Abstract
The receptor protein tyrosine phosphatase T (PTPRT/PTPρ) is the most frequently mutated tyrosine phosphatase in human cancer. PTPρ mediates homophilic cell-cell aggregation. In its extracellular region, PTPρ has cell adhesion molecule-like motifs, including a MAM domain, an immunoglobulin domain and four fibronectin type III (FNIII) repeats. Tumor-derived mutations have been identified in all of these extracellular domains. Previously, we determined that tumor-derived mutations in the MAM and immunoglobulin domains of PTPρ reduce homophilic cell-cell aggregation. In this manuscript, we describe experiments in which we evaluated the contribution of the FNIII repeats to PTPρ-mediated cell-cell adhesion. Our results demonstrate that deletion of the FNIII repeats of PTPρ result in defective cell-cell aggregation. Furthermore, all of the tumor-derived mutations in the FNIII repeats of PTPρ also disrupt cell-cell aggregation. These results further support the hypothesis that mutational inactivation of PTPρ may lead to cancer progression by disrupting cell-cell adhesion.
Keywords: Receptor protein tyrosine phosphatase, PTPRT, PTPρ, fibronectin type III repeat, cell-cell aggregation, cell-cell adhesion
1. Introduction
Cancer progression and metastasis is in part influenced by changes in cell adhesion and tyrosine phosphorylation mediated by cell adhesion molecules (CAMs) (Paschos et al. 2009; Wheelock et al. 2008), as well as tyrosine kinases and tyrosine phosphatases, respectively (Freiss and Vignon 2004; Ostman et al. 2006). Receptor protein tyrosine phosphatases (RPTPs) comprise a family of proteins that have extracellular CAM domains coupled to cytoplasmic tyrosine phosphatase domains, and thus can impact cancer progression by regulating both cell adhesion and tyrosine phosphorylation (Brady-Kalnay 2001; Ostman et al. 2006).
The type IIB PTPμ-like subfamily of RPTPs includes PTPμ, PTPρ, PTPκ, and PCP-2 (PTPλ) (Aricescu et al. 2008; Brady-Kalnay 2001). The extracellular segment of the PTPμ-like RPTPs contain motifs found in CAMs, including a MAM domain, an immunoglobulin (Ig) domain and four fibronectin type III (FNIII) repeats (Aricescu et al. 2008; Brady-Kalnay 2001; Ostman et al. 2006). PTPμ subfamily members bind homophilically (i.e. the “ligand” for PTPμ is an identical PTPμ molecule on an adjacent cell) (Brady-Kalnay et al. 1993; Cheng et al. 1997; Gebbink et al. 1993; Sap et al. 1994; Yu et al. 2008; Zondag et al. 1995). The Ig domain of PTPμ is responsible for promoting homophilic interactions (Brady-Kalnay and Tonks 1994), and proper cell surface localization (Del Vecchio and Tonks 2005). However, both the MAM and Ig domains as well as the first two FNIII repeats are required for efficient cell-cell adhesion of PTPμ (Aricescu et al. 2006; Aricescu et al. 2007; Aricescu et al. 2008; Brady-Kalnay and Tonks 1994; Cismasiu et al. 2004; Zondag et al. 1995). PTPρ and PTPκ also mediate homophilic cell-cell adhesion (Sap et al. 1994; Yu et al. 2008). However, PCP2 has not yet been demonstrated to mediate cell-cell adhesion or aggregation although in vitro binding studies indicate it can bind homophilically (Cheng et al. 1997).
All type IIB RPTPs are potential tumor suppressor genes (Burgoyne et al. 2009a; Flavell et al. 2008; McArdle et al. 2001; Yan et al. 2006). Notably, PTPρ is the most frequently mutated phosphatase gene in human cancers including colon, lung, skin and stomach cancers (Forbes et al. 2008; Wang et al. 2004). Defective adhesion mediated by PTPρ may be at least one mechanism of cancer progression. In fact, cancer derived mutations in the MAM and Ig domains of PTPρ are defective in mediating cell-cell aggregation (Yu et al. 2008).
In addition to the tumor-derived mutations in the MAM and Ig domains of PTPρ previously described (Wang et al. 2004; Yu et al. 2008), the majority of extracellular domain mutations are located in the FNIII repeats of PTPρ (Wang et al. 2004). Given the importance of FNIII repeats in PTPμ-mediated adhesion and aggregation (Aricescu et al. 2007), we tested the contribution of the FNIII repeats of PTPρ to PTPρ-mediated adhesion. We determined that deletion constructs where each of the extracellular domains of PTPρ is removed, including the FNIII repeats, are defective in mediating cell-cell aggregation of Sf9 cells. Furthermore, when we engineered the cancer-derived mutations in the FNIII repeats of these PTPρ mutant proteins, they also result in defective cell-cell aggregation. These results demonstrate the importance of the FNIII repeats in contributing to proper cell-cell adhesion and suggest that adhesion mediated by PTPρ is likely important for the tumor suppressor activity of PTPρ.
2. Materials and methods
2.1 Baculovirus generation
Baculoviruses were generated by transfecting cells with the BaculoGold™ Linearized Baculovirus DNA (BD Biosciences, San Diego, CA) according to the manufacturer’s protocol. The baculovirus for wild-type PTPρ was previously described (Yu et al. 2008).
