Abstract
Background
Cytochrome P450 2A6 (CYP2A6) is the human enzyme responsible for the majority of nicotine’s metabolism. CYP2A6 genetic variants contribute to the inter-individual and inter-ethnic variation in nicotine metabolism. We examined the association between the CYP2A6*1B variant and nicotine’s in vivo metabolism.
Methods
Intravenous infusions of deuterium-labeled nicotine were administered to 292 volunteers, 163 of whom were White and did not have common CYP2A6 variants, other than CYP2A6*1B.
Results
We discovered three novel CYP2A6*1B variants in the 3′-flanking region of the gene that can confound genotyping assays. We found significant differences between CYP2A6*1A/*1A, CYP2A6*1A/*1B and CYP2A6*1B/*1B groups in total nicotine clearance (17.2±5.2, 19.0±6.4 and 20.4±5.9, P < 0.02), nonrenal nicotine clearance (16.4±5.0, 18.5±6.2 and 19.8±5.7, P < 0.01) and the plasma 3HC/COT ratio (0.26±0.1, 0.26±0.1 and 0.34±0.1, P < 0.001). There were also differences in total nicotine (29.4±12.9, 25.8±0.12.9 and 22.4±12.4, P < 0.01), cotinine (29.2±8.1, 32.2±9.1 and 33.0±6.6, P < 0.01) and trans-3′-hydroxcotinine (32.4±9.1, 34.2±12.3 and 41.3±11.3, P < 0.001) excreted in the urine.
Conclusion
We report evidence that CYP2A6*1B genotype is associated with faster nicotine clearance in vivo, which will be important to future CYP2A6 genotype association studies.
INTRODUCTION
Cytochrome P450 2A6 (CYP2A6) is a human hepatic enzyme that metabolizes clinically and toxicologically relevant substrates. It is responsible for the metabolism of pharmaceuticals such as coumarin1, the platelet activating factor antagonist SM-125022 and the chemotherapeutic pro-drug tegafur.3 CYP2A6 also activates pro-carcinogens such as aflatoxin-B14 and tobacco-specific nitrosamines.5, 6 Perhaps the most notable substrate of CYP2A6 is nicotine, which is the psychoactive and addictive compound implicated in tobacco dependence.7In vivo, approximately 80% of absorbed nicotine is inactivated to cotinine via C-oxidation8 and CYP2A6 mediates 90% of this reaction.9, 10 CYP2A6 is also responsible for 100% of cotinine’s hydroxylation to trans-3′-hydroxycotinine11; making the 3HC/COT ratio a good proxy measure of in vivo CYP2A6 function.12
Inter-individual and inter-ethnic differences in the effect and toxicity of CYP2A6 substrates can result from variability in the activity and/or amount of the enzyme13 due to CYP2A6 genetic variants.14 In fact, CYP2A6 genotype has been associated with smoking behavioral phenotypes (i.e. the number of cigarettes consumed)15, 16 and the likelihood of having tobacco-related cancers.17, 18 Currently 22 numbered allelic variants have been discovered in this highly polymorphic gene (http://www.cypalleles.ki.se/cyp2a6.htm), although not all are functionally characterized.
Not all of the CYP2A6 variants affect the enzyme’s structure and function. Those that do not change the coding region of the gene can potentially alter the amount of enzyme expressed. For example, the single nucleotide polymorphism (SNP) in the regulatory TATA box (CYP2A6*9) causes a 50% decrease in transcription in vitro.19 We have found that CYP2A6*9 is associated with lower in vivo CYP2A6 activity and nicotine clearance.20 Another variant that does not alter the CYP2A6 enzyme structure or function is the 58 bp CYP2A7 gene conversion in the 3′-untranslated region (3′-UTR) of the CYP2A6 gene, commonly referred to as CYP2A6*1B.21 CYP2A6*1B is thought to increase the stability of the mRNA transcript resulting in a greater amount of the CYP2A6 enzyme.22 The CYP2A6*1B alleles are numbered from 1–13 to distinguish between haplotype differences in SNPs located in the 5′-flanking region, the 3′-flanking region, the introns as well as synonymous SNPs in the exons; all CYP2A6*1B have the 58 bp CYP2A7 gene conversion.
In this report we present findings of three novel variants in the 3′-flanking region of CYP2A6, which are in linkage with CYP2A6*1B, and that confound existing genotyping assays. In addition, we investigate the association between CYP2A6*1B and the plasma kinetics of nicotine, as well as on the pattern of nicotine metabolites excreted in the urine, after intravenous infusion of deuterium-labeled nicotine in a White population.
