Abstract
Classically Y chromosomes are thought to originate from X chromosomes through a process of degeneration and gene loss. Now, the availability of 12 Drosophila genomes provides the opportunity to study the origin and evolution of Y chromosomes in an informative phylogenetic context. Surprisingly, the majority of Drosophila Y-linked genes are recent acquisitions from autosomes, and Y chromosome gene gains are more frequent than gene losses. Moreover, the D. pseudoobscura Y chromosome lacks homology with the Y of most Drosophila species. Thus the Drosophila Y has a different evolutionary history from canonical Y chromosomes (such as the mammalian Y), and it also might have a different origin.
Sex chromosome origins
Sex chromosome evolution provides a particularly coherent and satisfying blend of empirical data and theory. According to canonical theory, X and Y chromosomes (for the sake of simplicity we called the “W” chromosome of birds and butterflies as “Y” [1]) originate from an autosomal pair via a three-step process that begins with the acquisition of one or more strong sex-determining genes by one autosome, giving rise to nascent X and Y chromosomes. Natural selection then favors the suppression of recombination between the two chromosomes. The lack of recombination, together with the joint effects of mutation, natural selection, and genetic drift, then leads to progressive degeneration and loss of Y chromosome genes, until only the sex-determining gene(s) and a few relic genes survive [1–5]. As the X chromosome become progressively haploid in males (“hemizygous”), natural selection favors increased transcription of X-linked genes in males, through several dosage compensation mechanisms [1,2]. In the later stages, the Y usually becomes heterochromatic, accumulating large amounts of repetitive DNA. It also frequently acquires male-specific genes from the autosomes [6,7] (or female-specific genes in the case of the W chromosome, where ZW is female and ZZ is male). Empirical data in a variety of organisms including plants and birds support this scenario, the evidence being particularly clear in mammals. The most compelling evidence is provided by the observation that among the 27 different proteins encoded by the human Y, 18 have an ancestral, close counter-part on the X chromosome [6]. This generality, coupled with the beautiful fit between theory and data, has led to the widespread assumption that all Y chromosomes arise through this path (or by the related neo-Y pathway; Box 1), invariably being homologous to X chromosomes.
Box 1. Neo-Y chromosomes.
X0/XX systems are believed to arise from an XY system, by degeneration and loss of the Y [1]. A Y chromosome can be “regenerated” in these species when an autosome fuses to the X chromosome, and the new arrangement is fixed within a population (Fig. 1, panel B). During male meiosis the free homolog of the fused autosome continues to pair with the newly added portion of the X (termed “neo-X”) and move to the opposite pole of the cell, behaving as a Y chromosome (hence its name, “neo-Y”). Neo-Y chromosomes are male-restricted and the same evolutionary factors that cause the degeneration of Y chromosomes are expected to act on them. Empirical data confirm that they accumulate repetitive DNA and that their genes degenerate. Indeed Drosophila neo-Y systems are popular models to study Y-chromosome degeneration [12,48]. There are important differences between canonical Y and neo-Y evolution [38]; nonetheless, in both cases the Y originated through degeneration (of the X or the neo-X).
Fusions between the sex chromosomes (either the X or Y) and autosomes also occur in XY/XX species. D. pseudoobscura and its close relatives exemplify the former case: their X chromosomes stem from a fusion between the ancestral X and an autosome [1]. Following fixation of the X-autosome fusion the ancestral lineage must have passed through a “X/Y/neo-Y” stage, in which three chromosomes pair during male meiosis (the female meiosis is normal). Not surprisingly, such an event causes meiotic problems, and a similar system in another Drosophila species generates 1–3% of sex-chromosome aneuploidy [49]. This intermediate stage usually is short lived, and in a few million years only one Y chromosome remains; it is believed that the neo-Y is lost or fused to the ancestral Y [1]. However, the outcome was surprisingly different in the D. pseudoobscura lineage: the ancestral Y was incorporated into an autosome and was replaced by a new Y chromosome, possibly derived from the neo-Y [8].
Prior to 2000, an extensive identification of Y-linked genes had been done only in humans [7]. The genome sequencing of D. melanogaster in 2000, followed by D. pseudoobscura (2003) and now by 10 additional Drosophila genomes (2007) uncovered several unexpected phenomena, including the lack of X-Y shared genes, a wholesale replacement of the Y chromosome in the D. pseudoobscura lineage, and the preponderance of gene gains (in comparison to gene losses) in the 12 sequenced Drosophila species. Here we review these data and explore their consequences in the broader context of Diptera sex-chromosome evolution. We concentrate on single copy genes on the Drosophila Y, with an emphasis on results [8,9] appearing since the last review on this subject [10]. Readers interested on the role of repetitive DNA in the evolution of the Drosophila Y chromosome and on evolutionary models of Y-degeneration should consult references [4,11–13].
