko D, Bennett D, Sano M, Ernesto C, Thomas R, Grundman M, Ferris S. Alzheimer Dis Assoc Disord. 1997;11 Suppl 2:S33–S39. [PubMed] [Google Scholar]
- 6.Brookmeyer R, Gray S, Kawas C. Am J Public Health. 1998;88:1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 8.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glenner GG, Wong CW. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 10.Robakis NK, Ramakrishna N, Wolfe G, Wisniewski HM. Proc Natl Acad Sci U S A. 1987;84:4190–4194. doi: 10.1073/pnas.84.12.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 12.Checler F. J Neurochem. 1995;65:1431–1444. doi: 10.1046/j.1471-4159.1995.65041431.x. [DOI] [PubMed] [Google Scholar]
- 13.Sambamurti K, Greig NH, Lahiri DK. Neuromolecular Med. 2002;1:1–31. doi: 10.1385/NMM:1:1:1. [DOI] [PubMed] [Google Scholar]
- 14.Anderson JP, Esch FS, Keim PS, Sambamurti K, Lieberburg I, Robakis NK. Neurosci Lett. 1991;128:126–128. doi: 10.1016/0304-3940(91)90775-o. [DOI] [PubMed] [Google Scholar]
- 15.Esch FS, Keim PS, Beattie EC, Blacher RW, Culwell AR, Oltersdorf T, McClure D, Ward PJ. Science. 1990;248:1122–1124. doi: 10.1126/science.2111583. [DOI] [PubMed] [Google Scholar]
- 16.Seubert P, Vigo-Pelfrey C, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher M, Whaley J, Swindlehurst C, McCormack R, Wolfert R, Selkoe D, Lieberburg I, Schenk D. Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 17.Seubert P, Oltersdorf T, Lee MG, Barbour R, Blomquist C, Davis DL, Bryant K, Fritz LC, Galasko D, Thal LJ, Lieberburg I, Schenk DB. Nature. 1993;361:260–263. doi: 10.1038/361260a0. [DOI] [PubMed] [Google Scholar]
- 18.Estus S, Golde TE, Kunishita T, Blades D, Lowery D, Eisen M, Usiak M, Qu XM, Tabira T, Greenberg BD, Younkin S. Science. 1992;255:726–728. doi: 10.1126/science.1738846. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki N, Cheung TT, Cai XD, Odaka A, Otvos L, Jr, Eckman C, Golde TE, Younkin SG. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 20.Passer B, Pellegrini L, Russo C, Siegel RM, Lenardo MJ, Schettini G, Bachmann M, Tabaton M, D'Adamio L. J Alzheimers Dis. 2000;2:289–301. doi: 10.3233/jad-2000-23-408. [DOI] [PubMed] [Google Scholar]
- 21.Pinnix I, Musunuru U, Tun H, Sridharan A, Golde T, Eckman C, Ziani-Cherif C, Onstead L, Sambamurti K. J Biol Chem. 2001;276:481–487. doi: 10.1074/jbc.M005968200. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, Misonou H, Sato T, Dohmae N, Takio K, Ihara Y. J Biol Chem. 2001;276:35235–35238. doi: 10.1074/jbc.C100357200. [DOI] [PubMed] [Google Scholar]
- 23.Yu C, Kim SH, Ikeuchi T, Xu H, Gasparini L, Wang R, Sisodia SS. J Biol Chem. 2001;276:43756–43760. doi: 10.1074/jbc.C100410200. [DOI] [PubMed] [Google Scholar]
- 24.Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C. EMBO Rep. 2001;2:835–841. doi: 10.1093/embo-reports/kve180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy AM, Newman S, McCaddon A, Ball J, Roques P, Mullan M, Hardy J, Chartier-Harlin MC, Frackowiak RS, Warrington EK, Rossor MN. Brain. 1993;116(Pt 2):309–324. doi: 10.1093/brain/116.2.309. [DOI] [PubMed] [Google Scholar]
- 27.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu Y-H, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 28.Mullan M. Bmj. 1992;305:1108–1109. doi: 10.1136/bmj.305.6862.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murrell J, Farlow M, Ghetti B, Benson MD. Science. 1991;254:97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 30.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin J-F, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HAR, Haines JL, Pericak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St George-Hyslop PH. Nature. 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 31.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 32.Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, Lannfelt L, Selkoe DJ. Nat Med. 1995;1:1291–1296. doi: 10.1038/nm1295-1291. [DOI] [PubMed] [Google Scholar]