METHODS
Study participants
A complete description of the study’s methods is available elsewhere.20, 23, 24 Genetic assessment was performed in all 292 participants from the volunteer-based North California Twin Registry (n = 222 White, n = 36 Hispanic, n = 15 Asian, n = 10 Black, and n = 9 of unknown ethnic origin). Allele frequencies were determined from one randomly selected co-twin from each of the monozygotic or dizygotic White twin pairs (n = 105). The White siblings (n = 12) were excluded from allele frequency analysis. Kinetic analyses herein were restricted to 163 White participants (119 monozygotic twins, 34 dizygotic twins and 10 siblings) assessed as having none of the following variant alleles CYP2A6*2, *4A&D, *7, *9, *10 *12 and *14. This restriction did not exclude the possibility of novel or non-tested CYP2A6 alleles. The 163 study participants had a mean age of 38 ± 1 years, 31% male (n = 51) and 16.6% were current smokers (n = 27). Of the 27 smokers, the number of cigarettes per day was available for 22 participants and the mean number of cigarettes smoked per day was 12 ± 2 with a range of 1 to 40. This study was approved by the Committees on Human Research at the University of California, San Francisco (San Francisco, California), SRI International (Menlo Park, California) and the University of Toronto (Toronto, Canada) and was conducted in accordance with the Declaration of Helsinki.
Nicotine and nicotine metabolite kinetics
The details of the experimental procedure are published elsewhere.20, 23, 24 Briefly, study participants received simultaneous 30-minute infusions of deuterium-labeled nicotine-d2 (3′,3′-dideuteronicotine) and cotinine-d4 (2,4,5,6-tetradeuterocotinine). The doses of nicotine-d2 and cotinine-d4 were individualized, and based on cotinine blood levels before the infusion. If baseline plasma cotinine was ≤ 50 ng/ml (nonsmokers or light smokers) participants received 0.5 μg/kg/min of nicotine-d2 and cotinine-d4 each, if plasma cotinine levels were 50 to 150 ng/ml (moderate smokers) participants received 1.0 μg/kg/min of nicotine-d2 and cotinine-d4 and if plasma cotinine levels were ≥ 150 ng/ml (heavy smokers) participants received 2.0 μg/kg/min of nicotine-d2 and cotinine-d4. Both twins, within a twin pair, received the same dose that was based on the lower plasma cotinine level. Of the total sample 93% received the lowest dose, 5% received the middle dose and 2% received the highest dose. The distribution of infusion doses did not vary by genotype group and the mean values for clearance did not vary by infusion dose. In addition to a baseline sample, after the start of the infusion ten blood samples were collected over 480 minutes for the measurement of plasma nicotine-d2 and cotinine-d4 and then daily four days after. Nicotine concentrations in the plasma were measured by gas chromatography–mass spectrometry (GC–MS), cotinine and trans-3′-hydroxycotinine were measured by GC-MS or by liquid chromatography–tandem mass spectrometry. Trans-3′-hydroxycotinine was only measured at the 180-minute time-point. All urine was collected over the eight-hour protocol. Concentrations of nicotine, cotinine, trans-3′-hydroxycotine and their respective glucuronides were measured in urine as the difference in analyte concentrations after and before enzymatic hydrolysis, as previously described.20
Pharmacokinetics Analysis
Pharmacokinetic parameters were estimated by use of model-independent methods 20 and are describe below:
Urine metabolites were analyzed as the percent of total molar nicotine and metabolites recovered in an eight-hour collection.
Sequencing
The full CYP2A6 gene and surrounding flanking regions were specifically amplified via polymerase chain reaction (PCR). The 9.2 kb PCR product was subcloned into a pCR-4 TOPOXL vector (TOPO TA Cloning XL® Kit for Sequencing, Invitrogen Canada Inc.) and sequenced using a walk-on strategy (The Centre for Applied Genomics, Toronto, Canada).
Genotyping
Based on sequencing results and the reference genomic DNA sequence (GenBank Accession number NG_000008.7), we developed allele-specific PCR assays to detect each of the three novel variants identified, all of which are in linkage with CYP2A6*1B and termed from here forward as CYP2A6*1B14, *1B15 and *1B16. In addition, we modified the CYP2A6*1B assay as the novel variants are situated where the 2A6R2 primer (used in previous CYP2A6*1B assays25) anneals.