2.2 Sf9 cell culture
Insect Sf9 cells from (BD Biosciences) were maintained at 27°C in Graces Insect Medium (BD Biosciences). The medium was supplemented with 10% FBS from (Hyclone, Logan, UT) and Gentamicin (MP Biomedicals, Solon, OH). Viruses were added to Sf9 cell medium to induce the expression of proteins of interest. Cells were harvested 30 hours post-infection for further analysis.
2.3 Generation of PTPRT deletion constructs
The PTPρ-ΔMAM, PTPρ-ΔIg, PTPρ-ΔFNIII-1, PTPρ-ΔFNIII-2, PTPρ-ΔFNIII-3 and PTPρ-ΔFNIII-4 constructs were generated by fusion PCR using the full-length PTPRT pVL1393-V5-His vector as the template. The primers 5’-GAAGATCTATGGCGAGCCTCGCCGCG-3’ and 5’-CCGGAATTCGGATCCTGGGGCGCTCTGAGCCCGGG-3’ were used to generate the PTPρ-ΔMAM construct; the primers 5’-GCGTGGGAGGCTCTCTGCATGGATGAGCAAGGAC-3’ and 5’-CAATGGGTCTTGCTCATCCATGCAGAGAGCCTCCCACGCCCATTG-3’ were used to generate the PTPρ-ΔIg construct; the primers 5’-CTGCACACTTGGTCCTGGTTTTCACGATCAGCTCCGCGTA-3’ and 5’-TACGCGGAGCGTCGTGAAAACCAGGACCAAGTGTGCAG-3’ were used to generate the PTPρ-ΔFNIII-1 construct; the primers 5’-CCGCCAGGGCCTCCCCTCACCGAGGAAGACGTTCCAGGAG-3’ and 5’-CTCCTGGAACGTCTTCCTCGGTGAGGGGAGGCCCTGGCGG-3’ were used to generate the PTPρ-ΔFNIII-2 construct; the primers 5’-GAGGAGCTGGTGGTGCAGACTAAAATTTCAGCTCCATCCA-3’ and 5’-TGGATGGAGCTGAAATTTTAGTCTGCACCACCAGCTCCTC-3’ were used to generate the PTPρ-ΔFNIII-3 construct and the 5’-GTCACCACTCGGATTGCCACCGGTGCCTCCACCCAGAATT-3’ and 5’-AATTCTGGGTGGAGGCACCGGTGGCAATCCGAGTGGTGAC-3’ were used to generate the PTPρ-ΔFNIII-4 construct.
2.4 Immunoblotting
Sf9 cells were lysed in a standard lysis buffer (20 mM Tris·HCl, pH 7.5, 1% Triton X-100, 150 mM sodium chloride, 2 mM Benzamidine, 2.5μg/ml Aprotinin, 2.5μg/ml Leupeptin, and 0.5μg/ml Pepstatin). Samples were solubilized in 2X SDS loading buffer and separated by SDS-PAGE. Samples were transferred to a nitrocellulose membrane (Whatman, Piscataway, NJ) and immunoblotted with SK18 (Brady-Kalnay et al. 1993), which cross-reacts with PTPρ.
2.5 Cell surface expression of PTPρ
Cell surface expression of PTPρ protein in baculovirus infected Sf9 cells was determined using a modified biotin-avidin reaction with the Cell Surface Protein Isolation Kit (catalog# 89991, Pierce, Rockford, IL), as previously described (Yu et al. 2008). Briefly, Sf9 cell media was removed and the flasks were incubated with rocking for 30 minutes at 4°C in 5ml of 0.25mg/ml Sulfo-NHS-SS-Biotin in 1X PBS. The cells were mechanically removed from the flask, centrifuged, washed, and lysed according to the manufacturer’s protocols. To isolate cell surface expressed PTPρ the cell lysate was added to a NeutrAvidin Gel column and incubated for 60 min at room temperature (RT) with rocking prior to washing and eluting proteins with 200μl of sample buffer containing a final concentration of 55mM DTT for 60min at RT. The flow through was run on 6% SDS-PAGE gels and transferred to nitrocellulose. Total cell surface protein levels were normalized by immunoblotting cell lysates with an anti-actin antibody (JLA20). The normalized cell surface protein was run on 6% SDS-PAGE gels, transferred to nitrocellulose and immunoblotted with antibodies to PTPμ that also recognized PTPρ (SK18) followed by secondary antibody conjugated to HRP. The HRP signal was detected using a Fluor-S MAX MultiImager (Bio-Rad Laboratories Inc., Hercules, CA).
2.6 Aggregation Assays
Sf9 cells were harvested by trituration. The cell suspensions were added to glass scintillation vials and incubated at 27°C at 90 rpm in a gyratory shaker for 30 minutes. For a given condition, the cell suspension was transferred to a 100mm petri dish either prior to (0 time point) or 30 minutes after aggregation. Images were captured at 10X magnification with a Nikon TE-200 inverted fluorescent microscope (Tokyo, Japan). To quantify cell aggregates, pictures of four randomly selected fields were taken per dish. The area of each object (single cells and aggregates) was determined by the Metamorph software (Molecular Devices) using an auto-threshold setting for light objects and appropriate size filters that allowed the counting of cells and aggregates but not debris. The four replicates were combined to yield average readings per condition. The percent aggregation was then calculated as the average aggregate area at 30 minutes minus the average aggregate area at zero minutes divided by the average aggregate area at 30 minutes [(N30-N0)/N30]. A minimum of three independent experiments was performed per condition. Statistical significance was determined in Microsoft Excel using Student’s T-test. Error bars indicate standard error.