- 33.Cai XD, Golde TE, Younkin SG. Science. 1993;259:514–516. doi: 10.1126/science.8424174. [DOI] [PubMed] [Google Scholar]
- 34.Sambamurti K, Suram A, Venugopal C, Prakasam A, Zhou Y, Lahiri DK, Greig NH. Curr Alzheimer Res. 2006;3:81–90. doi: 10.2174/156720506775697142. [DOI] [PubMed] [Google Scholar]
- 35.Chevallier N, Vizzavona J, Marambaud P, Baur CP, Spillantini M, Fulcrand P, Martinez J, Goedert M, Vincent JP, Checler F. Brain Res. 1997;750:11–19. doi: 10.1016/s0006-8993(96)01330-3. [DOI] [PubMed] [Google Scholar]
- 36.Brown AM, Tummolo DM, Spruyt MA, Jacobsen JS, Sonnenberg-Reines J. J Neurochem. 1996;66:2436–2445. doi: 10.1046/j.1471-4159.1996.66062436.x. [DOI] [PubMed] [Google Scholar]
- 37.Saftig P, Peters C, von Figura K, Craessaerts K, Van Leuven F, De Strooper B. J Biol Chem. 1996;271:27241–27244. doi: 10.1074/jbc.271.44.27241. [DOI] [PubMed] [Google Scholar]
- 38.Steinhilb ML, Turner RS, Gaut JR. J Biol Chem. 2001;276:4476–4484. doi: 10.1074/jbc.M008793200. [DOI] [PubMed] [Google Scholar]
- 39.Citron M, Diehl TS, Capell A, Haass C, Teplow DB, Selkoe DJ. Neuron. 1996;17:171–179. doi: 10.1016/s0896-6273(00)80290-1. [DOI] [PubMed] [Google Scholar]
- 40.Citron M, Teplow DB, Selkoe DJ. Neuron. 1995;14:661–670. doi: 10.1016/0896-6273(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 41.De Strooper B, Craessaerts K, Van Leuven F, Van Den Berghe H. J Biol Chem. 1995;270:30310–30314. doi: 10.1074/jbc.270.51.30310. [DOI] [PubMed] [Google Scholar]
- 42.Capell A, Meyn L, Fluhrer R, Teplow DB, Walter J, Haass C. J Biol Chem. 2002;277:5637–5643. doi: 10.1074/jbc.M109119200. [DOI] [PubMed] [Google Scholar]
- 43.Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Proc Natl Acad Sci U S A. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain I, Powell DJ, Howlett DR, Chapman GA, Gilmour L, Murdock PR, Tew DG, Meek TD, Chapman C, Schneider K, Ratcliffe SJ, Tattersall D, Testa TT, Southan C, Ryan DM, Simmons DL, Walsh FS, Dingwall C, Christie G. Mol Cell Neurosci. 2000;16:609–619. doi: 10.1006/mcne.2000.0884. [DOI] [PubMed] [Google Scholar]
- 45.Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, Carter DB, Tomasselli AG, Parodi LA, Heinrikson RL, Gurney ME. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 46.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 47.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson-Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, Zhao J, McConlogue L, John V. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 48.Sinha S, Knops J, Esch F, Moyer ED, Oltersdorf T. J Biol Chem. 1991;266:21011–21013. [PubMed] [Google Scholar]
- 49.Solans A, Estivill X, de La Luna S. Cytogenet Cell Genet. 2000;89:177–184. doi: 10.1159/000015608. [DOI] [PubMed] [Google Scholar]
- 50.Bennett BD, Babu-Khan S, Loeloff R, Louis JC, Curran E, Citron M, Vassar R. J Biol Chem. 2000;275:20647–20651. doi: 10.1074/jbc.M002688200. [DOI] [PubMed] [Google Scholar]
- 51.Farzan M, Schnitzler CE, Vasilieva N, Leung D, Choe H. Proc Natl Acad Sci U S A. 2000;97:9712–9717. doi: 10.1073/pnas.160115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan R, Munzner JB, Shuck ME, Bienkowski MJ. J Biol Chem. 2001;276:34019–34027. doi: 10.1074/jbc.M105583200. [DOI] [PubMed] [Google Scholar]
- 53.Basi G, Frigon N, Barbour R, Doan T, Gordon G, McConlogue L, Sinha S, Zeller M. J Biol Chem. 2003;278:31512–31520. doi: 10.1074/jbc.M300169200. [DOI] [PubMed] [Google Scholar]
- 54.Tomasselli AG, Qahwash I, Emmons TL, Lu Y, Leone JW, Lull JM, Fok KF, Bannow CA, Smith CW, Bienkowski MJ, Heinrikson RL, Yan R. J Neurochem. 2003;84:1006–1017. doi: 10.1046/j.1471-4159.2003.01597.x. [DOI] [PubMed] [Google Scholar]
- 55.Dominguez D, Tournoy J, Hartmann D, Huth T, Cryns K, Deforce S, Serneels L, Camacho IE, Marjaux E, Craessaerts K, Roebroek AJ, Schwake M, D'Hooge R, Bach P, Kalinke U, Moechars D, Alzheimer C, Reiss K, Saftig P, De Strooper B. J Biol Chem. 2005;280:30797–30806. doi: 10.1074/jbc.M505249200. [DOI] [PubMed] [Google Scholar]
