Abstract
The objective of this study was to develop a symptom scoring system for use in clinical studies that differentiates children with cold symptoms who have an identifiable viral etiology for their upper respiratory tract infection (URI) from those in whom no virus is detected. Nasal swabs for PCR testing for identification of respiratory viruses were obtained on children aged 2–11 y at baseline and when parents thought their child was developing a cold. Parental-recorded severity of specific symptoms in children with and without a documented viral URI were compared. Nasal swabs were obtained on 108 children whose parents reported their child was developing a cold. A viral etiology was identified in 62 of 108 (57.4%) samples. Symptom measures that best differentiated children with a viral etiology from those without were significant runny nose and significant cough on days 1–4 of the illness. A URI symptom score was developed based on these symptoms, with a sensitivity of 81.4%, specificity of 61.9%, and accuracy of 73.3%. Parental impression is only a moderately accurate predictor of viral URI in children. Our URI symptom score provided a more accurate method for identifying children with viral URIs for clinical studies.
Main
The common cold is a ubiquitous feature of childhood. The average child develops up to 6–8 colds per year, each lasting 7–9 d (1). Cold symptoms are an extraordinarily common reason for health care utilization in pediatric patients (2,3). Beginning in the 1950s, investigators discovered rhinovirus and other viruses that cause the common cold (4,5). Subsequently, the term “viral upper respiratory tract infection” (viral URI) has become virtually synonymous with “cold” for describing the illness.
In many settings, differentiating a viral URI from other respiratory illnesses may be of trivial importance. However, for epidemiologic studies of URI or randomized controlled trials of various cold remedies, it may be critically important to more accurately differentiate patients with an actual viral infection from those with other respiratory conditions. The “gold standard” for diagnosing a viral URI, PCR (6,7), is expensive and requires access to sophisticated laboratory facilities, which limit its applicability for large clinical or epidemiologic studies. Thus, other methodologies for identifying patients with viral URI may be more practical.
Over a half century ago, Jackson et al. (8) developed a symptom scoring system to differentiate adults with viral URIs from other conditions. For that study, adult volunteers were randomized to have either saline solution or saline solution containing filtered nasal secretions from individuals with “typical colds” instilled in their noses. For the next 14 d, the study participants rated the severity of 12 symptoms of URI on a 0–3 scale. Those who received the filtered nasal secretions had significantly more severe symptoms than those receiving saline alone; a total symptom severity score of ≥14 was found to be the best cut-off value for defining a URI. This symptom scoring system, commonly referred to as the “Jackson score,” continues to be used in studies on URIs in adults (9). Because the presence of some of the symptoms in the score, such as chilliness and myalgia, may be difficult to detect in preverbal children, the use of the Jackson score in pediatric studies may be limited. Other investigators have characterized symptom patterns in children and adults with known viral URIs (6,10,11). There has been little effort to differentiate symptoms among pediatric patients with a documented viral URI from those of children who have some features of a URI but in whom no virus is detected. Thus, there is a need for a validated symptom checklist, or prediction rule, that accurately predicts the presence of a community-acquired viral URI in children enrolled in clinical studies.
As a precursor to a planned randomized controlled trial of the efficacy of Echinacea purpurea in preventing viral URIs in pediatric patients, we collected respiratory samples using nasal swabs on children with a clinical cold for PCR testing against respiratory viruses. Parents of these children completed comprehensive symptom diaries during the episode. Our goal was to identify a set of symptoms that could be used to develop a prediction rule that differentiated children with a viral URI from those with negative viral testing.
METHODS
A prospective observational study was conducted by the Puget Sound Pediatric Research Network (PSPRN) and Bastyr University. PSPRN is a regional practice-based research network of community pediatric practices in the Puget Sound area; for this project, seven offices participated. Bastyr University is an academic alternative medical school located in Kenmore, WA. At PSPRN practices, participating providers discussed the study with parents of potentially eligible children; if the parent was interested, contact information was sent to the research assistants who explained the study in detail and completed the enrollment procedures. Bastyr University used a generalized recruitment strategy to enroll patients by advertisements.