D. melanogaster Y chromosome: a lack of X-Y homology
The D. melanogaster Y chromosome is comprised mainly of repetitive DNA, and is heterochromatic. It does not determine sex; instead, in Drosophila sex determination is basically accomplished by a count of the number of X chromosomes [14,15]. However, males devoid of Y chromosomes (“X0 males”) are sterile [16], and formal genetic studies identified 6 genes that are essential for male fertility in the D. melanogaster Y [17–20]. Now, genome sequencing and the development of proper bioinformatics methods allows a thorough molecular identification of the Y chromosome gene content [21–23]. Despite its large size (~40 Mbp), it contains few single-copy protein-coding genes: 12 are currently known, and indirect evidence suggests an upper bound of ~20 genes [21–24]. These genes are unusually large, due to Mbp-sized introns composed of repetitive DNA [25]. D. melanogaster Y-linked genes have two additional important features: many (and probably all) have male-related functions (e.g., encoding sperm flagella motor proteins), and all arose by duplication from autosomal genes [9,21–24]. The initial step of their origin must have been a gene duplication that created a mutant Y carrying a copy of an autosomal gene. Then in some cases the mutant Y became fixed in the population, either by genetic drift or because it conferred a selective advantage to males that carried it. Given their redundancy, the autosomal copy could have pseudogenized and disappeared, the new Y-linked copy could have disappeared, or the two copies could have retained functionality, possibly by diverging to perform different functions [26]. Different Y-linked genes illustrate each of these steps. The flagrante delicto Y gene (FDY), which is present only in D. melanogaster (estimated age: ~2 Myr) is the youngest gene we identified. It arose from a genomic duplication of the autosomal gene CG11844 gene to the Y, and the flanking sequences remain clearly recognizable [27]. The autosomal and the Y-linked copies are still present and functional. By contrast, the WD40 Y gene (WDY) and the male fertility factor kl5 gene (kl-5) arrived to the Y chromosome ~50 Myr ago and in these cases the autosomal copies were lost [9,23]. However, the autosomal copy does not appear to be lost in all old transpositions: the Protein Phosphatase 1 Regulatory Subunit Y gene (Ppr-Y), for example, arrived on the Y chromosome more than 63 Myr ago, and this gene as well as the putative autosomal parent gene CG13125 have been retained [9,22]. The D. melanogaster Y chromosome does not share any single-copy genes with the X. This unusual feature is made clear when the D. melanogaster and human Y chromosomes are compared (Fig 1): the “X-ancestral” genes, which comprise the majority of Y-linked genes in mammals and are the hallmark of the X-Y shared ancestry, are absent in D. melanogaster Y. As Drosophila Y-linked genes came from the autosomes, one might expect some inter-specific variation, but at what time scale? Years ago we cautiously wrote that “the genic content of the Y chromosome would be fluid and may differ among (…) species [of the Drosophilidae family]” [22]. As described in the next section, when the 12 Drosophila genomes became available it was found that Y chromosome gene content differs even among species of the same group [8,9].
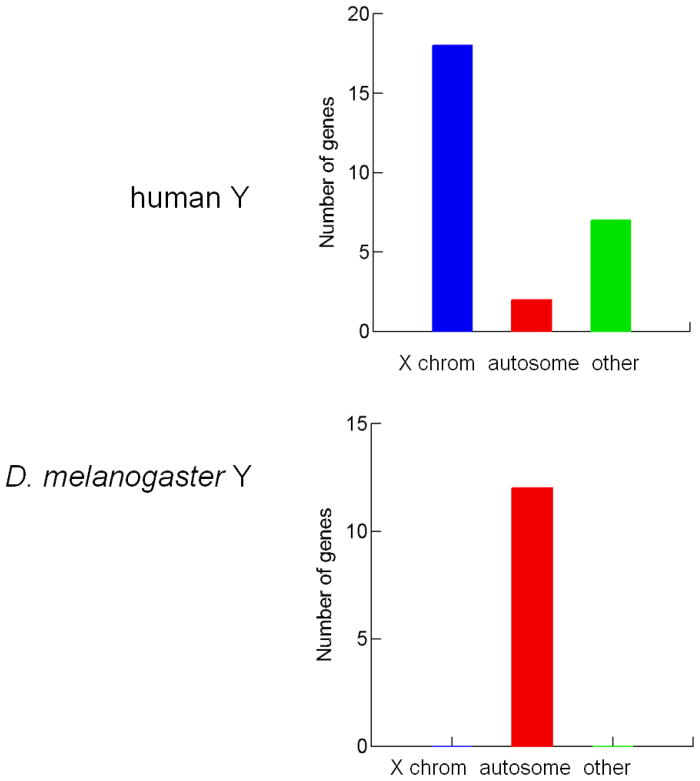
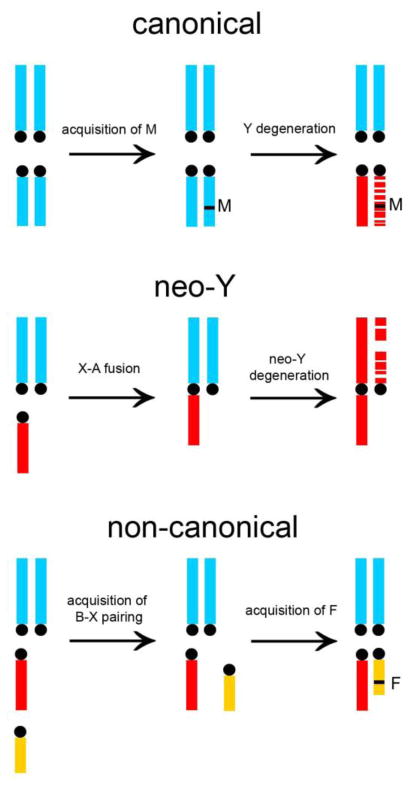
FIGURE 1. Origins of Y chromosomes.
Panel A. Y-linked genes in humans and D. melanogaster. Genes ancestrally shared with the X are shown in blue bars, and those acquired from autosomes, in red bars. Genes with unknown origin or that are later additions to both the X and the Y are grouped in the “other” class (green bar). Genes on the human Y chromosome encode 27 different proteins. The majority of these genes are ancestrally shared with the X-chromosome, indicating that these chromosomes are homologous [6,7]. This class of genes is absent among the 12 known single-copy genes in the D. melanogaster Y chromosome, suggesting that X and Y are not homologous [9]. Several human Y-linked genes are multi-copy; we counted them only once.