- 56.Johnston JA, Cowburn RF, Norgren S, Wiehager B, Venizelos N, Winblad B, Vigo-Pelfrey C, Schenk D, Lannfelt L, O'Neill C. FEBS Lett. 1994;354:274–278. doi: 10.1016/0014-5793(94)01137-0. [DOI] [PubMed] [Google Scholar]
- 57.Gruninger-Leitch F, Schlatter D, Kung E, Nelbock P, Dobeli H. J Biol Chem. 2002;277:4687–4693. doi: 10.1074/jbc.M109266200. [DOI] [PubMed] [Google Scholar]
- 58.Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 59.Turner RT, 3rd, Koelsch G, Hong L, Castanheira P, Ermolieff J, Ghosh AK, Tang J. Biochemistry. 2001;40:10001–10006. doi: 10.1021/bi015546s. [DOI] [PubMed] [Google Scholar]
- 60.Schechter I, Berger A. Biochem Biophys Res Commun. 1968;32:898–902. doi: 10.1016/0006-291x(68)90326-4. [DOI] [PubMed] [Google Scholar]
- 61.Tang J, He X, Huang X, Hong L. Curr Alzheimer Res. 2005;2:261–264. doi: 10.2174/1567205053585800. [DOI] [PubMed] [Google Scholar]
- 62.He X, Li F, Chang WP, Tang J. J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- 63.He X, Zhu G, Koelsch G, Rodgers KK, Zhang XC. Tang J. Biochemistry. 2003;42:12174–12180. doi: 10.1021/bi035199h. [DOI] [PubMed] [Google Scholar]
- 64.Hong L, Koelsch G, Lin X, Wu S, Terzyan S, Ghosh AK, Zhang XC, Tang J. Science. 2000;290:150–153. doi: 10.1126/science.290.5489.150. [DOI] [PubMed] [Google Scholar]
- 65.Kornacker MG, Lai Z, Witmer M, Ma J, Hendrick J, Lee VG, Riexinger DJ, Mapelli C, Metzler W, Copeland RA. Biochemistry. 2005;44:11567–11573. doi: 10.1021/bi050932l. [DOI] [PubMed] [Google Scholar]
- 66.Hong L, Tang J. Biochemistry. 2004;43:4689–4695. doi: 10.1021/bi0498252. [DOI] [PubMed] [Google Scholar]
- 67.Hong L, Turner RT, 3rd, Koelsch G, Shin D, Ghosh AK, Tang J. Biochemistry. 2002;41:10963–10967. doi: 10.1021/bi026232n. [DOI] [PubMed] [Google Scholar]
- 68.Turner RT, 3rd, Hong L, Koelsch G, Ghosh AK, Tang J. Biochemistry. 2005;44:105–112. doi: 10.1021/bi048106k. [DOI] [PubMed] [Google Scholar]
- 69.Creemers JW, Ines Dominguez D, Plets E, Serneels L, Taylor NA, Multhaup G, Craessaerts K, Annaert W, De Strooper B. J Biol Chem. 2001;276:4211–4217. doi: 10.1074/jbc.M006947200. [DOI] [PubMed] [Google Scholar]
- 70.Ermolieff J, Loy JA, Koelsch G, Tang J. Biochemistry. 2000;39:12450–12456. doi: 10.1021/bi001494f. [DOI] [PubMed] [Google Scholar]
- 71.Seidah NG, Chretien M. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 72.Zhou A, Webb G, Zhu X, Steiner DF. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 73.Pinnix I, Council JE, Roseberry B, Onstead L, Mallender W, Sucic J, Sambamurti K, Faseb J. 2001;15:1810–1812. doi: 10.1096/fj.00-0891fje. [DOI] [PubMed] [Google Scholar]
- 74.Vetrivel KS, Thinakaran G. Neurology. 2006;66:S69–S73. doi: 10.1212/01.wnl.0000192107.17175.39. [DOI] [PubMed] [Google Scholar]
- 75.Vetrivel KS, Zhang YW, Xu H, Thinakaran G. Mol Neurodegener. 2006;1:4. doi: 10.1186/1750-1326-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walter J, Fluhrer R, Hartung B, Willem M, Kaether C, Capell A, Lammich S, Multhaup G, Haass C. J Biol Chem. 2001;276:14634–14641. doi: 10.1074/jbc.M011116200. [DOI] [PubMed] [Google Scholar]
- 77.Benjannet S, Elagoz A, Wickham L, Mamarbachi M, Munzer JS, Basak A, Lazure C, Cromlish JA, Sisodia S, Checler F, Chretien M, Seidah NG. J Biol Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 78.Haniu M, Denis P, Young Y, Mendiaz EA, Fuller J, Hui JO, Bennett BD, Kahn S, Ross S, Burgess T, Katta V, Rogers G, Vassar R, Citron M. J Biol Chem. 2000;275:21099–21106. doi: 10.1074/jbc.M002095200. [DOI] [PubMed] [Google Scholar]
- 79.Huse JT, Pijak DS, Leslie GJ, Lee VM, Doms RW. J Biol Chem. 2000;275:33729–33737. doi: 10.1074/jbc.M004175200. [DOI] [PubMed] [Google Scholar]
- 80.Capell A, Steiner H, Willem M, Kaiser H, Meyer C, Walter J, Lammich S, Multhaup G, Haass C. J Biol Chem. 2000;275:30849–30854. doi: 10.1074/jbc.M003202200. [DOI] [PubMed] [Google Scholar]