2.7 Site-directed mutagenesis
The I395V, Y412F, R453C, Q479E, S492F, N510K, T605M, V648G, A707T, and L708P tumor-derived mutations (Wang et al. 2004) were generated by fusion PCR using the full-length PTPRT pVL1393-V5-His vector as the template. The primers 5’-TGGCCCACAGAACGTGGAAGTCGTAGACATCAGAGCC-3’ and 5’-CTTCCACGTTCTGTGGGCCA-3’ were used as the mutagenic primers for the I395V mutant; the primers 5’-CAGTGGGAGCCCTTCGGCTTCGCGGTGACCCGCTGCCAT-3’ and 5’-AAGCCGAAGGGCTCCCACTG-3’ were used as the mutagenic primers for the Y412F mutant; the primers 5’-CTACACCCTGCGAGGCCTGTGCCCCTTCATGACCATCCG-3’ and 5’-ACAGGCCTCGCAGGGTGTAG-3’ were used as the mutagenic primers for the R453C mutant; the primers 5’-GAGCGAGGAGCTGGTGGTGGAGACTGAGGAAGACGTTCC-3’ and 5’-CCACCACCAGCTCCTCGCTC-3’ were used as the mutagenic primers for the Q479E mutant; the primers 5’-TGTTCCTCTAGAATTCATCCAAGGGGGGCCCT TT-3’ and 5’-ATGAATTCTAGAGGAACA-3’ were used as the mutagenic primers for the S492F mutant; the primers 5’-TCCAGTGGAAACCTCCCAAGGAGACCAATGGGGTCATCA-3’ and 5’-CTTGGGAGGTTTCCACTGGA-3’ were used as the mutagenic primers for the N510K mutant; the primers 5’-CCATTGAATGAGACAGACATGACC ATCACAGTGATGCTG3-’ and 5’-ATGTCTGTCTCATTCAATGG-3’ were used as the mutagenic primers for the T605M mutant; the primers 5’-ATTATTGAGTGCTTTTC GGGGCCCGTGAGCTATCGGAAT-3’ and 5’-CCCGAAAAGCACTCAATAAT-3’ were used as the mutagenic primers for the V648G mutant; the primers 5’-CTACAGCATCTACTTCCAGACACTCAGCAAAGCCAATGG-3’ and 5’-TCTGGAAGTAGATGCTGTAG-3’ were used as the mutagenic primers for the A707T mutant; the primers 5’-AGCATCTACTTCCAGGCACCCAGCAAAGCCAATGGAGAG-3’ and 5’-GGTGCCTGGAAGTAGATGCT-3’ were used as the mutagenic primers for the L708P mutant.
2.8 Mapping of PTPρ mutations onto the PTPμ crystal structure
The sequence of PTPμ and PTPρ were aligned using CLUSTALW. Mutations probed in this study were mapped using this sequence alignment onto the PTPμ structure (PDB ID: 2V5Y). The figure was prepared utilizing PyMOL 1.2r1 (http://pymol.org).
3. Results
3.1 Deletions in the MAM, Ig, and FNIII repeat domains of PTPρ result in defective cell-cell aggregation
To test whether a protein mediates cell-cell aggregation or adhesion, one has to express the protein of interest in the non-adhesive cells. Otherwise, one cannot measure the aggregation or adhesion of a single molecule in the sea of other adhesion molecules naturally expressed in adhesive cells. This is the main reason that the non-adhesive Sf9 or Drosophila S2 cells are widely used models for such studies. To our knowledge, nearly all cell lines of epithelial origin are adhesive cells and thus are not suitable for these studies. Introduction of putative adhesion molecules into non-adhesive Drosophila S2 insect cells has been used to demonstrate adhesive functions for fasciclin II, connectin, Dtrk, ARK, Neuroglian, and Capricious molecules directly (Bellosta et al. 1995; Hortsch et al. 1995; Nose et al. 1992; Pulido et al. 1992; Shinza-Kameda et al. 2006; Snow et al. 1989). In a similar approach, we and others demonstrated that the full-length form of PTPμ induced aggregation, via a homophilic binding mechanism, when expressed in nonadhesive Sf9 insect cells, which are derived from the Fall army-worm Spodoptera frugiperda (Brady-Kalnay et al. 1993; Gebbink et al. 1993; Zondag et al. 1995). We have also used this system for analyzing PTPRT/PTP rho (Yu et al., 2008). Examples of other cell adhesion molecules that have been studied in Sf9 cells include L1 (Gouveia et al. 2008), Galectin-3 (Inohara and Raz 1995) and C-CAM1 (Phan et al. 2001). Insect S2 and Sf9 cells are extremely useful systems to analyze adhesive mechanisms of cell adhesion molecules.