All four assays use a common first PCR reaction (PCR-I) where a gene-specific region (2.9 kb) from intron 6 to the 3′-flanking region was amplified using the forward and reverse primers 2A6In6F1 5′-ATTTCCTGCTCTGAGACC-3′ and 2A6R0 5′-AGGTCATCTAGATTTTCTCCTACA-3′. The 25 μl reaction mixture consisted of: 1 X PCR Buffer (10 mM Tris pH8.8, 50 mM KCl), 0.2 mM of each dNTP, 1.5 mM MgCl2, 0.25 μM of each primer, 1.25 U of Taq polymerase (Fermentas, Life Sciences), 50 ng of genomic DNA and H2O. Initial denaturation was performed at 95°C for 1 min followed by 30 cycles, each consisting of denaturation at 95°C for 15 sec, annealing at 54°C for 20 sec and extension at 72°C for 180 sec (PTC-200 Peltier Thermal Cyclers). The second reaction for CYP2A6*1B (PCR-II) amplified an allele-specific region within the 3′-flanking region using the forward primers 2A6*1Bwt 5′-ACTGGGGGCAGGATGGC-3′ 25 and 2A6*1Bmut 5′-AATGGGGGGAAGATGCG-3′ 25 in combination with the reverse primer 2A6R4 5′-GCTTTTTAAGAATCTGTCTAGAA-3′ and produced a product of approximately 380 bp. The second PCR reactions for CYP2A6*1B14, *1B15 and *1B16 all amplified allele-specific regions from intron 7 to the 3′-region. CYP2A6 *1B14,*1B15 and *1B16 assays all used a common forward primer 2A6in7F 5′-ACCCACATTAGAAGCTTTCTAGA-3′ in combination with two reverse primers. CYP2A6*1B14 PCR-IIused the primers 2A6R14 5′-CAGAGGTTTGTGGCAATTAG-3′ and 2A7R14 5′-AGGTTTGTGGCAATTAAGTG-3′ and produced a 1.8 kb product. CYP2A6*1B15 used the primers 2A6R2 5′-AAAATGGGCATGAACGCCC-3′ 26 and 2A7R2 5′-AAAATGGGCATGAACGCTT-3′ and produced a 1.8 kb product. CYP2A6*1B16 used the primers 2A6R1-21 5′-GTTTTGTGAGACATCAGAGAC-3′ and 2A7R1-21 5′-TTTGTGAGACATCAGATAGAG-3′ and produced a 1.9 kb product. The four assays all had the same 25 μl PCR-II reaction mixtures which consisted of: 1 X PCR Buffer, 0.1 mM of each dNTP, 1.0 mM MgCl2, 0.25 μM of each primer, 0.3 U of Taq polymerase, 0.8 μl of undiluted PCR-I template and H2O. Initial denaturation was performed at 95°C for 1 min followed by 20 cycles, each consisting of denaturation at 95°C for 15 sec, annealing at 56°C, 60°C, 57°C and 57°C, respectively, for 20 sec, and extension at 72°C for 45 sec (for CYP2A6*1B) or 180 sec (for CYP2A6*1B14, *1B15 and *1B16). Aliquots (15 μl) of the PCR-II products were analyzed by electrophoresis using 1.2% agarose gel (OnBio, Richmond Hill, Canada) stained with ethidium bromide. The assays were developed using cosmids containing genomic clones of CYP2A6 as a positive control and CYP2A7 and CYP2A13 as negative controls.27, 28
Statistics
Chi-square was used to assess Hardy-Weinberg equilibrium for the CYP2A6*1B genotype. Kinetic parameter means were compared between CYP2A6 genotype groups (CYP2A6*1A/*1A, CYP2A6*1A/*1B and CYP2A6*1B/*1B) by use of the generalized linear models procedure in Stata software Release 9.29 All analyses controlled for the nonindependence of data in twin pairs by using a robust estimator for standard errors.30 This resulted in larger mean standard errors and minimized false-positive results. Where the main effect of genotype group was significant, the pairwise comparison test was accomplished by comparing the means of CYP2A6*1A/*1B and CYP2A6*1B/*1B to the referent group CYP2A6*1A/*1A using the z-statistic, and the regression coefficients of CYP2A6*1A/*1B and CYP2A6*1B/*1B were compared using Wald’s χ2-square test. To address the possible confounding effects of smoking status (current smokers versus former and never smokers), gender and age at the time of assessment, they were included as covariates in the models. Prior to analyses, all variables were log transformed. Levene’s test was used to assess for equality of variances across genotype groups.
RESULTS
We identified three novel CYP2A6*1B variants. These variants contained CYP2A7 gene conversions in the 3′-flanking region of CYP2A6 that were in linkage with the 58 bp CYP2A7 gene conversion in the 3′-UTR that is found in the CYP2A6*1B allele. The novel variant alleles were termed CYP2A6*1B14, *1B15 and *1B16 and their DNA sequences are shown in Figure 1. We developed assays for each new CYP2A6*1B allele (i.e. CYP2A6*1B14, *1B15 and *1B16) and modified the CYP2A6*1B1-13 genotyping assay. All 292 individuals in the study were genotyped for these alleles. The allele frequencies of the novel CYP2A6*1B alleles, as well as CYP2A6*1B1-13, were calculated from one randomly selected co-twin of each monozygotic or dizygotic White twin pair (n = 105) (siblings (n = 12) were excluded from genetic analyses) and are shown in Table I. In addition, two monozygotic co-twins of the eight Asian twin pairs were homozygous for the CYP2A6*1B16 allele. Among 18 Hispanic and 5 Black twin pairs no CYP2A6*1B14-16 alleles were found.
Fig 1.
Alignment of the 3′ regions of CYP2A6, CYP2A6*1B, the three novel haplotypes, CYP2A6*1B14-16 and the CYP2A7 pseudogene. Capital letters are amino acid coding region. Highlighted in grey are the regions of gene conversion. Bolded nucleotides differ between CYP2A6 and CYP2A7. Arrows are regions where primers anneal. Thick lines are regions of mRNA stabilization in the 3′-UTR.
Table I.