- 81.Helenius A. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charlwood J, Dingwall C, Matico R, Hussain I, Johanson K, Moore S, Powell DJ, Skehel JM, Ratcliffe S, Clarke B, Trill J, Sweitzer S, Camilleri P. J Biol Chem. 2001;276:16739–16748. doi: 10.1074/jbc.M009361200. [DOI] [PubMed] [Google Scholar]
- 83.Westmeyer GG, Willem M, Lichtenthaler SF, Lurman G, Multhaup G, Assfalg-Machleidt I, Reiss K, Saftig P, Haass C. J Biol Chem. 2004;279:53205–53212. doi: 10.1074/jbc.M410378200. [DOI] [PubMed] [Google Scholar]
- 84.Parsons RB, Austen BM. Biochem Soc Trans. 2005;33:1091–1093. doi: 10.1042/BST20051091. [DOI] [PubMed] [Google Scholar]
- 85.Sareneva T, Pirhonen J, Cantell K, Julkunen I. Biochem J. 1995;308(Pt 1):9–14. doi: 10.1042/bj3080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cole SL, Grudzien A, Manhart IO, Kelly BL, Oakley H, Vassar R. J Biol Chem. 2005;280:18755–18770. doi: 10.1074/jbc.M413895200. [DOI] [PubMed] [Google Scholar]
- 87.Zhou Y, Suram A, Venugopal C, Prakasam A, Lin S, Su Y, Li B, Paul SM, Sambamurti K. Faseb J. 2008;22:47–54. doi: 10.1096/fj.07-8175com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Refolo LM, Malester B, LaFrancois J, Bryant-Thomas T, Wang R, Tint GS, Sambamurti K, Duff K, Pappolla MA. Neurobiol Dis. 2000;7:321–331. doi: 10.1006/nbdi.2000.0304. [DOI] [PubMed] [Google Scholar]
- 89.Hartmann T, Kuchenbecker J, Grimm MO. J Neurochem. 2007;103 Suppl 1:159–170. doi: 10.1111/j.1471-4159.2007.04715.x. [DOI] [PubMed] [Google Scholar]
- 90.Fischer F, Molinari M, Bodendorf U, Paganetti P. J Neurochem. 2002;80:1079–1088. doi: 10.1046/j.0022-3042.2002.00806.x. [DOI] [PubMed] [Google Scholar]
- 91.Russo C, Schettini G, Saido TC, Hulette C, Lippa C, Lannfelt L, Ghetti B, Gambetti P, Tabaton M, Teller JK. Nature. 2000;405:531–532. doi: 10.1038/35014735. [DOI] [PubMed] [Google Scholar]
- 92.Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- 93.Fukumoto H, Rosene DL, Moss MB, Raju S, Hyman BT, Irizarry MC. Am J Pathol. 2004;164:719–725. doi: 10.1016/s0002-9440(10)63159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ge YW, Maloney B, Sambamurti K, Lahiri DK. Faseb J. 2004;18:1037–1039. doi: 10.1096/fj.03-1379fje. [DOI] [PubMed] [Google Scholar]
- 95.Lahiri DK, Maloney B, Ge YW. J Mol Neurosci. 2006;29:65–80. doi: 10.1385/JMN:29:1:65. [DOI] [PubMed] [Google Scholar]
- 96.Sambamurti K, Kinsey R, Maloney B, Ge YW, Lahiri DK. Faseb J. 2004;18:1034–1036. doi: 10.1096/fj.03-1378fje. [DOI] [PubMed] [Google Scholar]
- 97.Christensen MA, Zhou W, Qing H, Lehman A, Philipsen S, Song W. Mol Cell Biol. 2004;24:865–874. doi: 10.1128/MCB.24.2.865-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nowak K, Lange-Dohna C, Zeitschel U, Gunther A, Luscher B, Robitzki A, Perez-Polo R, Rossner S. J Neurochem. 2006;96:1696–1707. doi: 10.1111/j.1471-4159.2006.03692.x. [DOI] [PubMed] [Google Scholar]
- 99.Bourne KZ, Ferrari DC, Lange-Dohna C, Rossner S, Wood TG, Perez-Polo JR. J Neurosci Res. 2007;85:1194–1204. doi: 10.1002/jnr.21252. [DOI] [PubMed] [Google Scholar]
- 100.Sastre M, Dewachter I, Rossner S, Bogdanovic N, Rosen E, Borghgraef P, Evert BO, Dumitrescu-Ozimek L, Thal DR, Landreth G, Walter J, Klockgether T, van Leuven F, Heneka MT. Proc Natl Acad Sci U S A. 2006;103:443–448. doi: 10.1073/pnas.0503839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pushkarsky T, Yurchenko V, Vanpouille C, Brichacek B, Vaisman I, Hatakeyama S, Nakayama KI, Sherry B, Bukrinsky MI. J Biol Chem. 2005;280:27866–27871. doi: 10.1074/jbc.M503770200. [DOI] [PubMed] [Google Scholar]
- 102.Zhou S, Zhou H, Walian PJ, Jap BK. Proc Natl Acad Sci U S A. 2005 102;:7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tong Y, Zhou W, Fung V, Christensen MA, Qing H, Sun X, Song W. J Neural Transm. 2005;112:455–469. doi: 10.1007/s00702-004-0255-3. [DOI] [PubMed] [Google Scholar]
- 104.Patil S, Sheng L, Masserang A, Chan C. Neurosci Lett. 2006;406:55–59. doi: 10.1016/j.neulet.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 105.Tamagno E, Parola M, Bardini P, Piccini A, Borghi R, Guglielmotto M, Santoro G, Davit A, Danni O, Smith MA, Perry G, Tabaton M. J Neurochem. 2005;92:628–636. doi: 10.1111/j.1471-4159.2004.02895.x. [DOI] [PubMed] [Google Scholar]