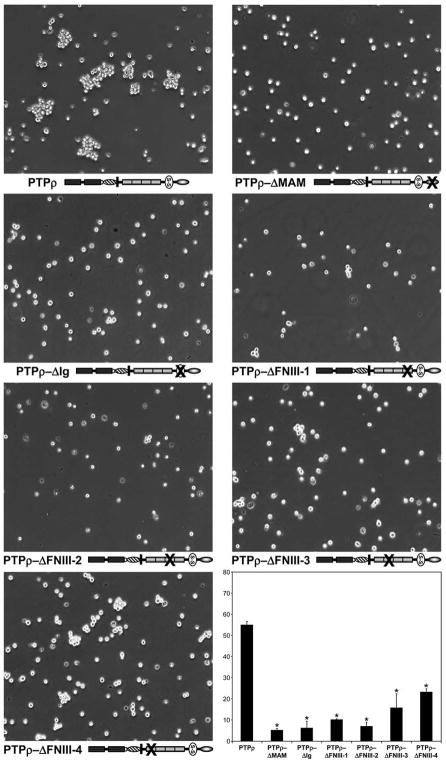
The MAM domain, Ig domain, and first two-FNIII repeats are essential for PTPμ cell-cell aggregation (Brady-Kalnay and Tonks 1994; Zondag et al. 1995). To test whether these domains are the minimal essential regions required for PTPρ cell-cell aggregation, baculoviral PTPρ deletion constructs of the MAM domain, Ig domain, and each of the four individual FNIII repeats were made and expressed in non-adhesive Sf9 insect cells (Figure 1). All the PTPρ deletion constructs were expressed and appropriately trafficked to the cell surface (Figure 1). To test whether these domains contribute to PTPρ-dependent adhesion, Sf9 cells were evaluated for their ability to mediate cell-cell aggregation when expressing the deletion constructs. Aggregation assays with Sf9 cells expressing the deletion constructs were conducted alongside aggregation assays using Sf9 cells expressing wild-type PTPρ, which mediates cell-cell aggregation (Figure 2) and serves as a positive control (Yu et al. 2008). Deletion of any of the extracellular domains of PTPρ resulted in defective cell-cell aggregation compared to aggregation of wild-type PTPρ expressing Sf9 cells (Figure 2). Deletion of the fourth FNIII repeat was the least severe where ~23% aggregation was observed. Deletion of the MAM domain, Ig domain and FNIII repeats 1, 2 or 3 resulted in 5–16% aggregation. When the level of aggregation was quantified, we determined that all of the deletion constructs resulted in a large and statistically significant reduction in cell-cell aggregation (Figure 2, p-value<0.005). Together with our previous study, these results demonstrate that all of the extracellular domains of PTPρ are necessary for PTPρ-mediated cell-cell aggregation.
Figure 1.
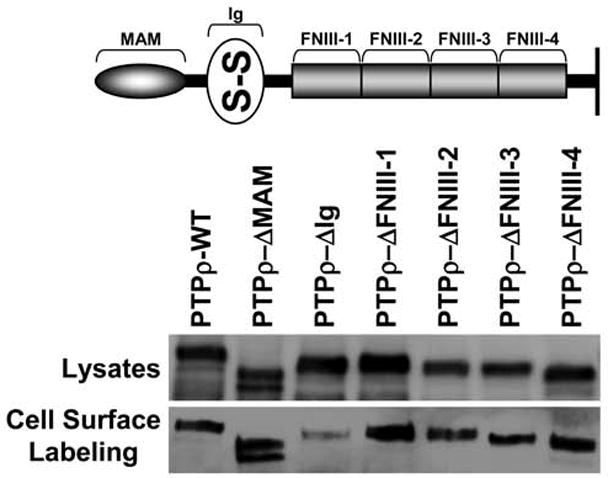
PTPρ deletion constructs are all expressed at the cell surface. Constructs containing deletions of one extracellular domain of PTPρ each were generated and expressed in Sf9 cells. Total cellular protein and cell surface protein were isolated and immunoblotted with the SK18 antibody.
Figure 2.
Deletion of any of the extracellular domains of PTPρ disrupts cell-cell aggregation. Sf9 cells expressing deletion constructs of the individual extracellular domains of PTPρ were allowed to aggregate for 30 minutes prior to imaging. Scale bar equals 100μm. The percentage of Sf9 cell-cell aggregation for each deletion protein was calculated from a minimum of three experiments. An asterisk indicates a statistically significant reduction in aggregation (p<0.005).
3.2 Tumor-Derived PTPρ Mutations in the FNIII repeats result in defective cell-cell aggregation
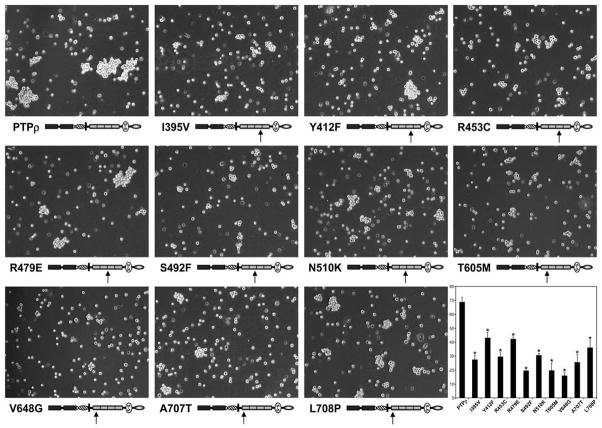
We next tested the effects of human tumor-derived mutations in the FNIII repeats of PTPρ on PTPρ-mediated cell-cell aggregation. We recently demonstrated that tumor-derived mutations in the MAM and Ig domains of PTPρ are defective in cell-cell aggregation using the Sf9 aggregation assay (Yu et al. 2008), and given the above results, we hypothesized that mutations in the FNIII domains would likewise result in defective aggregation. Ten human tumor-derived mutations are located in the second, third and fourth FNIII repeats of PTPρ (Wang et al. 2004). To test whether these mutations also affect cell-cell aggregation, we engineered the ten human mutations by site-directed mutagenesis using the full-length PTPρ baculoviral construct as a template (see Figure 3). The ten mutations are expressed in Sf9 cells at the expected molecular weight and at similar levels to full-length human PTPρ (Figure 3). All of these mutants are expressed at the cell surface (Figure 3). When these mutant PTPρ proteins were expressed in Sf9 cells and tested in aggregation assays, we observed that all of the mutant PTPρ proteins resulted in fewer and smaller aggregates than wild type PTPρ (Figure 4). When the level of aggregation was quantified, we determined that all of the FNIII mutations significantly reduced cell-cell aggregation (16–43%) compared to cells expressing wild type PTPρ (68% aggregation, p-value<0.008, Figure 4).