CYP2A6 allele frequencies in White subjects
| n alleles | % | |
|---|---|---|
| CYP2A6*1A | 126 | 60.0 |
| CYP2A6*1B1-13 | 76 | 36.2 |
| CYP2A6*1B14 | 1 | ‘0.5 |
| CYP2A6*1B15 | 7 | 3.3 |
| CYP2A6*1B1-15 | 84 | 40.0 |
| Total | 210 | 100.0 |
Of the 222 White subjects in this study, allele frequencies were determined from one randomly chosen twin of each pair (n = 105) and all siblings were excluded (n = 12). Genotype frequencies were consistent with Hardy-Weinberg equilibrium.
CYP2A6*1B16 was not found in White subjects.
The CYP2A6*1B15 and *1B16 have nucleotide insertions in regions where previous genotyping primers were designed to anneal (Figure 1). These two primers, 2A6R1 and 2A6R2, were designed by Oscarson and colleagues26 and have been used in the CYP2A6*1X2, *1B, *4, *5, *7 and *10 genotyping assays in a number of publications (e.g.25, 27, 31–33). The novel alleles confound genotyping assays as follows. If a variant is in linkage with the novel gene conversions in CYP2A6*1B15 or CYP2A6*1B16 (such as the 58 bp gene conversion in the 3′-UTR) then its frequency will be underestimated in assays that use the primers 2A6R1 and/or 2A6R2. Conversely, variants on the opposite allele will be overestimated. Essentially, based on the genotype frequency found in this population, in 5.7% of Whites, one allele would not amplify in assays where the 2A6R2 primer is used due to CYP2A6*1B15, resulting in an underestimation of CYP2A6*1B and an overestimation of the alternate allele.
The novel gene conversions in the alleles CYP2A6*1B14-16 are not in the coding region or in the 3′-UTR and therefore are not predicted to have further functional impact on the activity of the CYP2A6*1B allele. CYP2A6*1A/*1B15 (n = 8) were similar to the CYP2A6*1A/*1B1-13 (n = 77) with respect to their disposition kinetics of nicotine. For example, the mean total nicotine clearance was 20.5 ± 9.7 mL/min/kg for CYP2A6*1B15 heterozygotes compared to 18.9 ± 6.0 mL/min/kg for CYP2A6*1B1-13 heterozygotes.
To investigate the postulated difference between kinetic parameters by CYP2A6 ‘wildtype’ genotype groups, namely CYP2A6*1A/*1A, CYP2A6*1A/*1B and CYP2A6*1B/*1B, we examined all CYP2A6*1A (those without the 3′-UTR 58 bp gene conversion) versus CYP2A6*1B (those with the 3′-UTR 58 bp gene conversion). There were significant group differences between CYP2A6*1A/*1A, *1A/*1B and *1B/*1B in the disposition kinetic profiles of nicotine (Table II). Total nicotine clearance (Figure 2A), nonrenal nicotine clearance and the 3HC/COT ratio (measure of CYP2A6 activity) (Figure 2B) showed an increase in activity with the number of CYP2A6*1B alleles. Both the total and the nonrenal clearance of nicotine were about 20% higher in the fastest group, CYP2A6*1B/*1B compared to CYP2A6*1A/*1A, whereas the 3HC/COT ratio was about 30% higher. Consistent with this the half-life of nicotine tended to decrease as the clearance of nicotine increased, although this did not reach statistical significance. The heterozygous CYP2A6*1B group tended to demonstrate intermediate pharmacokinetic parameters between the two homozygous groups.
Table II.
Disposition kinetics of nicotine by CYP2A6 wildtype genotypes
| CYP2A6 genotype | n | Total CLNIC | Renal CLNIC | Nonrenal CLNIC | CLNIC→COT | fc | t1/2 | Vss | 3HC/COT |
|---|---|---|---|---|---|---|---|---|---|
| *1A/*1A | 44–45§ | 17.2 (5.2) | 0.7 (0.9) | 16.4 (5.0) | 13.5 (4.9) | 0.78 (0.1) | 119.8 (41.0) | 2.5 (0.7) | 0.26 (0.1) |
| *1A/*1B | 83–85§ | 19.0 (6.4)† | 0.5 (0.5) | 18.5 (6.2)† | 15.4 (6.6) | 0.80 (0.1) | 110.1 (35.6) | 2.5 (0.8) | 0.26 (0.1) |
| *1B/*1B | 30–33§ | 20.4 (5.9)† | 0.5 (0.5) | 19.8 (5.7)† | 14.7 (5.4) | 0.72 (0.1) | 106.7 (33.0) | 2.7 (0.7) | 0.34 (0.1)†‡ |
| Overall P value | 0.02 | 0.65 | 0.01 | 0.33 | 0.09 | 0.06 | 0.32 | 0.001 |
Data are presented as mean (SD). Kinetic analysis was restricted to 163 ‘wildtype’ Whites. Total CLNIC – Clearance of nicotine-d2 (mL/min/kg); Renal CLNIC – Renal clearance of nicotine-d2 (mL/min/kg); Nonrenal CLNIC – Nonrenal clearance of nicotine-d2 (mL/min/kg); CLNIC→COT – Clearance of nicotine-d2 to cotinine-d2 (mL/min/kg); fc– Percent nicotine-d2 converted to cotinine-d2; t1/2 – half-life of nicotined2 (min); Vss – Steady state volume of distribution of nicotine-d2 (L/kg); 3HC/COT-d2 – plasma trans-3′-hydroxycotinine/cotinine ratio from nicotine-d2. The *1A/*1B and *1B/*1B groups contain the novel gene conversion alleles identified in this study (i.e. *1B14 and *1B15).