Study participants.
Eligible children were aged 2–11 y, in good health, and with no history of chronic lung disease. Eligibility was based on the inclusion/exclusion criteria planned for a subsequent randomized trial of Echinacea. Thus, patients with a history of asthma, allergic rhinitis, autoimmune disease, or allergy to Echinacea or related species were not enrolled. Each participant was in the study until she/he developed a cold or completed a 120-d observation period without developing cold symptoms. Study data were collected from November 2007 to May 2008. The study was approved by the Seattle Children's Hospital Institutional Review Board and the Bastyr University Institutional Review Board. Written informed consent was obtained from the parents of study children; written assent was obtained from children aged 7 y and older.
Study design.
At enrollment, parents completed a study form that included demographic information, use of daycare > 20 h/wk, school attendance, and exposure to cigarette smokers. In addition, a baseline nasal swab for PCR testing was collected. For patients with URI symptoms at the baseline visit, the baseline swab collection was rescheduled at least 14 d later. At enrollment, parents were given a cold symptom diary and asked to notify the research team as soon as they thought that their child was developing a cold.
Research assistants telephoned parents of study patients every 14–17 d to remind them of the study procedures and ask about any cold symptoms in the child. If the parent indicated that the child had developed a cold, but that he/she had not contacted the study team, the patient was considered to have a “missed” cold and excluded from further analysis.
Parents notified a research assistant when they believed their child was developing a cold. The research assistant specifically inquired about the presence of four respiratory symptoms: runny nose, nasal congestion, cough, and sneezing. If the research assistant confirmed the child had one or more symptoms, he/she was considered to have a clinical cold, and arrangements were made to collect an “acute” nasal swab for PCR testing within 48 h.
Once the research assistant determined that a study child had met criteria for a clinical cold, the parent was asked to begin completing the daily symptom diary. The scoring system used in the diary was based on the Jackson score and other cold severity rating systems and included measures for 12 separate symptoms for each day of the cold, for up to 14 d (8,10,11). Symptoms included sneezing, runny nose, nasal congestion, cough, fever, headache, malaise, chilliness, scratchy throat, sore throat, hoarseness, and myalgias. Each morning, for as long as symptoms persisted, parents were asked to rate the severity of each of these symptoms using the scale: 0 = none, 1 = mild, 2 = moderate, and 3 = severe. An additional possible response was “can't tell,” which was added because it was felt that it would be difficult for parents of young children to rate the presence and severity of some of these symptoms. In an attempt to standardize the reporting, a guide for rating severity of symptoms including examples of symptoms warranting a particular severity was provided to all parents of study children. The guide for rating symptoms is shown in Table 1.
Table 1.
Symptom severity guide distributed to parents of study children
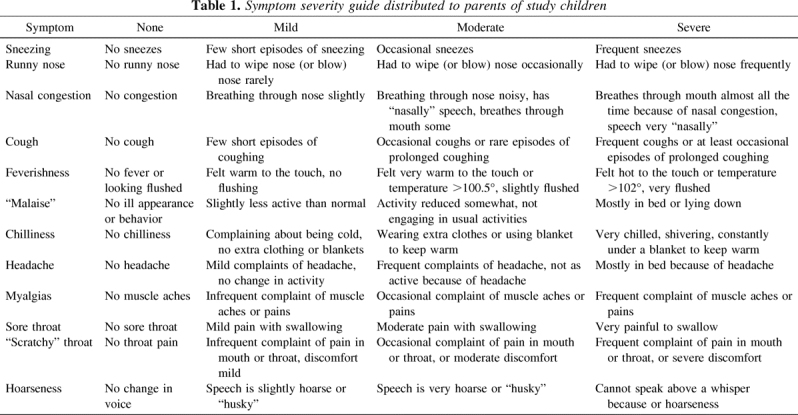
Initially, we planned to include data from only the first 3 d of recorded symptoms to develop a prediction rule for identifying a viral URI in a child. Ultimately, data from the first 4 d of symptoms were used to develop the prediction rule because inclusion of information from day 4 on symptoms improved the precision of the rule. Inclusion of data after day 4 did not increase the accuracy of the prediction rule.