Figure 3.
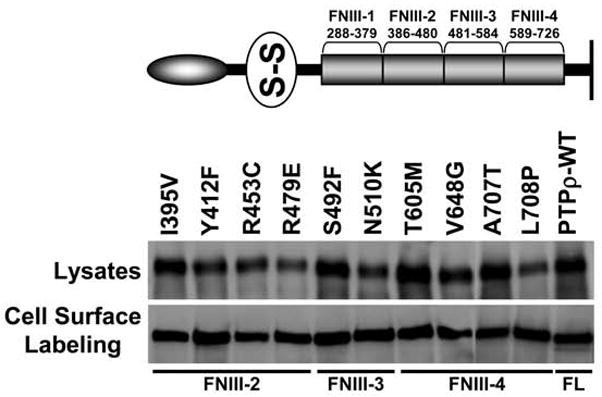
PTPρ proteins containing tumor-derived mutations in their FNIII domains are expressed at the cell surface. Ten mutations in the FNIII domains of PTPρ were generated by site-directed mutagenesis and expressed in Sf9 cells. Total cellular and cell surface proteins were isolated and analyzed by immunoblotting with the SK18 antibody.
Figure 4.
Tumor derived mutations in FNIII domains of PTPρ are defective in cell-cell aggregation. Sf9 cells expressing the mutated PTPρ proteins were allowed to aggregate for 30 minutes prior to imaging. Scale bar equals 100μm. The percentage of Sf9 cell-cell aggregation for each mutant protein was calculated from a minimum of three experiments. An asterisk indicates a statistically significant reduction in aggregation (p<0.008).
4. Discussion
PTPRT/PTPρ is the most highly mutated tyrosine phosphatase in human colon carcinomas. We and others found that PTPRT/ PTPρ is also mutated in lung, gastric and skin (melanoma) cancers (Forbes et al. 2008; Wang et al. 2004). The spectrum of mutations, which included nonsense mutations and frameshifts, suggested that these mutations were inactivating (Wang et al. 2004). Biochemical analyses demonstrated that missense mutations in the catalytic domains of PTPRT diminished its phosphatase activity and overexpression of PTPRT inhibited colon cancer cell growth. Moreover, we showed previously that the tumor-derived mutations in the MAM and Ig domains of PTPρ are defective in cell-cell adhesion (Yu et al. 2008). All these data suggest that PTPRT/PTPρ normally functions as a tumor suppressor gene. The experiments presented in this manuscript evaluate the contribution of the individual extracellular domains of PTPρ to PTPρ-mediated adhesion. We demonstrate that in addition to the MAM and Ig domains, the FNIII repeats are required for PTPρ-mediated adhesion. Importantly, we also show that PTPρ-mediated adhesion is impaired by human tumor-derived mutations in those FNIII repeats. These studies indicate that all the tumor-derived mutations located in the extracellular domain of PTPRT are loss-of-function mutations, thereby providing further evidence that PTPRT is a tumor suppressor. This notion is further supported by our recent study showing that PTPRT knockout mice are highly susceptible to colon specific carcinogen azoxymethane (AOM)-induced colon cancer (Zhao et al.).
PTPμ-mediated cell-cell aggregation requires the MAM domain, Ig domain, and the first two FNIII repeats (Aricescu et al. 2006; Aricescu et al. 2007; Brady-Kalnay and Tonks 1994; Cismasiu et al. 2004; Zondag et al. 1995). The entire extracellular domain of PTPμ is predicted to make a rigid conformation with extensive interfaces between each of the domains (Aricescu et al. 2006). The minimal adhesive unit for functional cell-cell aggregation is the MAM domain, Ig domain and the first two FNIII repeats (Aricescu et al. 2006; Brady-Kalnay and Tonks 1994; Zondag et al. 1995). However, the FNIII repeats appear to play a role in spacing the key homophilic interaction domain(s) a certain distance from the plasma membrane. Deletion of the FNIII repeats changed the intercellular spacing of two apposing membranes (Aricescu et al. 2007). Cell-cell aggregation assays of the FNIII deletion constructs were not performed in that study (Aricescu et al. 2007). The amino acid residues of the interfaces between the extracellular domains are highly conserved between members of the type IIB RPTP subfamily (Aricescu et al. 2006; Aricescu et al. 2007) therefore it is likely that the PTPρ adhesive interface resembles that of PTPμ. Together our data support the hypothesis that the essential regions for PTPρ-mediated adhesion resemble those of PTPμ.