P < 0.05 compared with CYP2A6*1A/*1A
P < 0.05 compared with CYP2A6*1A/*1B
Indicates some missing values
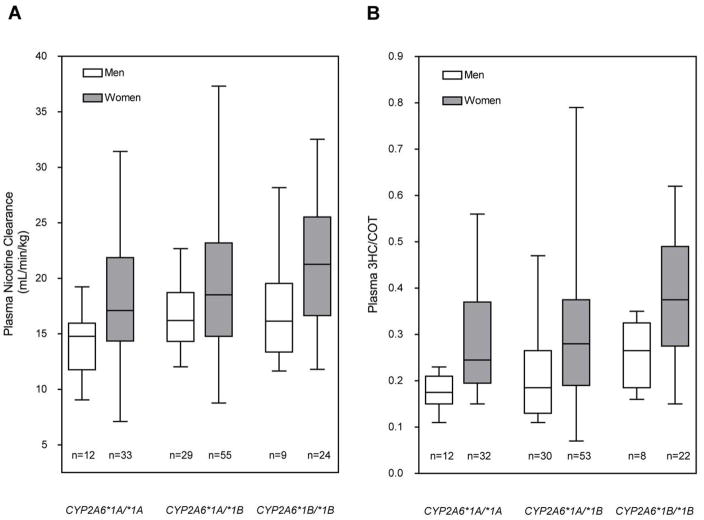
Fig 2.
CYP2A6*1B genotype-phenotype associations for (A) total plasma clearance of nicotine and (B) the 3HC/COT ratio. The box plot provides a visual summary of the distribution of the data. The box represents the interquartile range, which contains 50% of the values. The whiskers are lines that extend from the box to the highest and lowest values. The line across the box indicates the median. Genotype groups were split by gender to illustrate the faster total nicotine clearance and higher 3HC/COT ratio in women compared to men, as has been previously demonstrated.35 There was no statistically significant gender by genotype interaction.
The mean of the urine nicotine metabolite levels, according to genotype group, are shown in Table III. There were significant group differences in urinary excretion of total nicotine and its proximal metabolites, total cotinine and total trans-3′-hydroxycotinine, between groups. On average, CYP2A6*1A/*1A subjects excreted the highest levels of nicotine and the lowest levels of cotinine and trans-3′-hydroxycotinine. Conversely, but as expected based on the plasma data, the CYP2A6*1B/*1B group had the lowest levels of nicotine, the highest cotinine and the most trans-3′-hydroxycotinine. The difference between genotype groups in the percent recovery of nicotine and metabolites from urine is consistent with the finding that nicotine metabolism becomes faster with the number of CYP2A6*1B alleles.
Table III.
Nicotine urine metabolites by CYP2A6 wildtype genotype: percent recovery of nicotine and metabolites
| CYP2A6 genotype | n | Nicotine | Nicotine glucuronide |
Total Nicotine | Cotinine | Cotinine glucuronide |
Total Cotinine | 3HC | 3HC glucuronide |
Total 3HC glucuronide |
|---|---|---|---|---|---|---|---|---|---|---|
| *1A/*1A | 44 | 22.0 (14.6) | 7.3 (4.5) | 29.4 (12.9) | 20.4 (7.1) | 8.8 (6.0) | 29.2 (8.1) | 28.0 (8.2) | 4.4 (2.4) | 32.4 (9.1) |
| *1A/*1B | 85 | 18.1 (13.1) | 7.7 (4.9) | 25.8 (12.9) | 24.1 (8.3)† | 8.2 (4.5) | 32.2 (9.1)† | 29.9 (11.2) | 4.3 (3.6) | 34.2 (12.3) |
| *1B/*1B | 33 | 16.6 (12.3) | 5.9 (3.0) | 22.4 (12.4)† | 24.2 (8.3)† | 8.8 (4.1) | 33.0 (6.6)† | 36.3 (9.8)†‡ | 5.0 (2.9) | 41.3 (11.3)†‡ |
| Overall P value | 0.10 | 0.56 | 0.01 | 0.04 | 0.26 | 0.01 | 0.001 | 0.36 | 0.001 |
Data are presented as mean (SD) and are based on recovery of d2-labeled nicotine and metabolites in urine over an 8-hour period after the start of the infusion.
P < 0.05 compared with CYP2A6*1A/*1A
P < 0.05 compared with CYP2A6*1A/*1B
DISCUSSION
Here we present the identification of three novel CYP2A7 gene conversions in the 3′-flanking region of the CYP2A6 gene that are in linkage with the CYP2A6*1B allele. Two of them can confound genotyping assays that require the widely used primers 2A6R1 and 2A6R2.26 In addition, we present novel data on the disposition kinetics and metabolism of intravenous nicotine in individuals who, in most previous published studies, have been considered to have ‘wildtype’ or similar nicotine metabolizing capacities. Ours is the first explicit demonstration that the in vivo clearance of nicotine is faster in individuals with the CYP2A6*1B allele compared to the CYP2A6*1A allele.