Sample collection and testing.
A deep nasal swab, obtained by a small flexible nylon flocked swab (Copan Diagnostics, Corona, CA) inserted approximately one half the distance between the nares and bridge of the nose, was used for all sample collections. After collecting the specimen, the swab was rinsed vigorously in 0.5 mL of lysis buffer. Samples were stored at room temperature until testing (12). Total nucleic acid was extracted from 200 μL of swab specimen in lysis buffer as described previously (13). One thousand copies/reaction of an external control was added to identify false-negative PCR results (14). Nasal swab samples were tested by quantitative PCR for respiratory syncytial virus A and B, human metapneumovirus (hMPV), influenza A and B, parainfluenza types 1, 2, 3, and 4, adenovirus, rhinovirus, and coronavirus (subtypes 229E, HKU1, NL63, and OC43) (7,13–16).
Nasal swabs were also tested for bocavirus. During the course of the study, evidence emerged that bocavirus does not seem to be an etiologic agent of viral URIs in children older than 2 y (17,18). Therefore, we did not include bocavirus in our analysis.
A study patient was classified as having a viral URI if one or more viruses were detected that had not been found in the child at the baseline swab. If the same virus was detected at the baseline and acute swab, the child was classified as having a viral URI only if at least a 2 log increase in viral load was present in the acute swab.
Data analysis.
An iterative process was used to develop the prediction rule for accurately classifying whether a study child had a viral URI (19,20). Ratings for each measured symptom for the first 4 d of the illness were coded into two variables. The first measure of each symptom was whether it was present, corresponding to scores of 0 versus scores of 1–3. The second measure was whether the symptom was present and of “significant severity,” corresponding to scores of 0–1 versus 2–3. With this procedure, 96 different variables defining the presence or absence of symptoms were created (12 symptoms × 2 measures/symptom × 4 d of assessment).
The rate of presence of each symptom measure in children with a PCR confirmed viral URI and those with a negative PCR (“no URI”) was then compared using χ2 tests. Those symptom measures for which rates in the two groups (viral URI or no URI) were different (p < 0.20) were classified as candidate symptom measures and included in the next step. For symptoms in which both severity measures (e.g., any runny nose and significant runny nose) were different in patients with and without viral URI, only the measure with the more significant difference was included. In addition, candidate symptom measures in which greater than 10% of parents indicated “can't tell” regarding the presence of the symptom were excluded.
Stepwise forward logistic regression was used to identify combinations of symptom measures that differentiated children with and without viral URIs. The final regression model included all candidate symptom measures associated with the outcome of viral URI (p < 0.30). To develop a “URI symptom score,” 1 point was given for the presence of each of the identified symptom measures; the total URI symptom score in a given patient was the sum of these points. Receiver-operating-characteristic (ROC) analysis was done to choose the cut-off score that maximized the accuracy of the cold score in predicting the presence of a viral URI. “Accuracy” was defined as the proportion of children with a viral URI with a “positive” URI symptom score plus the proportion with no URI with a negative score.
Before collecting study data, a decision was made to develop a URI symptom score measure that had “sensibility.” This term is used to describe a decision rule that has face validity to clinicians caring for children (19,20). For example, if a symptom measure on days 2 and 4 of illness was associated with the outcome of a viral URI in the regression model, it would be sensible to also include scores for that symptom on days 1 and 3. Thus, additional models for the URI symptom score were developed by including symptom measures that we judged to increase the sensibility of the prediction rule. Alternative models of the score were developed in which one or more symptom measures were removed until the final model for the URI symptom score with the highest accuracy in differentiating children with and without a viral URI was derived. A final model, using the Jackson score, (8) was also constructed. For the Jackson score, the sum of the severity of eight symptom measures for 6 d is calculated; a score ≥14 is considered as indicative of a viral URI.