Panel B. Main paths for the origin of Y chromosomes. Only male karyotypes are shown (autosomes in blue, mature sex-chromosomes in red, and extra chromosomes such as a B chromosome in yellow). The canonical path (top) starts when an autosome acquires a strong male determining gene M, becoming a nascent Y (its homologous became a nascent X). Degeneration of the nascent Y and the evolution of dosage compensation in the X originate mature sex chromosomes [1–5]. The neo-Y path (middle; shown here in a species with X0/XX sex-determination) starts with a X-autosome fusion, transforming the fused autosome into a neo-X. The free homolog became a neo-Y, which then degenerates [1]. The non-canonical path (bottom) starts when a parasitic B chromosome (which usually do not pair) acquires the capacity to pair with the X. Improvement of B-X pairing and the acquisition of male-fertility genes (F) originate a chromosome that will be termed as Y [10,34–36]. However, in contrast with the two other paths, these non-canonical Y chromosomes do not share any homologous region with the X, and is not formed by degeneration. Note that only canonical Y chromosomes are expected to have male-determining function (imparted by the M gene), and that the neo-Y and non-canonical paths can be distinguished because the former reduces the number of autosomes [1].
The 12 Drosophila Y chromosomes: a prominent role of gene gains
Before examining the results of Koerich et al. [9], it is worth mentioning a caveat. Ideally one should have the complete gene set of the Y chromosomes in the 12 species before starting the comparative analysis, as has been done (at least approximately) with the euchromatic portion of the other chromosomes. However, given the notorious difficulties in sequencing and assembling heterochromatic regions and the Y [27,28], Koerich et al. [9] investigated Y-linkage in the Drosophila orthologs of the D. melanogaster Y-linked genes. The ensuing ascertainment bias should be corrected when estimating gene gain and loss rates [9]. The special case of two species (D. pseudoobscura and the closely related D. persimilis) will be discussed in the next section, because the changes in their Y-chromosome gene content were caused by a wholesale replacement of the Y, rather than by individual gains and losses of genes [8].
In many cases the orthologs of the D. melanogaster Y-linked genes were autosomal in other species. This change in chromosomal location can be explained either by an autosome to Y transposition in the D. melanogaster lineage (i.e., a gene gain in the Y) or by a Y to autosome transposition in the other lineage (i.e., a gene loss in the Y). Additional data based on synteny, however, provided unambiguous answers (Fig 2). Among the 12 known D. melanogaster Y-linked genes, 7 were acquired by the Y in the D. melanogaster lineage less than 63 Myr ago [9]. This pattern contrasts sharply with the rest of the genome, where ~95% of the genes remained on the same chromosome arm across the 12 species [29]. However, the absolute number of gene movements is similar in the Y and in the other chromosomes. So why is the Y-linked gene content so poorly conserved? The most likely explanation is that the other chromosomes had thousands of genes in the ancestor of the 12 sequenced species, whereas the Y chromosome had a very low number of genes (we know of five: male fertility factor kl2 (kl-2), male fertility factor kl3 (kl-3), Ppr-Y, Polycystine-Related-Y (PRY) and Occludin-Related-Y (ORY); Fig. 2). This condition, coupled with a small and similar number of gene movements in the Y and the other chromosomes, would produce the present pattern of low conservation of gene content in the Y chromosome and high conservation in the other chromosomes. This pattern dates back to mosquitoes, which diverged from Drosophila ~260 Myr ago: although there are clear homologies in other chromosomes between these Diptera [30,31], all orthologs of D. melanogaster Y-linked genes are autosomal in mosquitoes. Thus, the gene content of the Drosophila Y is younger than the other chromosomes.
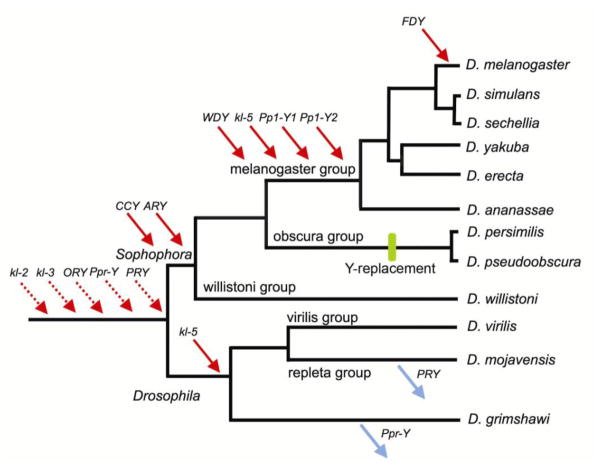
FIGURE 2.
Gene movements in the Drosophila Y chromosome. Gene gains by the Y chromosome are marked with red arrows and gene losses by blue arrows. The genes assigned with dashed red arrows are probable gains, which could not yet be confirmed due to lack of close outgroups [9]. The green bar in the D. pseudoobscura/D. persimilis lineage marks the incorporation of the ancestral Y into an autosome, and its replacement by a new Y chromosome [8]. Due to the experimental design, gene gains basically can only be detected in the D. melanogaster lineage, and gene losses only in the other lineages. After correction of this ascertainment bias it was found that the rate of gene gain is 11-fold higher than the rate of gene loss [9]. Note that the majority of D. melanogaster Y-linked genes (7 out 12) were acquired after the split between the Drosophila and Sophophora subgenera, which happened ~ 63 Myr ago [64]. Modified from ref [9], with permission.