There is high homology between the neighboring CYP2 genes particularly between CYP2A6 and the CYP2A7 pseudogene that shared 95% genomic nucleotide sequence identity. The similarity adds complexity to the technical aspects of genotyping and is the reason for the gene-specific first steps in our PCR genotyping assays. This report highlights an example where, as our knowledge of the CYP2A6 gene and its variants increased, our techniques evolve to improve the accuracy of the genotyping. In addition, our findings provide further evidence for the existence of domain swaps between the CYP2A6 and CYP2A7 genes. Of note, the three novel CYP2A7 gene conversions in linkage with the CYP2A6*1B allele, described here, are reciprocals of recently described CYP2A6 gene conversions found in the CYP2A7 gene.34
The apparent gender-genotype interaction (Figure 2) is an interesting observation, where it appeared that there may be a much smaller effect of the CYP2A6*1B allele in men compared to women (not statistically significant). In a prior study we have found that women have faster nicotine clearance compared to men, which has been attributed to greater CYP2A6 activity.35 Moreover, women on oral estrogen contraceptives have even faster nicotine metabolism.35 This suggests that hormones may function in the regulation of the gene. If estrogen is associated with greater mRNA levels in the women compared to men, then even if the fold increase in CYP2A6 expression due to the CYP2A6*1B allele is similar, the absolute effect size in the women would be larger. This, and the fact that there were more women in the study, would give more power to detect the difference between the CYP2A6*1A/*1A, CYP2A6*1A/*1B and CYP2A6*1B/*1B groups in women. Thus women show the gene-dose effect in both the total clearance of nicotine and 3HC/COT, while the data from the men may be somewhat underpowered. This gene-gender effect should be considered in gene association studies as well as in pharmacokinetic studies as the proportion of females may influence the findings.
As the rate of nicotine clearance becomes faster, the number of cigarettes and/or the amount of nicotine extracted per cigarette by the smoker increases.36 Here we show in vivo evidence of faster nicotine clearance in White individuals that have either one or two CYP2A6*1B alleles. Several lines of indirect evidence support our findings of faster nicotine clearance in individuals with the CYP2A6*1B allele. First, the 3′-UTR and the secondary structures of the 3′-UTR forms are involved in the posttranscriptional regulation of CYP2A.37 A number of proteins have been found in human hepatocytes that specifically interact with the 3′-UTR and stabilize CYP2A6 mRNA.38 For instance, the heterogenous nuclear ribonucleoprotein (hnRNP) A1 binds to an AG rich site within the UTR (Figure 1).38 Second, the 58 bp CYP2A7 gene conversion increased mRNA stability and gene expression in an in vitro luciferase reporter construct system.22 Third, there are higher mRNA, enzyme level and faster catalytic activity in the formation of 7-hydroxycoumarin from coumarin (CYP2A6 mediates 100% of coumarin’s 7-hydroxylation1) in CYP2A6*1B human liver microsomes.22 Finally, CYP2A6*1B heterozygotes have a higher mean cotinine/nicotine ratio (a proxy measure of CYP2A6 activity) and 7-hydroxycoumarin formation in vivo in Asian populations39 as well as higher 3HC/COT in individuals with the CYP2A6*1B allele.40
Our data explains, in part, some of the considerable variation in the traditional ‘wildtype’ group. The impact of CYP2A6*1B on nicotine clearance is relatively modest, as there was only a 20% difference between the CYP2A6*1A/*1A and CYP2A6*1B/*1B groups. However, it is the predominate variant allele in all ethnoracial groups.16 The CYP2A6*1B allele has been associated with greater tobacco dependence in White Brazilians41, an increased likelihood of being a smoker in Japanese42 and higher cigarette consumption in Whites.43 Our results provide a biological rationale for these findings.
Acknowledgments
This study was supported by a Canada Institute for Health research grant MOP53248, the Centre for Addiction and Mental Health, a Canada Research Chair in Pharmacogenetics and a SPICE scholarship (molecular identification of novel variants, assay design, genotyping, related analyses, data interpretation and preparation of the manuscript). Funding was also from Public Health Service grants DA11170, DA02277, DA12393 and DA20830 (design and conduct of the pharmacokinetic/pharmacogenetic study, analytical laboratory support and statistical analyses) awarded by the National Institute on Drug Abuse, and carried out in part at the General Clinical Research Center at San Francisco General Hospital with support of the Division of Research Resources NIH RR00083.
Footnotes
DISCLOSURE
Dr. Benowitz has been a paid consultant for several pharmaceutical companies that market smoking cessation medications. Dr. Tyndale holds shares in Nicogen Inc., a company focused on creating novel smoking cessation treatments. No funding for this study was received from Nicogen. The other authors have no potential conflicts to reports.
