Abstract
Many actin-binding proteins contain calponin homology (CH) domains, but the manner in which these domains interact with F-actin has been controversial. Crystal structures have shown the tandem CH domains of α-actinin to be in a compact, closed conformation, but the interpretations of complexes of such tandem CH domains with F-actin have been ambiguous. We show that the tandem CH domains of α-actinin bind to F-actin in an open conformation, explaining mutations that cause human diseases, and suggesting that the opening of these domains may be one of the main regulatory mechanisms for proteins with tandem CH domains.
Many actin-binding proteins have a modular architecture, and the calponin homology (CH) domain is found in some actin-binding proteins in one, two or four copies. But the manner in which these domains interact with F-actin has been controversial1, 2. α-Actinin is an actin filament crosslinking protein that was originally found in skeletal muscle. It is now known that α-actinin plays a crucial role in many cell types in forming the actin cytoskeleton and is involved in cell-cell and cell-matrix interactions, lamellipodia and stress fibers, among other cellular sites. Not surprisingly, mutations in α-actinin have been linked to a number of human diseases3. The actin-binding domain (ABD) of α-actinin contains two CH domains. Multiple crystal structures4, 5 have shown these CH domains to be in a compact, closed conformation (where there is a substantial interface between the two domains), as have crystal structures of tandem CH domains from other proteins such as utrophin6, dystrophin7 and filamin8, although in some of these crystals the compact conformation is maintained by domain swapping of CH1 and CH2 between two different extended molecules. Solution studies4 have shown that the CH domains of α-actinin adopt a closed conformation independent of crystal packing. An electron microscopy (EM) study at 20 Å resolution of two-dimensional crystals of α-actinin revealed that both closed and open (where the two CH domains are substantially separated) tandem CH domains were present9. In complexes of tandem CH domains with F-actin, however, most EM reconstructions have suffered from limited resolution, and there has been a debate as to whether the density bound to actin arises from CH domains in an open or closed conformation1, 2, 5. A three-dimensional reconstruction of the α-actinin tandem CH domains bound to F-actin was made10 before atomic structures were available for any CH domain. That low-resolution reconstruction has been subsequently interpreted as showing that the two CH domains are in a compact or closed conformation1, 5.
We have used both EM of negatively stained samples and cryo-EM of unstained, frozen-hydrated samples (see Supplementary Methods) to examine complexes of the α-actinin ABD with F-actin (Fig. 1). Since previous controversies involving CH domains bound to F-actin have used either cryo-EM or negative stain, it is important to understand whether the different specimen preparation methods and imaging techniques can account for any of the differences.
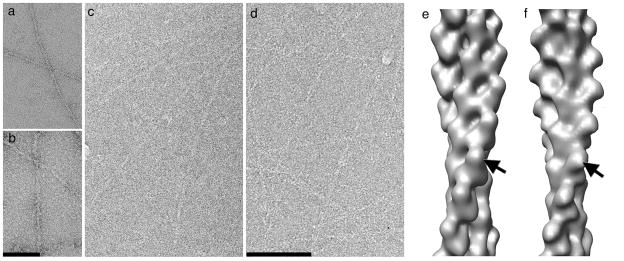
Figure 1.
EM of complexes. (a,b) Both negative stain and (c,d) cryo-electron microscopy (cryo-EM) have been used to look at control naked F-actin (a,c) and F-actin decorated with the actin-binding domain (ABD) of α-actinin (b,d). Three-dimensional reconstructions filtered to a common 23 Å resolution from negative stain (e) and cryo-EM (f) are similar, establishing that at this limited resolution the two techniques yield similar results.
Reconstructions generated with the iterative helical real space reconstruction (IHRSR) method from negatively stained specimens (Fig. 1e) and frozen-hydrated specimens (Fig. 1f), when filtered to the limiting resolution in negative stain (~ 23 Å), are similar, and show a single mass of additional density bound to each subunit in the actin filament (arrows, Fig. 1e,f). These reconstructions are similar to a previous reconstruction10 of F-actin decorated with the α-actinin ABD, and full decoration is seen without the need for any additional sorting or enhancement of the bound density. This contrasts with the binding of fimbrin’s ABD1 (containing CH1 and CH2) to F-actin, where very weak decoration was seen11, 12, or the binding of utrophin’s ABD to F-actin where both weak and polymorphic binding was seen2. Even with full decoration, at this limited resolution ambiguities exist in the interpretation of such a volume. We have therefore used the cryo-EM reconstruction at a resolution of 16 Å (Fig. 2a) for model building. At this resolution the actin subunits must actually have a more closed nucleotide-binding cleft than in the more than 20 existing crystal structures of G-actin. When the threshold of the surface is chosen to enclose an atomic model of actin, the additional mass bound to F-actin can only accommodate a single CH domain. Lowering the threshold (Supplementary Figure 1) shows some density at higher radius, but is inconsistent with closed tandem CH domains bound to actin. Given the separation by ~25 Å of CH domains within the same molecule in the crystal of the utrophin ABD6, a second CH domain in the open state that is not bound to F-actin could be at a considerable radius from F-actin and not seen after helical averaging. For comparison, when the volume12 of F-actin decorated with fimbrin’s ABD2 (CH3-CH4) is filtered to the same 16 Å resolution and thresholded to enclose actin (Fig. 2b), the surface encloses two CH domains. A potential ambiguity exists at this resolution as to whether the additional mass is due to CH1 or CH2 from α-actinin, given the high level of structural similarity between these domains. Since it has been shown13 that the main binding site within α-actinin for actin is contained within CH1, we think it reasonable to fit CH1 into this volume. At the available resolution there is no ambiguity in docking CH1 to actin, either manually or with automated procedures, with respect to any translational or rotational degrees of freedom.