To gain insight into how the point mutations may alter the adhesive function of PTPρ, we modeled the human tumor derived PTPρ mutations onto the crystal structure of PTPμ (Figure 5). The crystal structure of PTPμ includes only the first three FNIII repeats (Aricescu et al. 2007). Four of these mutations fall within the second FNIII (FNIII-2) repeat (I395V, Y412F, R453C, Q479E). It is important to note that FNIII-2 is required for efficient cell-cell adhesion (Aricescu et al. 2006). Two of the mutations fall in the third FNIII (FNIII-3) repeat (S492F, N510K). The mutation within the linker region between FNIII-1 and FNII-2 (Y412F) could alter the flexibility or positioning between the two domains. The other mutations (I395V R453C, Q479E, S492F, N510K) appear to be clustered in surface exposed regions and/or near the linker regions between FNIII-2 and FNIII-3 that could alter flexibility, position of individual FNIII repeats and/or protein-protein interactions. While these mutations do not correspond to the adhesive interface hypothesized for PTPμ based upon a single static crystal structure (Aricescu et al. 2007), some of the mutations fall within the FNIII-2 repeat which is required for adhesion, and all of these surface modifications would likely alter either the three-dimensional topology and/or cis/trans interactions of PTPρ.
Figure 5.
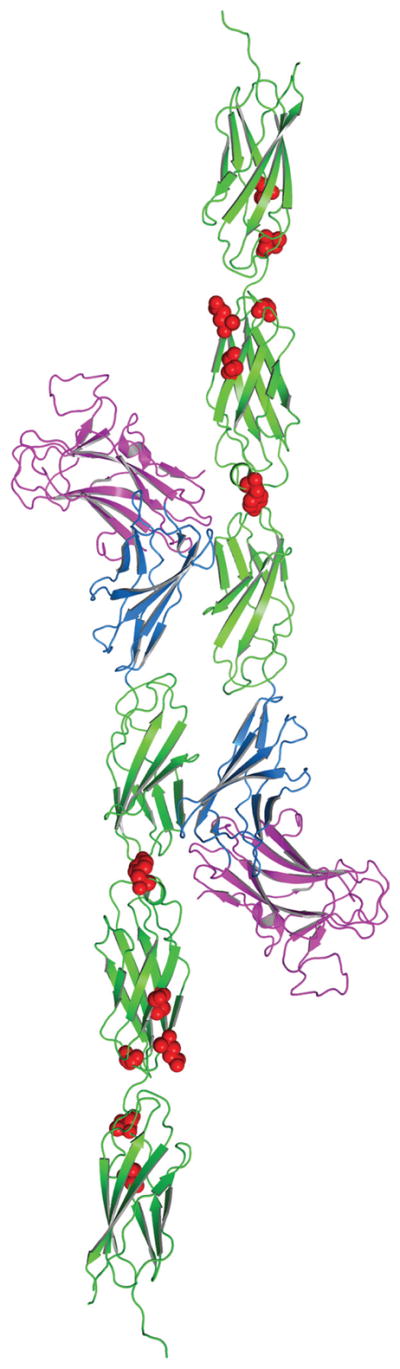
Structural modeling of the point mutations of PTPρ. The point mutations (indicated in red) evaluated in this manuscript were modeled onto the equivalent sites in the crystal structure of PTPμ. The PTPρ mutations likely alter cell-cell aggregation either by altering the three-dimensional topology and/or cis/trans interactions of PTPρ.
Other type IIB RPTPs also function as tumor suppressor genes, by regulating adhesion and/or tyrosine phosphorylation of effector proteins. Expression of PTPμ is reduced in human glioblastomas (GBM) and cell lines (Burgoyne et al. 2009a). Reduction in full-length PTPμ expression with a concomitant increase in a proteolytically processed cytoplasmic fragment of PTPμ is observed in GBM cells and both are correlated with decreased adhesion and increased migration observed in the disease (Burgoyne et al. 2009a; Burgoyne et al. 2009b). Over-expression of the full-length form of PTPμ suppressed migration of GBM cells and reduces their survival (Burgoyne et al. 2009b). Of note, at the same time that stable cell adhesion is reduced due to the loss of full-length PTPμ expression in GBM cells, a catalytically active cytoplasmic fragment of PTPμ is released and translocates to the nucleus (Burgoyne et al. 2009b). Catalytic activity of this fragment is also necessary for cell migration and viability of GBM cells (Burgoyne et al. 2009b). Therefore, both the tyrosine phosphatase activity of the cytoplasmic fragment of PTPμ and the loss of PTPμ-mediated adhesion may contribute to the invasiveness of GBM cells.
PTPκ is also implicated in tumor progression. PTPκ expression is reduced in melanoma cell lines and human tissue biopsies (McArdle et al. 2001), and in Hodgkin’s lymphoma cells (Flavell et al. 2008). Over-expression of PTPκ in Hodgkin lymphoma cells reduces cellular proliferation and survival (Flavell et al. 2008). Like PTPμ, PTPκ is proteolytically processed (Anders et al. 2006), potentially as a result of aberrant glycosylation (Kim et al. 2006). It is not clear whether adhesion or catalytic activity of PTPκ is required for its tumor suppressor activity (Flavell et al. 2008; Kim et al. 2006). Of note, the cleaved cytoplasmic domain fragment of PTPκ is catalytically active, translocates to the nucleus and promotes β-catenin-mediated transcription (Anders et al. 2006). Signaling via the PTPκ cytoplasmic fragment might augment the loss of stable cell-cell adhesion and may promote tumor progression.