Comparisons between gene gains and losses also yield interesting clues about the evolution of the Drosophila Y: the rate of gene gain is ~11 times higher than the rate of gene loss (P = 0.003; 95% confidence interval: 2.3–52.5; ref [9]). Thus, the Drosophila Y shows another unusual pattern (for a Y-chromosome): gene gains play a prominent role in its evolution, and at least in the last 63 Myr its gene content has indeed increased. This finding also suggests that the Drosophila Y is relatively young, because it is gaining genes and yet has few of them.
The D. pseudoobscura case: wholesale replacement of the Y chromosome
Given the phylogenetic proximity between D. melanogaster and D. pseudoobscura (they belong to the sister groups melanogaster and obscura) we expected their Y chromosomes to be very similar. Instead, they have nothing in common! The ancestral Drosophila Y chromosome became part of an autosome in the D. pseudoobscura lineage and was replaced by a new Y chromosome, possibly derived from a neo-Y [8]. This new Y chromosome is essential for male fertility [32], so it presumably acquired one or more fertility genes from other chromosomes, by the process outlined above for D. melanogaster (alternatively, some mechanical problem in meiosis might cause sterility in X0 males). We identified 15 genes and pseudogenes in the D. pseudoobscura Y chromosome, and none are shared with the D. melanogaster Y [8]. Hence, despite their functional and cytogenetic similarities (both are required for male fertility, pair with the X, and are heterochromatic), these Y chromosomes are not homologous. This unexpected finding has obvious implications for the origin and evolution of Y chromosomes, and raises the question of why it happened. A possible explanation follows. The incorporation of the ancestral Y into an autosome occurred only in species such as D. pseudoobscura and D. affinis which also have a known X-autosome fusion, whereas the more distantly related D. bifasciata and D. guanche (which do not have the X-autosome fusion) still carry the ancestral Y [8]. As detailed in Box 1, this association provides a clue: X-autosome fusions cause meiotic problems because three chromosomes (the ancestral Y, the X and the neo-Y) have to pair. Hence, the incorporation of the ancestral Y into an autosome may have been advantageous because it solved the meiotic problems caused by the X-autosome fusion.
For a long time, it was thought that the D. pseudoobscura Y chromosome was the ancestral Y, and that the neo-Y was lost or was incorporated into the ancestral Y [1]. Instead, the genomic data showed that the Y became part of an autosome. The data also suggest a possible origin for the current Y: it might be a degenerated neo-Y. The finding that 10 of the 15 known genes and pseudogenes have homologs on the neo-X (the autosome that became fused to the ancestral X) seems at first sight to confirm this hypothesis; but actually the evidence is at best weak, for two reasons. First, the 15 genes do not represent independent events because most of them are adjacent to each other in their original locations. They came from five different locations, and among these five, only two are homologous to the neo-X [8]. Hence the proper evidence in favor of a neo-Y origin of the D. pseudoobscura Y is not 10 out 15, but rather two out five. Second, and more importantly, if these 15 Y-linked genes are part of the neo-Y chromosome originated by the X-autosome fusion, at least some should be present in the Y chromosome of other species that share the same X-autosome fusion. However, in preliminary experiments done in two labs (we and Carlos Machado’s), the presence of none of the 15 genes could be confirmed by PCR-amplification even in the closely related D. persimilis. This failure, coupled with the high nucleotide identity (typically above 97%) between these 15 Y-linked genes and their autosomal paralogs, strongly suggest that they are recent acquisitions that occurred after the X-autosome fusion. The bottom line is that the neo-Y hypothesis provides a simple explanation for the origin of the current Y, but the question remains unsolved. Identification of more Y-linked genes, as well as identification of the pairing regions between the X and Y in D. pseudoobscura may provide an answer. But the data in hand already show that Drosophila Y chromosomes can be very labile; indeed, chromosomes that appear to be identical (e.g., those of D. melanogaster and D. pseudoobscura) can be non-homologous. These new results also hint that X-chromosome pairing and a determining role in male-fertility, characteristics that define a Drosophila Y chromosome, can evolve rapidly (the current D. pseudoobscura Y originated between 2 to 18 Myr ago; [8]) and provide little information about the origin of the chromosome itself.
How frequent is the phenomenon of wholesale replacement of the Y? Does it occur in every case of X-autosome fusion? Is it restricted to species with X-autosome fusion, as required by the “neo-Y origin” hypothesis? These three questions are amenable to direct experimental investigation, and already the results described in the previous section answered one of them. In particular, D. willistoni, which has undergone an independent X-autosome fusion, carries the ancestral Y chromosome [9]; therefore Y-autosome fusions do not necessarily follow X-autosome fusions. The remaining two questions are being addressed by an ongoing study of a large number Drosophila species. Preliminary data suggest that among the 310 tested species, 42 had their Y chromosomes replaced, owing to four independent events (the D. pseudoobscura lineage case, and three additional lineages). None of the three additional events (amounting to 34 species) has a X-autosome fusion, so the current Y of these species cannot be a degenerated neo-Y. Instead, they must have a different origin.