References
- 1.Miles JS, McLaren AW, Forrester LM, Glancey MJ, Lang MA, Wolf CR. Identification of the human liver cytochrome P-450 responsible for coumarin 7-hydroxylase activity. Biochem J. 1990;267:365–71. doi: 10.1042/bj2670365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nunoya K, et al. (+)-cis-3,5-dimethyl-2-(3-pyridyl) thiazolidin-4-one hydrochloride (SM-12502) as a novel substrate for cytochrome P450 2A6 in human liver microsomes. J Pharmacol Exp Ther. 1996;277:768–74. [PubMed] [Google Scholar]
- 3.Ikeda K, et al. Bioactivation of tegafur to 5-fluorouracil is catalyzed by cytochrome P-450 2A6 in human liver microsomes in vitro. Clin Cancer Res. 2000;6:4409–15. [PubMed] [Google Scholar]
- 4.Salonpaa P, et al. Retrovirus-mediated stable expression of human CYP2A6 in mammalian cells. Eur J Pharmacol. 1993;248:95–102. doi: 10.1016/0926-6917(93)90030-t. [DOI] [PubMed] [Google Scholar]
- 5.Patten CJ, Smith TJ, Friesen MJ, Tynes RE, Yang CS, Murphy SE. Evidence for cytochrome P450 2A6 and 3A4 as major catalysts for N′-nitrosonornicotine alpha-hydroxylation by human liver microsomes. Carcinogenesis. 1997;18:1623–30. doi: 10.1093/carcin/18.8.1623. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki H, Inui Y, Yun CH, Guengerich FP, Shimada T. Cytochrome P450 2E1 and 2A6 enzymes as major catalysts for metabolic activation of N-nitrosodialkylamines and tobacco-related nitrosamines in human liver microsomes. Carcinogenesis. 1992;13:1789–94. doi: 10.1093/carcin/13.10.1789. [DOI] [PubMed] [Google Scholar]
- 7.Benowitz NL. Drug therapy. Pharmacologic aspects of cigarette smoking and nicotine addition. N Engl J Med. 1988;319:1318–30. doi: 10.1056/NEJM198811173192005. [DOI] [PubMed] [Google Scholar]
- 8.Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56:483–93. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima M, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24:1212–7. [PubMed] [Google Scholar]
- 10.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;282:1608–14. [PubMed] [Google Scholar]
- 11.Yamanaka H, et al. Metabolic profile of nicotine in subjects whose CYP2A6 gene is deleted. Eur J Pharm Sci. 2004;22:419–25. doi: 10.1016/j.ejps.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Dempsey D, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Salguero P, et al. A genetic polymorphism in coumarin 7-hydroxylation: sequence of the human CYP2A genes and identification of variant CYP2A6 alleles. Am J Hum Genet. 1995;57:651–60. [PMC free article] [PubMed] [Google Scholar]
- 14.Daigo S, et al. A novel mutant allele of the CYP2A6 gene (CYP2A6*11) found in a cancer patient who showed poor metabolic phenotype towards tegafur. Pharmacogenetics. 2002;12:299–306. doi: 10.1097/00008571-200206000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11:400–9. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 16.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–26. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Ariyoshi N, et al. Genetic polymorphism of CYP2A6 gene and tobacco-induced lung cancer risk in male smokers. Cancer Epidemiol Biomarkers Prev. 2002;11:890–4. [PubMed] [Google Scholar]
- 18.Miyamoto M, et al. CYP2A6 gene deletion reduces susceptibility to lung cancer. Biochem Biophys Res Commun. 1999;261:658–60. doi: 10.1006/bbrc.1999.1089. [DOI] [PubMed] [Google Scholar]
- 19.Pitarque M, von Richter O, Oke B, Berkkan H, Oscarson M, Ingelman-Sundberg M. Identification of a single nucleotide polymorphism in the TATA box of the CYP2A6 gene: impairment of its promoter activity. Biochem Biophys Res Commun. 2001;284:455–60. doi: 10.1006/bbrc.2001.4990. [DOI] [PubMed] [Google Scholar]
- 20.Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80:457–67. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Ariyoshi N, et al. Structural characterization of a new variant of the CYP2A6 gene (CYP2A6*1B) apparently diagnosed as heterozygotes of CYP2A6*1A and CYP2A6*4C. Pharmacogenetics. 2000;10:687–93. doi: 10.1097/00008571-200011000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Pitarque M, Ingelman-Sundberg M. 3′-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem Biophys Res Commun. 2006;340:491–7. doi: 10.1016/j.bbrc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Swan GE, et al. Pharmacogenetics of nicotine metabolism in twins: methods and procedures. Twin Res. 2004;7:435–48. doi: 10.1375/1369052042335269. [DOI] [PubMed] [Google Scholar]