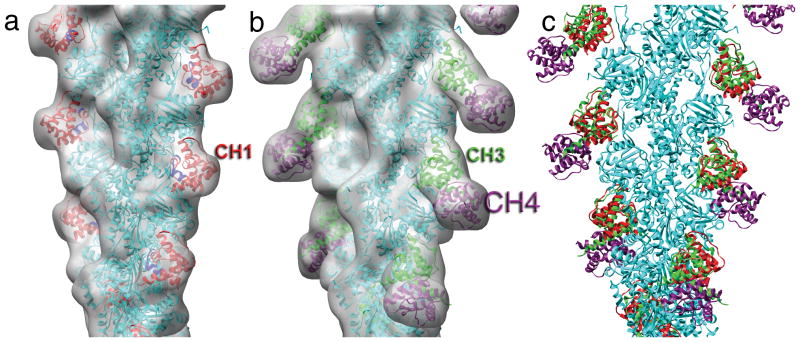
Figure 2.
Pseudo-atomic models. (a) A cryo-electron microscopy reconstruction of F-actin decorated with the actin-binding domain (ABD) of α-actinin at 16 Å resolution can be compared with (b) a reconstruction12 filtered to that same resolution of F-actin decorated with ABD2 from fimbrin. Actin is shown by the cyan ribbons, while calponin homology domain 1 (CH1) from α-actinin is shown in red (a), CH3 from fimbrin is shown in green (b), and CH4 from fimbrin is shown in magenta (b). Residues 133-147 are shown in blue, as the corresponding residues in smooth muscle α-actinin were shown to be making the dominant contact with actin13. A superposition of the models for the binding to F-actin of α-actinin’s CH1 and fimbrin’s CH3-CH4 (c) shows that the interaction between CH1 and actin is very similar to the interaction between CH3 and actin..
Our new model of α-actinin CH1 bound to F-actin is very similar to the model of fimbrin CH3 bound to F-actin (Fig. 2c), indicating that some features of this interactions are conserved. Using CH1 for alignment, superimposition of the compact α-actinin CH1-CH2 crystal structure upon CH1 bound to F-actin (Fig. 3a) results in a steric clash of CH2 with actin. Thus, the two domains of this closed CH1-CH2 ABD must either rearrange or separate to allow binding. In the case of fimbrin’s ABD2, the two domains undergo a small rearrangement with respect to each other compared to crystal structures, as steric clashes between CH4 and actin would have resulted in the absence of such rearrangments12. In a crystal of fimbrin’s actin-binding core (containing two tandem CH domains, CH1-CH2 and CH3-CH4; PDB ID 1PXY), a substantial difference in the CH1-CH2 interface was seen between two molecules in the asymmeric unit, while the CH3-CH4 interface was structurally very similar between the two molecules.
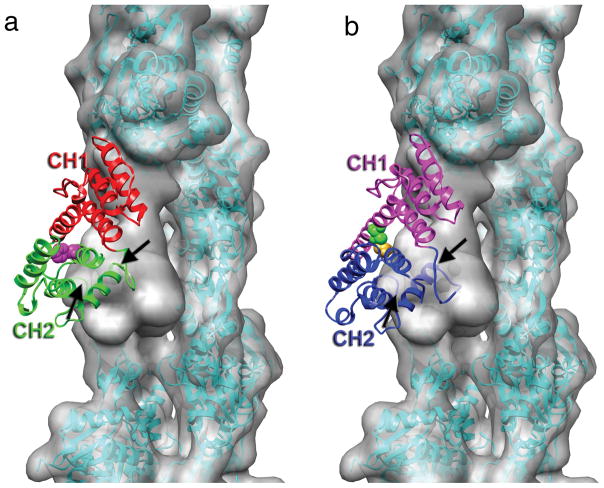
Figure 3.