An alternative hypothesis for the origin of the Drosophila Y
Given the many similarities between the Drosophila Y and other Y chromosomes (X-Y pairing, low gene density, heterochromatic state), and the seemingly universal validity of sex-chromosome evolution theory, it is easy to understand why the Drosophila Y is generally thought to have originated through the same canonical path. However, researchers have repeatedly uncovered contradictory evidence while studying this chromosome in detail. The Drosophila Y lacks a sex-determining gene [16], which, according to the canonical model, should be present. Furthermore Brosseau [17] (see also [33]) commented that “the Y has no genetic regions homologous to most of the X (…). This view is diametrically opposed to the notion that the Y is a degenerate X.” Finally, the recent genomic data [8, 9, 21–23] reviewed in the previous sections also do not fit well with the canonical model. Each of these discrepancies can be accommodated by an ad hoc assumption: the lack of X-Y shared genes can be explained by the assumption that all signs of X-Y homology were erased. The lack of a male sex-determining gene can be explained by the assumption that the degeneration erased even this gene (or by the Drosophila Y being a neo-Y; but see below). The odd D. pseudoobscura data could be rationalized by the assumption that its current Y is a neo-Y (although this explanation ignores the unexpected wholesale replacement of the Y). To explain the prominent role of gene gains across the 12 species, one could assume that the Drosophila Y arose from the degeneration of the X chromosome, and hence only more recently have gene gains become important. Although these arguments cannot be completely excluded as a possibility, it might be time to consider alternative hypotheses that generate new experimental questions rather than ad hoc explanations. In fact, these discrepancies can be explained parsimoniously by the hypothesis that the Drosophila Y chromosome is not a degenerated X, but rather that it originated from the addition of an extra chromosome (that evolved the ability to pair with the X) to an XX/X0 sex-chromosome system (Fig 1, panel B; [10,34]). Although this path might seem strange, it happened independently in two Homoptera species [35,36], whose Y chromosomes originated from “B chromosomes” that evolved accurate pairing with the X-chromosome (Box 2). B chromosomes are supernumerary dispensable chromosomes that are present in many species [1,37].
Box 2. Y chromosomes that originated from B chromosomes.
Nokkala and co-workers showed that the Y chromosomes of two Homoptera species (Rhinocola aceris and Cacopsylla peregrina) originated from “B chromosomes” [35,36], supernumerary dispensable chromosomes that are present in many species [1,37]. B chromosomes frequently have detrimental fitness effects, but, as in other cases of selfish genetic elements, their elimination from populations is prevented by meiotic drive. In some cases they evolve another mechanism that ensures their maintenance in populations: the ability to pair with X chromosomes in X0/XX species [37]. B chromosomes typically are heterochromatic and occur in variable numbers, both within and among populations; in the same population, individuals might carry 0, 1, 2, 3 or more copies. This characteristic provides the strongest evidence that the R. aceris Y originated from a B chromosome: in one population males and females carry a variable number of heterochromatic chromosomes which pair imperfectly with the X in males (making it clear that they are typical B chromosomes), whereas in other populations, a similar chromosome is present only in males, in a single copy, and nearly perfectly pairs with the X (like a bona fide Y chromosome). The foreknowledge that the basic sex chromosome system is X0/XX in the family Psylloidea [50], together with the maintenance of the number of autosomes in R. aceris, ruled out the possibility of a neo-Y origin. The lack of chiasmata between the X and Y of the two species (which are present in other Psylloidea species with neo-Y) provided additional evidence that their Y chromosomes originated from B chromosomes [35,36].
As B chromosomes are rare, (although not absent) in Drosophila [51], perhaps a more likely source of a non-canonical Y chromosome is the small “dot” chromosome (D. melanogaster chromosome 4; ref [34]). Indeed, trisomy for the dot chromosome is well tolerated [52], and it pairs with the X [53,54]. Of course the suggestion of a non-canonical origin of the Drosophila Y does not imply that the canonical evolutionary mechanism could not operate in Drosophila: neo-Y chromosomes are known to degenerate in these species too (e.g., [12,48]). Therefore, the important question is whether or not the canonical model provides the most likely explanation, given the available evidence.
How common are these non-canonical Y chromosomes? It is common practice to term as Y any chromosome which pairs with the X and is present only in males; moreover, it is implicitly assumed to be a degenerated X, even if it does not play a role in sex-determination (e.g., Drosophila). Thus, almost by definition non-canonical Y chromosomes will be overlooked. The unconventional origin of the Y chromosome of the Homoptera Rhinocola aceris was identified owing to the combination of incomplete fixation and high confidence of the ancestral state [35], which most likely reflects the fortuitous observation of a recent event. After the R. aceris study, a careful investigation of Cacopsylla peregrina revealed the same phenomenon [36]. It is likely that additional cases will be found, as researchers become aware of the possibility of non-canonical origin of Y-chromosomes, and tsetse flies, whose Y-chromosome has clear similarities with B chromosomes, may be an example (Box 3). As the XX/X0 sex-chromosome system is widespread among insects, and is almost certainly the ancestral system in several orders [38], there might be many additional cases of Y chromosomes arising from B chromosomes. Furthermore, although it is easier to envisage the origin of non-canonical Y chromosomes in species with X0/XX sex-chromosomes, B chromosomes are also able to pair with the sex-chromosomes in XY/XX species [39].
Box 3. The Y chromosome of tsetse flies.
Tsetse flies (genus Glossina) are considered to have XY/XX sex-chromosome system, yet their Y chromosome is not involved in sex-determination and shows irregular segregation with the X [55]. Indeed, aneuploidy is common in natural populations (e.g., X0, XY and XYY males co-exist [55]).
Furthermore, the Y seems to cause meiotic-drive in XXY females, as they produce twice as many XY as X eggs (Table 14.1 in [56]). Variable numbers and meiotic-drive are hallmarks of B chromosomes [1,37], and if not for the sterility of X0 males [56], the Glossina Y would perfectly fit the definition of a B chromosome. Indeed Glossina is known to have B chromosomes [57], and the close similarity between their B and Y chromosomes in meiotic behavior and C-band pattern led Amos and Dover [58] to propose that their B chromosomes originated from the Y (but note that the opposite relationship – Y originated from B - additionally explains the irregular X-Y segregation). The Glossina genome currently is being sequenced. The genomic data, together with studies aimed at uncovering the ancestral state of sex-chromosomes in related families, might show whether or not the Glossina Y chromosome actually is a specialized B-chromosome that acquired male-fertility genes.