After determining the symptom measures to be used in calculating each model of the URI symptom score, ROC analysis was done to determine the optimal cut-off value. Based on this cut-off value, the sensitivity, specificity, positive predictive value (PPV), negative predictive value, and accuracy of the model in predicting a viral URI were calculated; 95% confidence interval (CI) around the point estimates were also calculated. The sensitivity of the final URI symptom score in predicting the presence of viral URIs caused by rhinoviruses (versus no URI) was determined. A similar analysis was conducted for viral URIs caused by viruses other than rhinovirus.
RESULTS
A total of 150 children were enrolled in the study including 126 by PSPRN pediatricians and 24 by the research team at Bastyr University. Disposition of study patients is summarized in Figure 1. Acute nasal swabs for PCR analysis were obtained in 108 patients whose parents thought that their child was developing a cold and had at least one cold symptom. The rest of the analysis is focused on the 108 children in whom an acute nasal swab was obtained. Demographic characteristics for these 108 children are summarized in Table 2. No differences for any characteristics between children who had an acute nasal swab and those who did not were noted. There were also no statistically significant differences in demographic characteristics between the 108 children in whom acute swabs were obtained and the eight participants who had a “missed” cold.
Figure 1.

Disposition of children enrolled in the study.
Table 2.
Characteristics of 108 study children in whom a nasal swab was obtained for PCR testing during an acute illness

No virus was detected at baseline in 87 of the 108 children (80.6%) who had a nasal swab collected for an acute illness. Viruses detected at baseline in the remaining 21 children are shown in Table 3. Five patients had rhinovirus detected at both baseline and with the acute nasal swab. In one child, there was >2 log increase in viral load from baseline to the acute swab; this patient was classified as having a viral URI. The other four patients had no substantial increase in rhinovirus load and were categorized as having no URI. In all other “positive” acute swabs, the virus detected had not been present at baseline. Overall, a respiratory viral etiology was identified in 62 of the 108 children (57.4%, 95% CI: 47.5–66.9%) considered to be developing a cold with at least one respiratory symptom (Table 3). Rhinovirus was the most common etiologic agent, accounting for 49.2% of all viruses detected.
Table 3.
Viruses identified in 108 study children who had nasal swab samples obtained at baseline and during an acute illness

Completed symptom logs were returned by parents of 101 study children (93.5%). In addition, information from symptom logs was also collected by research assistants when acute swabs were obtained. Data on symptom severity were obtained on all 108 study children on day 1 of their cold symptoms, on 106 children (98.1%) on day 2, 104 (96.2%) on day 3, and 101 on day 4 (93.5%).
The presence of 96 separate symptom measures in study children were compared. Twenty-one symptom measures met criteria for inclusion as candidate symptom measures (Table 4). Differences in the rates of significant runny nose and significant cough were present during the first 4 d of the illness. The most significant difference between the two groups was in the rate of significant runny nose on day 4 (50.9% in those with viral URI and 9.5% in those with no URI, p < 0.001). Although children with a viral URI were less likely to have any headache or significant headache on day 1 than those with no URI (p = 0.035 and 0.048, respectively), parents of 14 children (age, 2.2–5.2 y) indicated they could not tell whether their child had this symptom. Two parents did not respond to this item on the symptom log, leading to missing headache data in 14.8% of study children. Similarly, 13 parents (12.3%) indicated they could not tell whether their child had a scratchy throat (or sore throat) on day 2. Because of the amount of missing data, neither headache nor scratchy throat was considered as candidate symptom measures.
Table 4.