- 106.Zuchner T, Perez-Polo JR, Schliebs R. J Neurosci Res. 2004;77:250–257. doi: 10.1002/jnr.20152. [DOI] [PubMed] [Google Scholar]
- 107.Puglielli L, Ellis BC, Saunders AJ, Kovacs DM. J Biol Chem. 2003;278:19777–19783. doi: 10.1074/jbc.M300466200. [DOI] [PubMed] [Google Scholar]
- 108.Yan R, Han P, Miao H, Greengard P, Xu H. J Biol Chem. 2001;276:36788–36796. doi: 10.1074/jbc.M104350200. [DOI] [PubMed] [Google Scholar]
- 109.Bennett BD, Denis P, Haniu M, Teplow DB, Kahn S, Louis JC, Citron M, Vassar R. J Biol Chem. 2000;275:37712–37717. doi: 10.1074/jbc.M005339200. [DOI] [PubMed] [Google Scholar]
- 110.Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G. J Biol Chem. 2005;280:32499–32504. doi: 10.1074/jbc.M506199200. [DOI] [PubMed] [Google Scholar]
- 111.Chyung JH, Selkoe DJ. J Biol Chem. 2003;278:51035–51043. doi: 10.1074/jbc.M304989200. [DOI] [PubMed] [Google Scholar]
- 112.Gandhi S, Refolo LM, Sambamurti K. J Mol Neurosci. 2004;24:137–143. doi: 10.1385/JMN:24:1:137. [DOI] [PubMed] [Google Scholar]
- 113.Sambamurti K, Sevlever D, Koothan T, Refolo LM, Pinnix I, Gandhi S, Onstead L, Younkin L, Prada CM, Yager D, Ohyagi Y, Eckman CB, Rosenberry TL, Younkin SG. J Biol Chem. 1999;274:26810–26814. doi: 10.1074/jbc.274.38.26810. [DOI] [PubMed] [Google Scholar]
- 114.Tun H, Marlow L, Pinnix I, Kinsey R, Sambamurti K. J Mol Neurosci. 2002;19:31–35. doi: 10.1007/s12031-002-0007-5. [DOI] [PubMed] [Google Scholar]
- 115.Huang XP, Chang WP, Koelsch G, Turner RT, 3rd, Lupu F, Tang J. J Biol Chem. 2004;279:37886–37894. doi: 10.1074/jbc.M402130200. [DOI] [PubMed] [Google Scholar]
- 116.Marlow L, Cain M, Pappolla MA, Sambamurti K. J Mol Neurosci. 2003;20:233–239. doi: 10.1385/JMN:20:3:233. [DOI] [PubMed] [Google Scholar]
- 117.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vidal R, Frangione B, Rostagno A, Mead S, Revesz T, Plant G, Ghiso J. Nature. 1999;399:776–781. doi: 10.1038/21637. [DOI] [PubMed] [Google Scholar]
- 119.Aguzzi A, Montrasio F, Kaeser PS. Nat Rev Mol Cell Biol. 2001;2:118–126. doi: 10.1038/35052063. [DOI] [PubMed] [Google Scholar]
- 120.Brown DR. Trends Neurosci. 2001;24:85–90. doi: 10.1016/s0166-2236(00)01689-1. [DOI] [PubMed] [Google Scholar]
- 121.Martins VR, Linden R, Prado MA, Walz R, Sakamoto AC, Izquierdo I, Brentani RR. FEBS Lett. 2002;512:25–28. doi: 10.1016/s0014-5793(02)02291-3. [DOI] [PubMed] [Google Scholar]
- 122.Dermaut B, Croes EA, Rademakers R, Van den Broeck M, Cruts M, Hofman A, van Duijn CM, Van Broeckhoven C. Ann Neurol. 2003;53:409–412. doi: 10.1002/ana.10507. [DOI] [PubMed] [Google Scholar]
- 123.Riemenschneider M, Klopp N, Xiang W, Wagenpfeil S, Vollmert C, Muller U, Forstl H, Illig T, Kretzschmar H, Kurz A. Neurology. 2004;63:364–366. doi: 10.1212/01.wnl.0000130198.72589.69. [DOI] [PubMed] [Google Scholar]
- 124.Parkin ET, Watt NT, Hussain I, Eckman EA, Eckman CB, Manson JC, Baybutt HN, Turner AJ, Hooper NM. Proc Natl Acad Sci U S A. 2007;104:11062–11067. doi: 10.1073/pnas.0609621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen TY, Liu PH, Ruan CT, Chiu L, Kung FL. Biochem Biophys Res Commun. 2006;342:266–272. doi: 10.1016/j.bbrc.2006.01.156. [DOI] [PubMed] [Google Scholar]
- 126.He W, Lu Y, Qahwash I, Hu XY, Chang A, Yan R. Nat Med. 2004;10:959–965. doi: 10.1038/nm1088. [DOI] [PubMed] [Google Scholar]
- 127.Spoelgen R, von Arnim CA, Thomas AV, Peltan ID, Koker M, Deng A, Irizarry MC, Andersen OM, Willnow TE, Hyman BT. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, Schaller C, Bujo H, Levey AI, Lah JJ. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 129.Ma QL, Teter B, Ubeda OJ, Morihara T, Dhoot D, Nyby MD, Tuck ML, Frautschy SA, Cole GM. J Neurosci. 2007;27:14299–14307. doi: 10.1523/JNEUROSCI.3593-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wickham L, Benjannet S, Marcinkiewicz E, Chretien M, Seidah NG. J Neurochem. 2005;92:93–102. doi: 10.1111/j.1471-4159.2004.02840.x. [DOI] [PubMed] [Google Scholar]