Will direct testing of the “non-canonical origin” hypothesis be possible in Drosophila? The Homoptera examples described above might provide some guidance. These cases were uncovered using cytogenetics (optical microscopy of chromosomes), and analogous work in the Drosophilidae sister families (e.g., Ephydridae) is badly needed (Box 4). Interestingly, the only Ephydridae species investigated so far has X0/XX sex-chromosomes [40]. As seen in R. aceris, uncovering recent events is critical as the mechanisms of Y origination have not yet been blurred by additional chromosomal changes. This forms part of the rationale for studying Y-linked gene content in ~300 Drosophila species; confirmation of the suspected cases of non-canonical Y origin in several Drosophila lineages would support a similar origin for the ancestral Drosophila Y. Finally, as the cost of genome sequencing continues to fall, it is certain that many Diptera genome sequences will become available during the next decade, shedding light on the origin and evolution of their sex chromosomes. For example, species in which the Y chromosome originated from a neo-Y should have a large block of additional genes (derived from the fused autosome) on the X chromosome, when compared to proper outgroups, even if the fusion is ancient. Some glimpses of this approach are already possible.
Box 4. Cytogenetics and the origin of the Drosophila Y.
Chromosome studies have provided important evidence for the origin of Y chromosomes in many species, and they are especially effective when the ancestral state, i.e. before the origin of the Y, is known [1,35,36]. The majority of Drosophila species have an XY/XX sex-chromosome system [59], so we must infer the ancestral state by looking outside the genus. The best outgroups for this purpose are the Steganinae subfamily (which belongs to the same Drosophilidae family) and sister families such as Ephydridae and Curtonotidae. Little is known about their chromosomes, and the limited data from more distant Acalyptrata families are contradictory, with one study suggesting that the Drosophilidae karyotype is highly derived [60], and another suggesting that it is ancestral [61]. An investigation of the karyotype of outgroups such as Ephydridae is clearly needed. This investigation should at least eliminate one mechanism from the list of possible origins of the Drosophila Y (X-degeneration, neo-Y, and non-canonical). For example, a X0/XX ancestral state would rule out the X-degeneration hypothesis. Neo-Y formation by X-autosome fusions can be detected by the reduction in the number of autosomes and (in many cases) by the transformation of a single-armed into a two-armed X [1]. The application of these ideas to the study of the origin of the Drosophila Y is hampered by the lack of data from the outgroups. The proposed cytogenetic approach also has some caveats. Neo-Y detection is straightforward in evolutionarily recent events with known ancestral state [1,50,62], but the Drosophila Y originated more than 63 Myr ago [9]; pending on the rate of chromosomal evolution, further changes that alter chromosome number and structure (such as chromosome fusions, fissions, and inversions) might have blurred the signs of the origin of the Y. Another limitation is that Y chromosomes that are non-homologous (e.g., D. melanogaster and D. pseudoobscura; ref [8]) can look nearly identical when using simple cytogenetics methods (e.g., orcein staining). However, the use of more elaborate cytogenetic methods, such as fluorescence in-situ hybridization (FISH) and chromosome painting (e.g., [63]), can alleviate these concerns.
Mosquito (Anopheles and Aedes) and Drosophila X chromosomes are homologous [30,31]; therefore the genesis of the X (as well as presumably of a canonical Y) occurred more than 260 Myr ago, and X chromosome identity is conserved across very distant Diptera families. This ancestral X is the sole major contributor to the Drosophila X [30,31], with no evidence of X-autosome fusions, which strongly suggests that the ancestral Drosophila Y is not a neo-Y. By contrast, half of the Aedes X chromosome is homologous to the Drosophila and Anopheles X, but the other half is homologous to a single autosome in each of these species [30], presumably due to a X-autosome fusion in the Aedes lineage. However, the Aedes Y is not a neo-Y: it is nearly identical to the X (except for the presence of the male-determining gene [41]), so additional changes happened. In contrast with mosquitoes, the X-chromosomes of the much more closely related families Tephritidae (Ceratitis capitata) and Muscidae (Musca domestica) are not homologous to the Drosophila X: they are completely heterochromatic, carry very few genes, and the genes orthologous to Drosophila X-linked genes that have been mapped are autosomal in these species [42,43]. Thus, in some Diptera families the X chromosome likely was replaced; such a possibility creates an interesting problem with the pre-existing dosage compensation mechanism. Namely, the former X chromosome (now an autosome) is present in equal dose in males and females in these Diptera families and hence its hyper-transcription in males should had been turned off [44]. Besides Muscidae and Tephritidae, this prediction may be tested in Aedes, which seems to have evolved to a system with very little differentiation between the X and the Y [41].
Concluding remarks
What is a Y chromosome? A good definition is that it is a chromosome that regularly segregates from the X chromosomes, irrespective of its origin. Many Y chromosomes will be degenerated Xs or neo-Ys, and a currently unknown number (particularly among insects) will be traced to a non-canonical origin. The canonical theory of sex-chromosome evolution, with its emphasis on X-Y homology, slow degeneration and gene loss, provides an incomplete conceptual framework to study the Drosophila Y chromosome, in which there is no sign of X-Y homology, several cases of wholesale replacement appear to have occurred, and gene gains play a prominent role. By considering the possibility of a non-canonical origin, the focus shifts to questions such as the origin of the Drosophila Y, its evolutionary lability, the sex-chromosomes of Ephydridae and other Diptera families, and to the close relationship between Y and B chromosomes in some species. Throughout this review we have emphasized that these questions are amenable to experimental investigation, and therefore will be answered eventually. As the Glossina case suggests, such a shift in focus might illuminate Y chromosome evolution far beyond Drosophila.