Rates of the presence of symptoms in children with, and without, a viral URI

All candidate variables were entered into stepwise forward logistic analysis; the following symptom measures were included in the final model: significant runny nose, day 1; significant cough, day 4; significant runny nose, day 4; and any fever, day 3. Based on these symptoms, three models for a URI symptom score were developed. For each model, a score was derived by assigning one point for the presence of each symptom included in the measure. ROC analysis was then used to determine the cut-off score that maximized the accuracy of the model. The properties of each model as a URI symptom score are summarized in Table 5. Model 1 included only the symptom measures included in the final forward regression analysis. For model 2, in which the “sensibility” of the prediction rule was improved, symptom measures for significant cough on days 1–3, significant runny nose on days 2 and 3, and any fever on days 1–2 and day 4 were included. Using systematic removal of different measures, the inclusion of any or all the fever symptom measures tended to decrease the precision of the prediction rule. Model 3, which included the symptom measures of significant runny nose on days 1–4 and significant cough on days 1–4, was the most accurate model for differentiating children with a viral URI from those with no URI and was chosen as the final URI symptom score. Model 4 was based on the Jackson score. However, because many parents responded “can't tell” for severity of certain symptoms, the score was calculated on only 65 children.
Table 5.
Diagnostic properties of different models for a URI symptom score to differentiate symptomatic children in whom a respiratory virus was identified from those in whom no viral etiology was detected

The overall sensitivity of the final URI symptom score was 81.4%, the specificity was 61.9%, and the accuracy was 73.3%. The sensitivity of the URI symptom score for viral URIs caused by rhinovirus was 76.7% (95% CI: 57.7–90.0%) and 86.2% (95% CI: 68.3–96.1%) for those caused by other viruses (p = 0.35).
DISCUSSION
In this study, nasal samples from children with clinical colds were collected to detect common respiratory viruses associated with viral URIs in pediatric patients. PCR techniques were used to identify viruses, a technique more sensitive than other detection or culture methods (6,7). Because children may have prolonged shedding with some respiratory viruses, particularly rhinovirus, in the absence of symptoms (21,22), we collected specimens on study participants both when they were well and when they were symptomatic. A child was classified as having a viral URI when either a new virus was isolated or a significant increase in viral load of a previously identified virus was detected in the presence of acute symptoms. With these techniques, we identified a virus in 57.4% of children whose parents thought their child was developing a cold and had at least one respiratory symptom.
Although it may seem surprising that a viral etiology was not identified in >40% of children with cold symptoms using the most sensitive molecular techniques currently available, the definition of a “cold” is actually somewhat nebulous (9). In one study of 215 adults who reported they were developing a cold, only 54% had at least one respiratory symptom and only 17% met Jackson criteria for a cold (23). Thus, the use of a “clinical cold” as the entry criterion for clinical trials on the efficacy of a potential viral URI treatment includes participants with heterogeneous clinical conditions and may bias findings toward the null if the therapy being tested is specifically acting by an antiviral mechanism. If a symptom scoring system could more reliably predict a viral URI, analysis of data from clinical studies could be conducted to assess efficacy in the subset of patients with a probable viral etiology. This approach would have the benefit of decreasing heterogeneity of the sample and eliminating the need for expensive laboratory testing to confirm viral etiology in clinical studies.
Viral URIs may be more difficult to identify in young children, who cannot articulate the presence of many symptoms. In our study, greater than 10% of parents indicated they could not tell whether their child had a headache or sore throat, both components of the Jackson score for defining a cold (8). Furthermore, the diagnostic properties in patients in whom the Jackson score could be calculated were not as good as our URI symptom score. This confirms our assertion that the Jackson score is not appropriate for clinical trials in children, and a new symptom scoring system is needed to identify viral URIs in pediatric patients.
With the use of our prediction rule, the PPV for identifying a child with a documented viral URI (75%) was similar to that found in other studies in both pediatric patients and adults using a variety of detection methods (6,24,25). In these previous studies, the estimated PPV may actually have been somewhat inflated because there was no baseline viral screening. Thus, in prior studies, a proportion of the viruses detected could potentially be related to prolonged shedding rather than as the cause of symptoms (26). In our study, 8.7% of viruses detected during an acute episode had been present at baseline without a significant increase in viral load.
Some study children may have been infected by a virus that is yet to be discovered leading to a false-negative test. It is also possible that some false-negative test results were caused by the technique used to obtain nasal secretions, although every effort was made to collect a proper sample. We may have detected more viruses if both culture and PCR testing had been used, although the sensitivity of PCR is known to exceed that of culture in most studies. For example, in one study, PCR tests consistently and substantially increased detection of multiple respiratory viruses compared with culture, and, in our laboratory, the use of PCR detected at least one respiratory virus in 53.4% of subjects compared with 38.3% who had virus detected by the use of direct immunofluorescence (7,27).