Although speculative, it is interesting to postulate that tyrosine phosphatase activity and or cleavage of PTPρ will also be an important element of its tumor suppressor activity, as has been demonstrated for PTPμ and PTPκ. Interestingly, we identified and validated STAT3 as a direct substrate of PTPRT (Zhang et al. 2007). STAT3 is latent transcription factor that translocates from the cytoplasm to the nucleus after being phosphorylated at the Y705 residue. It is possible that the cleaved intracellular fragment of PTPρ, which contains the phosphatase domains, translocates to nucleus and dephosphorylates STAT3. Besco and colleagues found that PTPRT associates with the adhesion molecule E-cadherin and its binding partner p120 catenin, which are PTPRT substrates (Besco et al. 2006). p120 also translocates to the nucleus and associates with the Kaiso transcription factor (Daniel and Reynolds 1999). Given the frequency and distribution of mutations in PTPRT/PTPρ in human colon and other cancers, a more thorough understanding of its biological function is warranted.
Acknowledgments
We thank Jianshi Yu and Scott Howell for technical assistance; Sara Lou for preparation of the figures and graphs. This research was supported by grants from the National Institutes of Health Grant R01-CA127590 (Z.W), RO1-NS051520 (S.B.K). Additional support was obtained from the R01-HG004722, HG004722-02S1, U54-CA116867, the V foundation (Z.W) and the Visual Sciences Research Center Core Grant PO-EY11373 from the National Eye Institute (S.B.K).
References
- Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, Haass C, Ullrich A. Furin-, ADAM 10-, and gamma-Secretase-Mediated Cleavage of a Receptor Tyrosine Phosphatase and Regulation of beta-Catenin’s Transcriptional Activity. Mol Cell Biol. 2006;26:3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, Hon WC, Siebold C, Lu W, van der Merwe PA, Jones EY. Molecular analysis of receptor protein tyrosine phosphatase mu-mediated cell adhesion. Embo J. 2006;25:701–12. doi: 10.1038/sj.emboj.7600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, Siebold C, Choudhuri K, Chang VT, Lu W, Davis SJ, van der Merwe PA, Jones EY. Structure of a tyrosine phosphatase adhesive interaction reveals a spacer-clamp mechanism. Science. 2007;317:1217–20. doi: 10.1126/science.1144646. [DOI] [PubMed] [Google Scholar]
- Aricescu AR, Siebold C, Jones EY. Receptor protein tyrosine phosphatase mu: measuring where to stick. Biochem Soc Trans. 2008;36:167–72. doi: 10.1042/BST0360167. [DOI] [PubMed] [Google Scholar]
- Bellosta P, Costa M, Lin DA, Basilico C. The receptor tyrosine kinase ARK mediates cell aggregation by homophilic binding. Mol Cell Biol. 1995;15:614–25. doi: 10.1128/mcb.15.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besco JA, Hooft van Huijsduijnen R, Frostholm A, Rotter A. Intracellular substrates of brain-enriched receptor protein tyrosine phosphatase rho (RPTPrho/PTPRT) Brain Res. 2006;1116:50–7. doi: 10.1016/j.brainres.2006.07.122. [DOI] [PubMed] [Google Scholar]
- Brady-Kalnay SM. Protein tyrosine phosphatases. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- Brady-Kalnay SM, Flint AJ, Tonks NK. Homophilic binding of PTP mu, a receptor-type protein tyrosine phosphatase, can mediate cell-cell aggregation. J Cell Biol. 1993;122:961–72. doi: 10.1083/jcb.122.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Tonks NK. Identification of the homophilic binding site of the receptor protein tyrosine phosphatase PTP mu. J Biol Chem. 1994;269:28472–7. [PubMed] [Google Scholar]
- Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, Robinson S, Sloan AE, Vogelbaum MA, Miller RH, et al. PTPmu suppresses glioma cell migration and dispersal. Neuro Oncol. 2009a;11:767–78. doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, Brady-Kalnay SM. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009b;69:6960–8. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Wu K, Armanini M, O’Rourke N, Dowbenko D, Lasky LA. A novel protein-tyrosine phosphatase related to the homotypically adhering κ and μ receptors. J Biol Chem. 1997;272:7264–7277. doi: 10.1074/jbc.272.11.7264. [DOI] [PubMed] [Google Scholar]
- Cismasiu VB, Denes SA, Reilander H, Michel H, Szedlacsek SE. The MAM (meprin/A5-protein/PTPmu) domain is a homophilic binding site promoting the lateral dimerization of receptor-like protein-tyrosine phosphatase mu. J Biol Chem. 2004;279:26922–31. doi: 10.1074/jbc.M313115200. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–23. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vecchio RL, Tonks NK. The conserved immunoglobulin domain controls the subcellular localization of the homophilic adhesion receptor protein-tyrosine phosphatase mu. J Biol Chem. 2005;280:1603–12. doi: 10.1074/jbc.M410181200. [DOI] [PubMed] [Google Scholar]