On the experimental side, we cannot emphasize enough the importance of an exhaustive identification of Y-linked genes. This is the main obstacle to the study of the Y chromosome because its richness of repetitive DNA precludes sequence assembly into large and easily studied scaffolds. Instead short Y-linked scaffolds (usually between 1 kb and 100 kb) must be individually identified among thousands of unmapped scaffolds that came from other heterochromatic regions, using computational methods followed by experimental verification of Y-linkage [8,21–23]. The experimental verification is necessary due to the rather high rate of false-positives and false-negatives of the current computational methods; this step is labor intensive (when applied to hundreds of scaffolds), and in practice is done only in the scaffolds that seem to encode genes [8,21–23]. Two recent developments will make this individual experimental verification unnecessary, and hence are likely to have a deep impact on the knowledge of Y chromosomes. We developed a method based on short-read sequences that allows massive identification of Y-linked sequences [45]. This method is suited for genomes that have already been sequenced using male-female mixed libraries (e.g., [46]). However, as proposed by Krzywinski et al. [47], the simple separation of male and female libraries before sequencing provides the most straightforward and powerful method to identify Y-linked sequences, at nearly zero additional cost. Currently several Anopheles genomes and the hemipteran Rhodnius prolixus are being sequenced in this way. We hope that this will become standard practice for future genome projects.
Acknowledgments
We thank B. Lemos, S. Nokkala, J. Lucchesi, Y. Tao, R. Arguello, and two anonymous reviewers for many valuable comments in the manuscript. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq, Coordenação de Aperfeiçoamento do Pessoal de Ensino Superior-CAPES, FAPERJ, FIC-NIH grant TW007604-02 (A.B.C.) and NIH grant GM64590 (A.G.C. and A.B.C.).
References
- 1.White MJD. Animal cytology and evolution. University Press; 1973. [Google Scholar]
- 2.Bull JJ. Evolution of sex determining mechanisms. Benjamin/Cummings Pub. Co., Advanced Book Program; 1983. [Google Scholar]
- 3.Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343. [Google Scholar]
- 4.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 6.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 7.Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho AB, Clark AG. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science. 2005;307:108–110. doi: 10.1126/science.1101675. [DOI] [PubMed] [Google Scholar]
- 9.Koerich LB, et al. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho AB. Origin and evolution of the Drosophila Y chromosome. Current Opinion in Genetics & Development. 2002;12:664–668. doi: 10.1016/s0959-437x(02)00356-8. [DOI] [PubMed] [Google Scholar]
- 11.Gvozdev VA, et al. The Y chromosome as a target for acquired and amplified genetic material in evolution. Bioessays. 2005;27:1256–1262. doi: 10.1002/bies.20321. [DOI] [PubMed] [Google Scholar]
- 12.Bachtrog D, et al. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemos B, et al. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 14.Cline TW. The Drosophila sex determination signal: how do flies count to two? Trends in Genetics. 1993;9:385–390. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- 15.Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biology. 2007;5:e332. doi: 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bridges CB. Non-disjunction as proof of the chromosome theory of heredity. Genetics. 1916;1:1–52. doi: 10.1093/genetics/1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brosseau GE. Genetic analysis of the male fertility factors on the Y chromosome of Drosophila melanogaster. Genetics. 1960;45:257–274. doi: 10.1093/genetics/45.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennison JA. The genetic and cytological organization of the Y chromosome of Drosophila melanogaster. Genetics. 1981;98:529–548. doi: 10.1093/genetics/98.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazelrigg T, et al. A cytogenetic analysis of X- ray induced male steriles on the Y chromosome of Drosophila melanogaster. Chromosoma. 1982;87:535–559. [Google Scholar]
- 20.Gatti M, Pimpinelli S. Cytological and genetic-analysis of the Y-chromosome of Drosophila melanogaster. 1 Organization of the fertility factors. Chromosoma. 1983;88:349–373. [Google Scholar]
- 21.Carvalho AB, et al. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci U S A. 2000;97:13239–13244. doi: 10.1073/pnas.230438397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho AB, et al. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:13225–13230. doi: 10.1073/pnas.231484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vibranovski MD, et al. Two new Y-linked genes in Drosophila melanogaster. Genetics. 2008;179:2325–2327. doi: 10.1534/genetics.108.086819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gepner J, Hays TS. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci U S A. 1993;90:11132–11136. doi: 10.1073/pnas.90.23.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reugels AM, et al. Mega-introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant threads loops in primary spermatocytes of Drosophila hydei. Genetics. 2000;154:759–769. doi: 10.1093/genetics/154.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends in Genetics. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho AB, Clark AG. Birth of a new gene on the Drosophila Y chromosome. Abstracts of the 44th Annual Drosophila Research Conference; 2003. p. 113. [Google Scholar]