- 131.Ghiso J, Revesz T, Holton J, Rostagno A, Lashley T, Houlden H, Gibb G, Anderton B, Bek T, Bojsen-Moller M, Wood N, Vidal R, Braendgaard H, Plant G, Frangione B. Amyloid. 2001;8:277–284. doi: 10.3109/13506120108993826. [DOI] [PubMed] [Google Scholar]
- 132.Bonifacino JS. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 133.von Arnim CA, Kinoshita A, Peltan ID, Tangredi MM, Herl L, Lee BM, Spoelgen R, Hshieh TT, Ranganathan S, Battey FD, Liu CX, Bacskai BJ, Sever S, Irizarry MC, Strickland DK, Hyman BT. J Biol Chem. 2005;280:17777–17785. doi: 10.1074/jbc.M414248200. [DOI] [PubMed] [Google Scholar]
- 134.Shiba Y, Katoh Y, Shiba T, Yoshino K, Takatsu H, Kobayashi H, Shin HW, Wakatsuki S, Nakayama K. J Biol Chem. 2004;279:7105–7111. doi: 10.1074/jbc.M311702200. [DOI] [PubMed] [Google Scholar]
- 135.Hattori C, Asai M, Oma Y, Kino Y, Sasagawa N, Saido TC, Maruyama K, Ishiura S. Biochem Biophys Res Commun. 2002;293:1228–1232. doi: 10.1016/S0006-291X(02)00351-0. [DOI] [PubMed] [Google Scholar]
- 136.Hebert SS, Bourdages V, Godin C, Ferland M, Carreau M, Levesque G. Neurobiol Dis. 2003;13:238–245. doi: 10.1016/s0969-9961(03)00035-4. [DOI] [PubMed] [Google Scholar]
- 137.Kuzuya A, Uemura K, Kitagawa N, Aoyagi N, Kihara T, Ninomiya H, Ishiura S, Takahashi R, Shimohama S. J Neurosci Res. 2007;85:153–165. doi: 10.1002/jnr.21104. [DOI] [PubMed] [Google Scholar]
- 138.Bush AI. Alzheimer Dis Assoc Disord. 2003;17:147–150. doi: 10.1097/00002093-200307000-00005. [DOI] [PubMed] [Google Scholar]
- 139.Bush AI, Masters CL, Tanzi RE. Proc Natl Acad Sci U S A. 2003;100:11193–11194. doi: 10.1073/pnas.2135061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Angeletti B, Waldron KJ, Freeman KB, Bawagan H, Hussain I, Miller CC, Lau KF, Tennant ME, Dennison C, Robinson NJ, Dingwall C. J Biol Chem. 2005;280:17930–17937. doi: 10.1074/jbc.M412034200. [DOI] [PubMed] [Google Scholar]
- 141.Landis GN, Tower J. Mech Ageing Dev. 2005;126:365–379. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 142.Magnani D, Solioz M. Biometals. 2005;18:407–412. doi: 10.1007/s10534-005-3715-9. [DOI] [PubMed] [Google Scholar]
- 143.Kametaka S, Shibata M, Moroe K, Kanamori S, Ohsawa Y, Waguri S, Sims PJ, Emoto K, Umeda M, Uchiyama Y. J Biol Chem. 2003;278:15239–15245. doi: 10.1074/jbc.M208611200. [DOI] [PubMed] [Google Scholar]
- 144.Turnbull J, Powell A, Guimond S. Trends Cell Biol. 2001;11:75–82. doi: 10.1016/s0962-8924(00)01897-3. [DOI] [PubMed] [Google Scholar]
- 145.Vasiljeva O, Dolinar M, Pungercar JR, Turk V, Turk B. FEBS Lett. 2005;579:1285–1290. doi: 10.1016/j.febslet.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 146.Leveugle B, Ding W, Durkin JT, Mistretta S, Eisle J, Matic M, Siman R, Greenberg BD, Fillit HM. Neurochem Int. 1997;30:543–548. doi: 10.1016/s0197-0186(96)00103-9. [DOI] [PubMed] [Google Scholar]
- 147.Beckman M, Holsinger RM, Small DH. Biochemistry. 2006;45:6703–6714. doi: 10.1021/bi052498t. [DOI] [PubMed] [Google Scholar]
- 148.Rogelj B, Mitchell JC, Miller CC, McLoughlin DM. Brain Res Rev. 2006;52:305–315. doi: 10.1016/j.brainresrev.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 149.Xie J, Guo Q. J Biol Chem. 2005;280:13824–13832. doi: 10.1074/jbc.M411933200. [DOI] [PubMed] [Google Scholar]
- 150.Li Q, Sudhof TC. J Biol Chem. 2004;279:10542–10550. doi: 10.1074/jbc.M310001200. [DOI] [PubMed] [Google Scholar]
- 151.Pastorino L, Ikin AF, Lamprianou S, Vacaresse N, Revelli JP, Platt K, Paganetti P, Mathews PM, Harroch S, Buxbaum JD. Mol Cell Neurosci. 2004;25:642–649. doi: 10.1016/j.mcn.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 152.Li Y, Liu T, Peng Y, Yuan C, Guo A. J Neurobiol. 2004;61:343–358. doi: 10.1002/neu.20048. [DOI] [PubMed] [Google Scholar]