- Flavell JR, Baumforth KR, Wood VH, Davies GL, Wei W, Reynolds GM, Morgan S, Boyce A, Kelly GL, Young LS, et al. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood. 2008;111:292–301. doi: 10.1182/blood-2006-11-059881. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10(Unit 10):11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiss G, Vignon F. Protein tyrosine phosphatases and breast cancer. Crit Rev Oncol Hematol. 2004;52:9–17. doi: 10.1016/j.critrevonc.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Gebbink MF, Zondag GC, Wubbolts RW, Beijersbergen RL, van Etten I, Moolenaar WH. Cell-cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J Biol Chem. 1993;268:16101–4. [PubMed] [Google Scholar]
- Gouveia RM, Gomes CM, Sousa M, Alves PM, Costa J. Kinetic analysis of L1 homophilic interaction: role of the first four immunoglobulin domains and implications on binding mechanism. J Biol Chem. 2008;283:28038–47. doi: 10.1074/jbc.M804991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortsch M, Wang YM, Marikar Y, Bieber AJ. The cytoplasmic domain of the Drosophila cell adhesion molecule neuroglian is not essential for its homophilic adhesive properties in S2 cells. J Biol Chem. 1995;270:18809–17. doi: 10.1074/jbc.270.32.18809. [DOI] [PubMed] [Google Scholar]
- Inohara H, Raz A. Functional evidence that cell surface galectin-3 mediates homotypic cell adhesion. Cancer Res. 1995;55:3267–71. [PubMed] [Google Scholar]
- Kim YS, Kang HY, Kim JY, Oh S, Kim CH, Ryu CJ, Miyoshi E, Taniguchi N, Ko JH. Identification of target proteins of N-acetylglucosaminyl transferase V in human colon cancer and implications of protein tyrosine phosphatase kappa in enhanced cancer cell migration. Proteomics. 2006;6:1187–91. doi: 10.1002/pmic.200500400. [DOI] [PubMed] [Google Scholar]
- McArdle L, Rafferty M, Maelandsmo GM, Bergin O, Farr CJ, Dervan PA, O’Loughlin S, Herlyn M, Easty DJ. Protein tyrosine phosphatase genes downregulated in melanoma. J Invest Dermatol. 2001;117:1255–60. doi: 10.1046/j.0022-202x.2001.01534.x. [DOI] [PubMed] [Google Scholar]
- Nose A, Mahajan VB, Goodman CS. Connectin: a homophilic cell adhesion molecule expressed on a subset of muscles and the motoneurons that innervate them in Drosophila. Cell. 1992;70:553–67. doi: 10.1016/0092-8674(92)90426-d. [DOI] [PubMed] [Google Scholar]
- Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- Paschos KA, Canovas D, Bird NC. The role of cell adhesion molecules in the progression of colorectal cancer and the development of liver metastasis. Cell Signal. 2009;21:665–74. doi: 10.1016/j.cellsig.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Phan D, Han E, Birrell G, Bonnal S, Duggan L, Esumi N, Gutstein H, Li R, Lopato S, Manogaran A, et al. Purification and characterization of human cell--cell adhesion molecule 1 (C-CAM1) expressed in insect cells. Protein Expr Purif. 2001;21:343–51. doi: 10.1006/prep.2000.1382. [DOI] [PubMed] [Google Scholar]
- Pulido D, Campuzano S, Koda T, Modolell J, Barbacid M. Dtrk, a Drosophila gene related to the trk family of neurotrophin receptors, encodes a novel class of neural cell adhesion molecule. Embo J. 1992;11:391–404. doi: 10.1002/j.1460-2075.1992.tb05067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sap J, Jiang YP, Friedlander D, Grumet M, Schlessinger J. Receptor tyrosine phosphatase R-PTP-κ mediates homophilic binding. Mol Cell Biol. 1994;14:1–9. doi: 10.1128/mcb.14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinza-Kameda M, Takasu E, Sakurai K, Hayashi S, Nose A. Regulation of layer-specific targeting by reciprocal expression of a cell adhesion molecule, capricious. Neuron. 2006;49:205–13. doi: 10.1016/j.neuron.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Snow PM, Bieber AJ, Goodman CS. Fasciclin III: a novel homophilic adhesion molecule in Drosophila. Cell. 1989;59:313–23. doi: 10.1016/0092-8674(89)90293-6. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–35. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- Yan HX, Yang W, Zhang R, Chen L, Tang L, Zhai B, Liu SQ, Cao HF, Man XB, Wu HP, et al. Protein-tyrosine phosphatase PCP-2 inhibits beta-catenin signaling and increases E-cadherin-dependent cell adhesion. J Biol Chem. 2006;281:15423–33. doi: 10.1074/jbc.M602607200. [DOI] [PubMed] [Google Scholar]
- Yu J, Becka S, Zhang P, Zhang X, Brady-Kalnay SM, Wang Z. Tumor-derived extracellular mutations of PTPRT/PTPrho are defective in cell adhesion. Mol Cancer Res. 2008;6:1106–13. doi: 10.1158/1541-7786.MCR-07-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo A, Yu J, Possemato A, Chen Y, Zheng W, Polakiewicz RD, Kinzler KW, Vogelstein B, Velculescu VE, et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci U S A. 2007;104:4060–4. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang X, Guda K, Lawrence E, Sun Q, Watanabe T, Iwakura Y, Asano M, Wei L, Yang Z, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.0914884107. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondag GC, Koningstein GM, Jiang YP, Sap J, Moolenaar WH, Gebbink MF. Homophilic interactions mediated by receptor tyrosine phosphatases mu and kappa. A critical role for the novel extracellular MAM domain. J Biol Chem. 1995;270:14247–50. doi: 10.1074/jbc.270.24.14247. [DOI] [PubMed] [Google Scholar]