Steric clash between calponin homology domain 2 (CH2) of α-actinin and actin. (a) The crystal structure of the actin-binding domain (ABD) of α-actinin (PDB 1TJT) has been docked onto F-actin so that CH1 is superimposed on the model for CH1 bound to F-actin in Fig. 2a. CH2 has a significant penetration into the actin volume, indicated by the black arrows. The position of residue 255 at the CH1-CH2 interface is indicated by the magenta space-filling atoms. The K255E mutation destabilizes this interface, and increases the affinity of α-actinin for actin14. (b) The binding of α-actinin’s ABD to F-actin may likely explain observations made about plectin19, another actin-binding protein containing tandem CH domains. When CH1 of plectin (PDB 1MB8) is aligned with CH1 of α-actinin, CH2 of plectin makes a steric clash with an actin protomer (black arrow). When the two plectin CH domains can separate, the affinity for F-actin is approximately three times greater than when the two CH domains are crosslinked (the disulfide crosslink is indicated by the space filling atoms at the CH1-CH2 interface) in a closed conformation19, suggesting that strain introduced into either F-actin or plectin to eliminate the steric clash reduces the binding affinity.
This new interpretation can be applied to previous results with α-actinin. A mutation (K255E) of a residue located at the interface between CH1 and CH2 in α-actinin (Fig. 3a) has been shown to be responsible for an inherited late-onset kidney failure. In vitro this mutant protein has a six-fold higher affinity for actin filaments (Kd=5μM) than the wild-type protein (Kd=32 μM). It was suggested14 that the mutation destabilizes the CH1-CH2 interface and exposes an additional actin-binding surface (ABS1) on CH1 that is buried in the closed CH1-CH2 ABD. This proposed ABS1 arose from peptide studies15 on dystrophin binding to actin, but such a binding site was not seen in a subsequent study using larger protein fragments from dystrophin16. Our results (Fig. 3a) suggest that no new actin-binding surface is exposed; rather, the presence of CH2 provides steric hindrance to binding F-actin, and opening of the CH1-CH2 interface is required for normal binding. Thus, destabilization of the CH1-CH2 interface enhances binding to F-actin. Strikingly, all of the α-actinin-4 mutations causing focal segmental glomerulosclerosis in humans map to residues at the CH1-CH2 interface14, and most of these have been shown to result in an increased affinity for binding to F-actin. The increased affinity undermines normal function, most likely by preventing the rapid exchange of α-actinin during cytoskeletal remodeling.
The notion that CH2 is a negative regulator in the binding of tandem CH domains to F-actin is consistent with the fact that while isolated CH1 domains bind to F-actin, isolated CH2 domains show no affinity for binding F-actin17. The ability of the two CH domains in α-actinin to open apart is supported by direct observations of a two-dimensional crystal of α-actinin dimers, where one molecule in each dimer exists with the CH domains in a closed conformation, while the other molecule has the CH domains in an open conformation9. It should be noted, however, that CH1 alone of α-actinin has been observed to bind to F-actin with lower affinity than a CH1-CH2 fragment17. CH2 may thus also serve as a positive regulator of binding of CH1 to F-actin, and the regulatory role of CH2 has previously been suggested based in part upon the fact that PIP2, a lipid that has been shown to regulate the bundling of F-actin by α-actinin, binds to CH218.
Our results suggest a reinterpretation of some previous observations with other actin-binding proteins. It was shown for plectin, which also contains tandem CH domains, that a disulfide crosslink which keeps CH1 and CH2 in a closed conformation still allows binding to F-actin, but reduces the affinity of such binding by a factor of about three19 (from Kd=23 μM in the absence of the crosslink to Kd=59 μM in the presence of the crosslink). When the closed CH1-CH2 crystal structure of plectin is superimposed on the model of α-actinin CH1 bound to F-actin (Fig. 3b), it can be seen that CH2 of plectin is making a steric clash with an actin subunit. We suggest that the opening of the two CH domains in plectin is required to eliminate this steric clash, and that the reduction in affinity for binding when the two domains are crosslinked is due to the strain imposed upon either the plectin ABD or F-actin. This contrasts with the interpretation that the opening of the two domains is required to reveal an actin-binding interface that is otherwise buried between the two CH domains19. Similarly, a mutation in human dystrophin (Y231N) that is found in Becker Muscular Dystrophy patients is buried7 at the interface of CH1 and CH2. Missense mutations within CH1 of filamin lead to a loss-of-function phenotype, while mutations that are within CH2 of filamin lead to increased affinity for actin and skeletal disorders8. One filamin mutation, S235P, which leads to boomerang dysplasia, is found at the CH1-CH2 interface. Two other filamin mutations buried in CH2 (where they could not interact with actin) but near the CH1-CH2 interface showed decreased thermostability and increased crystallographic temperature factors8, with increased affinity for F-actin (a shift of the Kd from 7 μM for the wt to either 2 μM or 0.6 μM for the mutants), completely consistent with our model that CH2 is a negative regulator whose destabilization would lead to increased binding to actin.