- 153.Hornsten A, Lieberthal J, Fadia S, Malins R, Ha L, Xu X, Daigle I, Markowitz M, O'Connor G, Plasterk R, Li C. Proc Natl Acad Sci U S A. 2007;104:1971–1976. doi: 10.1073/pnas.0603997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moechars D, Lorent K, De Strooper B, Dewachter I, Van Leuven F. Embo J. 1996;15:1265–1274. [PMC free article] [PubMed] [Google Scholar]
- 155.Fossgreen A, Bruckner B, Czech C, Masters CL, Beyreuther K, Paro R. Proc Natl Acad Sci U S A. 1998;95:13703–13708. doi: 10.1073/pnas.95.23.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lichtenthaler SF, Dominguez DI, Westmeyer GG, Reiss K, Haass C, Saftig P, De Strooper B, Seed B. J Biol Chem. 2003;278:48713–48719. doi: 10.1074/jbc.M303861200. [DOI] [PubMed] [Google Scholar]
- 157.Baisse B, Galisson F, Giraud S, Schapira M, Spertini O. BMC Evol Biol. 2007;7:166. doi: 10.1186/1471-2148-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kitazume S, Tachida Y, Oka R, Shirotani K, Saido TC, Hashimoto Y. Proc Natl Acad Sci U S A. 2001;98:13554–13559. doi: 10.1073/pnas.241509198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yoon IS, Pietrzik CU, Kang DE, Koo EH. J Biol Chem. 2005;280:20140–20147. doi: 10.1074/jbc.M413729200. [DOI] [PubMed] [Google Scholar]
- 160.Qiu S, Korwek KM, Weeber EJ. Neurobiol Learn Mem. 2006;85:16–29. doi: 10.1016/j.nlm.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 161.Mehta KD, Chen WJ, Goldstein JL, Brown MS. J Biol Chem. 1991;266:10406–10414. [PubMed] [Google Scholar]
- 162.Chen N, Pai S, Zhao Z, Mah A, Newbury R, Johnsen RC, Altun Z, Moerman DG, Baillie DL, Stein LD. Proc Natl Acad Sci U S A. 2005;102:146–151. doi: 10.1073/pnas.0408307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chabasse C, Bailly X, Sanchez S, Rousselot M, Zal F. J Mol Evol. 2006;63:365–374. doi: 10.1007/s00239-005-0198-9. [DOI] [PubMed] [Google Scholar]
- 164.Cheon HM, Shin SW, Bian G, Park JH, Raikhel AS. J Biol Chem. 2006;281:8426–8435. doi: 10.1074/jbc.M510957200. [DOI] [PubMed] [Google Scholar]
- 165.Wong HK, Sakurai T, Oyama F, Kaneko K, Wada K, Miyazaki H, Kurosawa M, De Strooper B, Saftig P, Nukina N. J Biol Chem. 2005;280:23009–23017. doi: 10.1074/jbc.M414648200. [DOI] [PubMed] [Google Scholar]
- 166.Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- 167.Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- 168.Sankaranarayanan S, Price EA, Wu G, Crouthamel MC, Shi XP, Tugusheva K, Tyler KX, Kahana J, Ellis J, Jin L, Steele T, Stachel SJ, Coburn C, Simon AJ. J Pharmacol Exp Ther. 2007 doi: 10.1124/jpet.107.130039. [DOI] [PubMed] [Google Scholar]
- 169.Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 170.Landgraf R, Fischer D, Eisenberg D. Protein Eng. 1999;12:943–951. doi: 10.1093/protein/12.11.943. [DOI] [PubMed] [Google Scholar]
- 171.Garlind A, Brauner A, Hojeberg B, Basun H, Schultzberg M. Brain Res. 1999;826:112–116. doi: 10.1016/s0006-8993(99)01092-6. [DOI] [PubMed] [Google Scholar]
- 172.Kuhn PH, Marjaux E, Imhof A, De Strooper B, Haass C, Lichtenthaler SF. J Biol Chem. 2007;282:11982–11995. doi: 10.1074/jbc.M700356200. [DOI] [PubMed] [Google Scholar]
- 173.Bowie A, O'Neill LA. J Leukoc Biol. 2000;67:508–514. doi: 10.1002/jlb.67.4.508. [DOI] [PubMed] [Google Scholar]
- 174.Fallon PG, Allen RL, Rich T. Trends Immunol. 2001;22:63–66. doi: 10.1016/s1471-4906(00)01800-7. [DOI] [PubMed] [Google Scholar]
- 175.Andrau D, Dumanchin-Njock C, Ayral E, Vizzavona J, Farzan M, Boisbrun M, Fulcrand P, Hernandez JF, Martinez J, Lefranc-Jullien S, Checler F. J Biol Chem. 2003;278:25859–25866. doi: 10.1074/jbc.M302622200. [DOI] [PubMed] [Google Scholar]
- 176.John V. Curr Top Med Chem. 2006;6:569–578. doi: 10.2174/156802606776743084. [DOI] [PubMed] [Google Scholar]
- 177.Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS. Nature. 2001;414:643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]
- 178.Laird FM, Cai H, Savonenko AV, Farah MH, He K, Melnikova T, Wen H, Chiang HC, Xu G, Koliatsos VE, Borchelt DR, Price DL, Lee HK, Wong PC. J Neurosci. 2005;25:11693–11709. doi: 10.1523/JNEUROSCI.2766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Garratt AN, Britsch S, Birchmeier C. Bioessays. 2000;22:987–996. doi: 10.1002/1521-1878(200011)22:11<987::AID-BIES5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 180.Lemke G. Sci STKE. 2006;2006:pe11. doi: 10.1126/stke.3252006pe11. [DOI] [PubMed] [Google Scholar]