One limitation of this study is that the diagnostic utility of our URI symptom score may be different in another population of children (28). It is also possible that the utility of the symptom score would be different at another time when the distribution of circulating respiratory viruses is different. Subsequent investigation is needed to validate the URI symptom score that we developed. However, we purposefully incorporated a number of features into the development of the score that were designed to maximize its generalizability. First, rather than including symptom measures based solely on a statistical analysis, we modified the URI symptom score to have clinical “sensibility.” The precision of the URI symptom score could have been increased in this particular population by weighting some symptoms (29). We chose the more conservative approach of giving equal weight to relevant symptoms to minimize the effects of statistical quirks of the data set. Finally, we only included symptom measures for which >90% of parents could adequately rate severity. Because measures included in the URI symptom score (significant runny nose and cough) are intuitively important, it is more likely that the score will perform similarly in other children than if a more complicated prediction rule had been developed.
The results of this study indicate that parental perception of a child developing a cold and the presence of respiratory symptoms is a moderately accurate predictor of a viral URI; a respiratory virus was detected in 57.4% of children who met these criteria. Predicting which child has a viral URI was substantially increased by using our URI symptom score. The use of this symptom score as an analytic criterion for children enrolled in trials of remedies to treat and/or prevent viral URIs should increase the chance of detecting the efficacy of study interventions in the future.
Glossary
- PPV
positive predictive value
- PSPRN
Puget Sound Pediatric Research Network
- URI
upper respiratory tract infection
Footnotes
Supported by the National Center for Complementary and Alternative Medicine (U01AT002400).
References
- 1.Dingle JH, Badger GF, Jordan WS Jr 1964 Common respiratory diseases. In: Dingle JH, Badger GF, Jordan WS Jr (eds) Illness in the Home. A Study of 25,000 Illnesses in a Group of Cleveland Families. Press of Western Reserve University, Cleveland, pp 33–96
- 2.Aitken M, Taylor JA. Prevalence of clinical sinusitis in young children followed up by primary care pediatricians. Arch Pediatr Adolesc Med. 1998;152:244–248. doi: 10.1001/archpedi.152.3.244. [DOI] [PubMed] [Google Scholar]
- 3.Taylor JA, Kwan-Gett TS, McMahon EM., Jr Effectiveness of a parental educational intervention in reducing antibiotic use in children: a randomized controlled trial. Pediatr Infect Dis J. 2005;24:489–493. doi: 10.1097/01.inf.0000164706.91337.5d. [DOI] [PubMed] [Google Scholar]
- 4.Monto AS. Epidemiology of viral respiratory infections. Am J Med. 2002;112:4S–12S. doi: 10.1016/S0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 5.Gwaltney JM. Viral respiratory infection therapy: historical perspectives and current trials. Am J Med. 2002;112:33S–41S. doi: 10.1016/S0002-9343(01)01062-2. [DOI] [PubMed] [Google Scholar]
- 6.Arruda E, Pitkaranta A, Witek TJ, Jr, Doyle CA, Hayden FG. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997;35:2864–2868. doi: 10.1128/jcm.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuypers J, Wright N, Ferrenberg J, Huang ML, Cent A, Corey L, Morrow R. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol. 2006;44:2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson GG, Dowling HF, Spiesman IG, Boand AV. Transmission of the common cold to volunteers under controlled conditions. I. The common cold as a clinical entity. AMA Arch Intern Med. 1958;101:267–278. doi: 10.1001/archinte.1958.00260140099015. [DOI] [PubMed] [Google Scholar]