It is clear that while the structural homology among all CH domains is great, the conserved structure does not dictate conserved interactions with F-actin. The CH domain within the eponymous protein calponin has been shown20 to never bind F-actin, just as CH2 of α-actinin does not interact with F-actin as has been shown previously13 and in this report. When both CH domains present in tandem CH domains do interact with F-actin, such as in ABD2 of fimbrin12, the interaction between CH4 and actin is very different from the interaction of CH3 with actin. Structural homology only really establishes that two proteins share common ancestry, and not that they have conserved interactions or functions. The CH domains found in many actin-binding proteins have thus evolved to have different functions, and the CH2 domains in proteins such as α-actinin, plectin and filamin appear to be negative regulatory elements.
Supplementary Material
Acknowledgments
This work was supported by NIH GM081303 (to E.H.E.) and by FWF P19060 to (K.Dj.C.).
Footnotes
Accession Codes
Protein Data Bank: Coordinates of the F-actin-CH1 model have been deposited with accession code 3LUE.
EM Data Bank: The three-dimensional reconstruction of the F-actin-CH1 complex has been deposited with accession code 5170.
Author Contributions
A.S. and A.O. performed sample preparations; A.O. did the electron microscopy; V.G. performed the image analysis and model building; V.G., A.O., E.H.E. and K.Dj.-C. prepared the manuscript.
Reference List
- 1.Lehman W, Craig R, Kendrick-Jones J, Sutherland-Smith AJ. J Muscle Res Cell Motil. 2004;25:351–358. doi: 10.1007/s10974-004-0690-7. [DOI] [PubMed] [Google Scholar]
- 2.Galkin VE, Orlova A, VanLoock MS, Egelman EH. J Mol Biol. 2003;331:967–972. doi: 10.1016/s0022-2836(03)00842-8. [DOI] [PubMed] [Google Scholar]
- 3.Broderick MJ, Winder SJ. Adv Protein Chem. 2005;70:203–246. doi: 10.1016/S0065-3233(05)70007-3. [DOI] [PubMed] [Google Scholar]
- 4.Lee SH, Weins A, Hayes DB, Pollak MR, Dominguez R. J Mol Biol. 2008;376:317–324. doi: 10.1016/j.jmb.2007.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrego-Diaz E, et al. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Keep NH, et al. Structure Fold Des. 1999;7:1539–1546. [Google Scholar]
- 7.Norwood FL, Sutherland-Smith AJ, Keep NH, Kendrick-Jones J. Structure Fold Des. 2000;8:481–491. doi: 10.1016/s0969-2126(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 8.Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. J Mol Biol. 2009;390:1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Taylor DW, Taylor KA. J Mol Biol. 2004;338:115–125. doi: 10.1016/j.jmb.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 10.McGough A, Way M, DeRosier D. J Cell Biol. 1994;126:433–443. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanein D, Matsudaira P, DeRosier DJ. J Cell Biol. 1997;139:387–396. doi: 10.1083/jcb.139.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galkin VE, Orlova A, Cherepanova O, Lebart MC, Egelman EH. Proc Natl Acad Sci U S A. 2008;105:1494–1498. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhlman PA, Hemmings L, Critchley DR. FEBS Lett. 1992;304:201–206. doi: 10.1016/0014-5793(92)80619-r. [DOI] [PubMed] [Google Scholar]
- 14.Weins A, et al. Proc Natl Acad Sci U S A. 2007;104:16080–16085. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine BA, Moir AJ, Patchell VB, Perry SV. FEBS Lett. 1990;263:159–162. doi: 10.1016/0014-5793(90)80728-2. [DOI] [PubMed] [Google Scholar]
- 16.Corrado K, Mills PL, Chamberlain JS. FEBS Lett. 1994;344:255–260. doi: 10.1016/0014-5793(94)00397-1. [DOI] [PubMed] [Google Scholar]
- 17.Way M, Pope B, Weeds AG. J Cell Biol. 1992;119:835–842. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young P, Gautel M. EMBO J. 2000;19:6331–6340. doi: 10.1093/emboj/19.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Alvarez B, Bobkov A, Sonnenberg A, de Pereda JM. Structure. 2003;11:615–625. doi: 10.1016/s0969-2126(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 20.Galkin VE, Orlova A, Fattoum A, Walsh MP, Egelman EH. J Mol Biol. 2006;359:478–485. doi: 10.1016/j.jmb.2006.03.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.