- 181.Rockenstein E, Mante M, Alford M, Adame A, Crews L, Hashimoto M, Esposito L, Mucke L, Masliah E. J Biol Chem. 2005;280:32957–32967. doi: 10.1074/jbc.M507016200. [DOI] [PubMed] [Google Scholar]
- 182.Cellerino A, Carroll P, Thoenen H, Barde YA. Mol Cell Neurosci. 1997;9:397–408. doi: 10.1006/mcne.1997.0641. [DOI] [PubMed] [Google Scholar]
- 183.Padovani A, Pastorino L, Borroni B, Colciaghi F, Rozzini L, Monastero R, Perez J, Pettenati C, Mussi M, Parrinello G, Cottini E, Lenzi GL, Trabucchi M, Cattabeni F, Di Luca M. Neurology. 2001;57:2243–2248. doi: 10.1212/wnl.57.12.2243. [DOI] [PubMed] [Google Scholar]
- 184.Rao JK, Erickson JW, Wlodawer A. Biochemistry. 1991;30:4663–4671. doi: 10.1021/bi00233a005. [DOI] [PubMed] [Google Scholar]
- 185.Geier G, Banaj HJ, Heid H, Bini L, Pallini V, Zwilling R. Eur J Biochem. 1999;264:872–879. doi: 10.1046/j.1432-1327.1999.00679.x. [DOI] [PubMed] [Google Scholar]
- 186.Tang J, James MN, Hsu IN, Jenkins JA, Blundell TL. Nature. 1978;271:618–621. doi: 10.1038/271618a0. [DOI] [PubMed] [Google Scholar]
- 187.Holm I, Ollo R, Panthier JJ, Rougeon F. Embo J. 1984;3:557–562. doi: 10.1002/j.1460-2075.1984.tb01846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tang J, Lin X. Curr Opin Biotechnol. 1994;5:422–427. doi: 10.1016/0958-1669(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 189.Borrelli L, De Stasio R, Filosa S, Parisi E, Riggio M, Scudiero R, Trinchella F. Gene. 2006;368:101–109. doi: 10.1016/j.gene.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 190.Nadeau JH, Sankoff D. Genetics. 1997;147:1259–1266. doi: 10.1093/genetics/147.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lynch M, Force A. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Litman GW, Cannon JP, Dishaw LJ. Nat Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Patthy L. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 194.Jean L, Long M, Young J, Pery P, Tomley F. Trends Parasitol. 2001;17:491–498. doi: 10.1016/s1471-4922(01)02030-x. [DOI] [PubMed] [Google Scholar]
- 195.Tcherepanova I, Bhattacharyya L, Rubin CS, Freedman JH. J Biol Chem. 2000;275:26359–26369. doi: 10.1074/jbc.M000956200. [DOI] [PubMed] [Google Scholar]
- 196.Cho WL, Fu TF, Chiou JY, Chen CC. Insect Biochem Mol Biol. 1997;27:351–358. doi: 10.1016/s0965-1748(97)00017-9. [DOI] [PubMed] [Google Scholar]
- 197.Harrop SA, Prociv P, Brindley PJ. Biochem Biophys Res Commun. 1996;227:294–302. doi: 10.1006/bbrc.1996.1503. [DOI] [PubMed] [Google Scholar]
- 198.Becker MM, Harrop SA, Dalton JP, Kalinna BH, McManus DP, Brindley PJ. J Biol Chem. 1995;270:24496–24501. doi: 10.1074/jbc.270.41.24496. [DOI] [PubMed] [Google Scholar]
- 199.Syntichaki P, Xu K, Driscoll M, Tavernarakis N. Nature. 2002;419:939–944. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 200.Kotani N, Kitazume S, Kamimura K, Takeo S, Aigaki T, Nakato H, Hashimoto Y. J Biochem. 2005;137:315–322. doi: 10.1093/jb/mvi034. [DOI] [PubMed] [Google Scholar]
- 201.Marchionni M, Gilbert W. Cell. 1986;46:133–141. doi: 10.1016/0092-8674(86)90867-6. [DOI] [PubMed] [Google Scholar]
- 202.Kersanach R, Brinkmann H, Liaud MF, Zhang DX, Martin W, Cerff R. Nature. 1994;367:387–389. doi: 10.1038/367387a0. [DOI] [PubMed] [Google Scholar]
- 203.Logsdon JM., Jr Curr Opin Genet Dev. 1998;8:637–648. doi: 10.1016/s0959-437x(98)80031-2. [DOI] [PubMed] [Google Scholar]
- 204.Long M, de Souza SJ, Rosenberg C, Gilbert W. Proc Natl Acad Sci U S A. 1998;95:219–223. doi: 10.1073/pnas.95.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rogozin IB, Lyons-Weiler J, Koonin EV. Trends Genet. 2000;16:430–432. doi: 10.1016/s0168-9525(00)02096-5. [DOI] [PubMed] [Google Scholar]
- 206.Tudge C. Oxford University Press; 2000. [Google Scholar]