- 24.Swan GE, Benowitz NL, Lessov CN, Jacob P, 3rd, Tyndale RF, Wilhelmsen K. Nicotine metabolism: the impact of CYP2A6 on estimates of additive genetic influence. Pharmacogenet Genomics. 2005;15:115–25. doi: 10.1097/01213011-200502000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Oscarson M, et al. Identification and characterisation of novel polymorphisms in the CYP2A locus: implications for nicotine metabolism. FEBS Lett. 1999;460:321–7. doi: 10.1016/s0014-5793(99)01364-2. [DOI] [PubMed] [Google Scholar]
- 26.Oscarson M, et al. Characterisation and PCR-based detection of a CYP2A6 gene deletion found at a high frequency in a Chinese population. FEBS Lett. 1999;448:105–10. doi: 10.1016/s0014-5793(99)00359-2. [DOI] [PubMed] [Google Scholar]
- 27.Rao Y, et al. Duplications and defects in the CYP2A6 gene: identification, genotyping, and in vivo effects on smoking. Mol Pharmacol. 2000;58:747–55. doi: 10.1124/mol.58.4.747. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman SM, Nelson DR, Keeney DS. Organization, structure and evolution of the CYP2 gene cluster on human chromosome 19. Pharmacogenetics. 2001;11:687–98. doi: 10.1097/00008571-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 29.StataCorp. Stata Statistical Software Release 9. 2005. [Google Scholar]
- 30.White H. Maximum likelihood estimation of misspecified models. Econometrica. 1982;50:1–25. [Google Scholar]
- 31.Xu C, et al. An in vivo pilot study characterizing the new CYP2A6*7, *8, and *10 alleles. Biochem Biophys Res Commun. 2002;290:318–24. doi: 10.1006/bbrc.2001.6209. [DOI] [PubMed] [Google Scholar]
- 32.Huang S, et al. CYP2A6, MAOA, DBH, DRD4, and 5HT2A genotypes, smoking behaviour and cotinine levels in 1518 UK adolescents. Pharmacogenet Genomics. 2005;15:839–50. doi: 10.1097/01213011-200512000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Nurfadhlina M, Foong K, Teh LK, Tan SC, Mohd Zaki S, Ismail R. CYP2A6 polymorphisms in Malays, Chinese and Indians. Xenobiotica. 2006;36:684–92. doi: 10.1080/00498250600715932. [DOI] [PubMed] [Google Scholar]
- 34.Fukami T, Nakajima M, Sakai H, McLeod HL, Yokoi T. CYP2A7 polymorphic alleles confound the genotyping of CYP2A6*4A allele. Pharmacogenomics J. 2006;6:401–12. doi: 10.1038/sj.tpj.6500390. [DOI] [PubMed] [Google Scholar]
- 35.Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther. 2006;79:480–8. doi: 10.1016/j.clpt.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Benowitz NL, Jacob P., 3rd Nicotine renal excretion rate influences nicotine intake during cigarette smoking. J Pharmacol Exp Ther. 1985;234:153–5. [PubMed] [Google Scholar]
- 37.Glisovic T, Ben-David Y, Lang MA, Raffalli-Mathieu F. Interplay between hnRNP A1 and a cis-acting element in the 3′ UTR of CYP2A5 mRNA is central for high expression of the gene. FEBS Lett. 2003;535:147–52. doi: 10.1016/s0014-5793(02)03893-0. [DOI] [PubMed] [Google Scholar]
- 38.Christian K, Lang M, Maurel P, Raffalli-Mathieu F. Interaction of heterogeneous nuclear ribonucleoprotein A1 with cytochrome P450 2A6 mRNA: implications for post-transcriptional regulation of the CYP2A6 gene. Mol Pharmacol. 2004;65:1405–14. doi: 10.1124/mol.65.6.1405. [DOI] [PubMed] [Google Scholar]
- 39.Peamkrasatam S, et al. In vivo evaluation of coumarin and nicotine as probe drugs to predict the metabolic capacity of CYP2A6 due to genetic polymorphism in Thais. Drug Metab Pharmacokinet. 2006;21:475–84. doi: 10.2133/dmpk.21.475. [DOI] [PubMed] [Google Scholar]
- 40.Johnstone E, et al. Determinants of the rate of nicotine metabolism and effects on smoking behavior. Clin Pharmacol Ther. 2006;80:319–30. doi: 10.1016/j.clpt.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Vasconcelos GM, Struchiner CJ, Suarez-Kurtz G. CYP2A6 genetic polymorphisms and correlation with smoking status in Brazilians. Pharmacogenomics J. 2005;5:42–8. doi: 10.1038/sj.tpj.6500290. [DOI] [PubMed] [Google Scholar]
- 42.Iwahashi K, Waga C, Takimoto T. Whole deletion of CYP2A6 gene (CYP2A6AST;4C) and smoking behavior. Neuropsychobiology. 2004;49:101–4. doi: 10.1159/000076418. [DOI] [PubMed] [Google Scholar]
- 43.Gambier N, Batt AM, Marie B, Pfister M, Siest G, Visvikis-Siest S. Association of CYP2A6*1B genetic variant with the amount of smoking in French adults from the Stanislas cohort. Pharmacogenomics J. 2005;5:271–5. doi: 10.1038/sj.tpj.6500314. [DOI] [PubMed] [Google Scholar]