- 28.Hoskins RA, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhutkar A, et al. Genome-scale analysis of positionally relocated genes. Genome Research. 2007;17:1880–1887. doi: 10.1101/gr.7062307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zdobnov EM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 32.Dobzhansky T. Further data on the variation of the Y chromosome in Drosophila pseudoobscura. Genetics. 1937;22:340–346. doi: 10.1093/genetics/22.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohe AR, et al. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackstein JH, et al. Is the Y chromosome of Drosophila an evolved supernumerary chromosome? Bioessays. 1996;18:317–323. doi: 10.1002/bies.950180410. [DOI] [PubMed] [Google Scholar]
- 35.Nokkala S, et al. Achiasmate segregation of a B chromosome from the X chromosome in two species of psyllids (Psylloidea, Homoptera) Genetica. 2000;108:181–189. doi: 10.1023/a:1004146118610. [DOI] [PubMed] [Google Scholar]
- 36.Nokkala S, et al. The origin of the achiasmatic XY sex chromosome system in Cacopsylla peregrina (Frst. ) (Psylloidea, Homoptera) Genetica. 2003;119:327–332. doi: 10.1023/b:gene.0000003757.27521.4d. [DOI] [PubMed] [Google Scholar]
- 37.Camacho JP. Preface. Cytogenetic and Genome Research. 2004;106:147–148. doi: 10.1159/000079308. [DOI] [PubMed] [Google Scholar]
- 38.Blackman RL. Sex determination in insects. In: Leather SR, Hardie J, editors. Insect reproduction. CRC press; 1995. [Google Scholar]
- 39.Nokkala S. The meiotic behaviour of B-chromosomes and their effect on the segregation of sex chromosomes in males of Hemerobius marginatus. Hereditas. 1986;105:221–227. [Google Scholar]
- 40.Heitz E. Die somatische Heteropyknose bei Drosophila melanogaster und ihre genetische Bedeutung. Cell and Tissue Research. 1933;20:237–287. [Google Scholar]
- 41.Clements AN. The Biology of Mosquitoes. Chapman & Hall; 1992. [Google Scholar]
- 42.Hiroyoshi T. Some new mutants and the revised linkage maps of the housefly, Musca domestica Japan. J Gen. 1977;52:275–288. [Google Scholar]
- 43.Stratikopoulos EE, et al. An integrated genetic and cytogenetic map for the Mediterranean fruit fly, Ceratitis capitata, based on microsatellite and morphological markers. Genetica. 2008;133:147–157. doi: 10.1007/s10709-007-9195-9. [DOI] [PubMed] [Google Scholar]
- 44.Larsson J, Meller VH. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 2006;14:417–431. doi: 10.1007/s10577-006-1064-3. [DOI] [PubMed] [Google Scholar]
- 45.Carvalho AB, Clark AG. Efficient identification of Drosophila Y-chromosome sequences by short read sequencing. 49th Annual Drosophila Research Conference; 2008. p. 124. [Google Scholar]
- 46.Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 47.Krzywinski J, et al. Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics. 2004;166:1291–1302. doi: 10.1534/genetics.166.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinemann M, et al. How Y chromosomes become genetically inert. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5737–5741. doi: 10.1073/pnas.90.12.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper KW. The mechanism of non-random segregation of sex chromosomes in male Drosophila miranda. Genetics. 1946;31:181–194. doi: 10.1093/genetics/31.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maryanska-Nadachowska A. A review of karyotype variation in jumping plant-lice (Psylloidea, Sternorrhyncha, Hemiptera) and checklist of chromosome numbers. Folia Biologica. 2002;50:135–152. [PubMed] [Google Scholar]
- 51.Ramachandra NB, Ranganath HA. Suprenumerary chromosomes in Drosophila nasuta albomicans. Experientia. 1985;41:680–681. doi: 10.1007/BF02007717. [DOI] [PubMed] [Google Scholar]
- 52.Lindsley DL, Grell EH. Genetic variations of Drosophila melanogaster. Carnegie Institute; 1972. [Google Scholar]
- 53.Hughes SE, et al. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genetics. 2009;5:e1000348. doi: 10.1371/journal.pgen.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandler L, Novitski E. Evidence for genetic homology between chromosomes I and IV in Drosophila melanogaster, with a proposed explanation for the crowding effect in triploids. Genetics. 1956;41:189–193. doi: 10.1093/genetics/41.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maudlin I. Chromosome polymorphism and sex determination in a wild population of tsetse. Nature. 1979;277:300–301. doi: 10.1038/277300a0. [DOI] [PubMed] [Google Scholar]
- 56.Southern DI. Chromosome diversity in Tsetse flies. In: Blackman RL, et al., editors. Insect cytogenetics. Blackwell Scientific Publications; 1980. [Google Scholar]
- 57.Southern DI, Pell PE. Chromosome relationships and meiotic mechanisms of certain morsitans group tsetse flies and their hybrids. Chromosoma. 1973;44:319–334. [Google Scholar]
- 58.Amos A, Dover G. The distribution of repetitive DNAs between regular and supernumerary chromosomes in species of Glossina (Tsetse): a two-step process in the origin of supernumeraries. Chromosoma. 1981;81:673–690. doi: 10.1007/BF00329579. [DOI] [PubMed] [Google Scholar]
- 59.Clayton FE. Published karyotypes of the Drosophilidae. Drosophila Information Service. 1998;81:5–125. [Google Scholar]
- 60.Boves JW, et al. Chromosomes of Richardiidae, Otitidae and Platystomatidae (Diptera: Acalyptratae) Genetica. 1973;44:553–571. [Google Scholar]
- 61.Block K. Chromosomal variation in Agromyzidae (Diptera). VI Comparative chromosome studies. Hereditas. 1976;84:177–212. [Google Scholar]
- 62.Patterson JT, Stone WS. Evolution in the genus Drosophila. The MacMillan Company; 1952. [Google Scholar]
- 63.Rens W, et al. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16257–16261. doi: 10.1073/pnas.0405702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura K, et al. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]