- 9.Barrett B, Brown R, Voland R, Maberry R, Turner R. Relations among questionnaire and laboratory measures of rhinovirus infection. Eur Respir J. 2006;28:358–363. doi: 10.1183/09031936.06.00002606. [DOI] [PubMed] [Google Scholar]
- 10.Gwaltney JM, Jr, Hendley JO, Simon G, Jordan WS., Jr Rhinovirus infections in an industrial population. II. Characteristics of illness and antibody response. JAMA. 1967;202:494–500. doi: 10.1001/jama.1967.03130190100014. [DOI] [PubMed] [Google Scholar]
- 11.Pappas DE, Hendley JO, Hayden FG, Winther B. Symptom profile of common colds in school-aged children. Pediatr Infect Dis J. 2008;27:8–11. doi: 10.1097/INF.0b013e31814847d9. [DOI] [PubMed] [Google Scholar]
- 12.Kunz AN, Englund JA, Kuypers J, Maranich A, Fairchok MP. Detection of multiple respiratory viruses by real-time polymerase chain reaction in infants attending an outpatient clinic. Eur J Clin Microbiol Infect Dis. 2008;27:1245–1248. doi: 10.1007/s10096-008-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuypers J, Wright N, Corey L, Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J Clin Virol. 2005;33:299–305. doi: 10.1016/j.jcv.2004.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuypers J, Wright N, Morrow R. Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J Clin Virol. 2004;31:123–129. doi: 10.1016/j.jcv.2004.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119:e70–e76. doi: 10.1542/peds.2006-1406. [DOI] [PubMed] [Google Scholar]
- 16.Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackay IM. Human bocavirus: multisystem detection raises questions about infection. J Infect Dis. 2007;196:968–970. doi: 10.1086/521311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longtin J, Bastien M, Gilca R, Leblanc E, de Serres G, Bergeron MG, Boivin G. Human bocavirus infections in hospitalized children and adults. Emerg Infect Dis. 2008;14:217–221. doi: 10.3201/eid1402.070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laupacis A, Sekar N, Stiell IG. Clinical prediction rules. A review and suggested modifications of methodological standards. JAMA. 1997;277:488–494. doi: 10.1001/jama.1997.03540300056034. [DOI] [PubMed] [Google Scholar]
- 20.Stiell IG 2001 The development of clinical decision rules for injury care. In: Rivara FP, Cummings P, Koepsell TD, Grossman DC, Maier RV (eds) Injury Control: A Guide to Research and Program Evaluation. Cambridge University Press, New York, pp 217–235
- 21.Alper CM, Doyle WJ, Winther B, Hendley JO. Upper respiratory virus detection without parent-reported illness in children is virus-specific. J Clin Virol. 2008;43:120–122. doi: 10.1016/j.jcv.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Zalm MM, van Ewijk BE, Wilbrink B, Uiterwaal CS, Wolfs TF, van der Ent CK. Respiratory pathogens in children with and without respiratory symptoms. J Pediatr. 2009;154:396–400. doi: 10.1016/j.jpeds.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zitter JN, Mazonson PD, Miller DP, Hulley SB, Balmes JR. Aircraft cabin air recirculation and symptoms of the common cold. JAMA. 2002;288:483–486. doi: 10.1001/jama.288.4.483. [DOI] [PubMed] [Google Scholar]
- 24.Legg JP, Warner JA, Johnston SL, Warner JO. Frequency of detection of picornaviruses and seven other respiratory pathogens in infants. Pediatr Infect Dis J. 2005;24:611–616. doi: 10.1097/01.inf.0000168747.94999.aa. [DOI] [PubMed] [Google Scholar]
- 25.Makela MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, Blomqvist S, Hyypia T, Arstila P. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 27.Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bleeker SE, Moll HA, Steyerberg EW, Donders AR, Derksen-Lubsen G, Grobbee DE, Moons KG. External validation is necessary in prediction research: a clinical example. J Clin Epidemiol. 2003;56:826–832. doi: 10.1016/S0895-4356(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 29.McIntosh MW, Pepe MS. Combining several screening tests: optimality of the risk score. Biometrics. 2002;58:657–664. doi: 10.1111/j.0006-341X.2002.00657.x. [DOI] [PubMed] [Google Scholar]


