Abstract
Density estimation for observational data plays an integral role in a broad spectrum of applications, e.g., statistical data analysis and information-theoretic image registration. Of late, wavelet-based density estimators have gained in popularity due to their ability to approximate a large class of functions, adapting well to difficult situations such as when densities exhibit abrupt changes. The decision to work with wavelet density estimators brings along with it theoretical considerations (e.g., non-negativity, integrability) and empirical issues (e.g., computation of basis coefficients) that must be addressed in order to obtain a bona fide density. In this paper, we present a new method to accurately estimate a non-negative density which directly addresses many of the problems in practical wavelet density estimation. We cast the estimation procedure in a maximum likelihood framework which estimates the square root of the density , allowing us to obtain the natural non-negative density representation . Analysis of this method will bring to light a remarkable theoretical connection with the Fisher information of the density and, consequently, lead to an efficient constrained optimization procedure to estimate the wavelet coefficients. We illustrate the effectiveness of the algorithm by evaluating its performance on mutual information-based image registration, shape point set alignment, and empirical comparisons to known densities. The present method is also compared to fixed and variable bandwidth kernel density estimators.
Index Terms: Density estimation, Fisher information, Hellinger divergence, image matching, image registration, modified Newton’s method, mutual information, shape matching, square root density, wavelets
I. INTRODUCTION
Density estimation is a well-studied field, encompassing a myriad of techniques and theoretical formulations all with the common goal of utilizing the observed data to discover the best approximation to the underlying density that generated them. Methods range from simple histogramming to more statistically efficient kernel-based Parzen window techniques [1], [2].Within the last 20 years, the widespread use of wavelet analysis in applied mathematics and engineering has also made its way into statistical applications. The use of wavelets as a density estimator was first explored in [3]. Wavelet bases have the desirable property of being able to approximate a large class of functions (𝕃2). Specifically for density estimation, wavelet analysis is often performed on normed spaces that have some notion of regularity like Besov, Hölder, and Sobolev. From an empirical point of view, the utility of representing a density in a wavelet basis comes from the fact that they are able to achieve good global approximation properties due to their locally compact nature—a key property when it comes to modeling densities that contain bumps and/or abrupt variations. It is well known that wavelets are localized in both time and frequency and this “compactness” is highly desirable in density estimation, as well.
The basic idea behind wavelet density estimation (for 1-D data) is to represent the density p as a linear combination of wavelet bases
| (1) |
where x ∈ ℝ, ϕ(x) and ψ(x) are the scaling (a.k.a. father) and wavelet (a.k.a. mother) basis functions respectively, and αj0,k and βj,k are scaling and wavelet basis function coefficients; the j-index represents the current level and the k-index the integer translation value (the translation range of k can be computed from the span of the data and basis function support size [4]). Our goal then is to estimate the coefficients of the wavelet expansion and obtain an estimator p̂ of the density. This should be accomplished in a manner that retains the properties of the true density—notably the density should be non-negative and integrate to one. Typically, wavelet density estimators (WDE) are classified as linear and nonlinear. The term linear estimator denotes the fact that the coefficients are obtained via a projection of the density’s distribution onto the space spanned by the wavelet basis. Nonlinear estimators threshold the estimated coefficients, both globally and locally, to obtain optimal convergence to the true density. These estimators, especially those with thresholding, often cannot guarantee that the resulting p̂ from the estimation process satisfies the aforementioned properties.
To guarantee these properties, one typically resorts to estimating as
| (2) |
which directly gives . Previous work on wavelet density estimation of [5], [6] stays within the projection paradigm of trying to estimate the coefficients as an inner product with the corresponding (orthogonal) basis. As such, they have to directly address the estimation of the scalar product involving the square root estimate and the appropriate basis function, e.g., estimating coefficient αj0,k requires finding an acceptable substitute for . In this work, we will show how to completely avoid this paradigm by casting the estimation process in a maximum likelihood framework. The wavelet coefficients of the expansion are obtained by minimizing the negative log likelihood over the observed samples with respect to the coefficients. Moreover, asymptotic analysis will illustrate a remarkable property of the Fisher information matrix of the density under this representation—which is 4I, where I is the identity matrix—and this is leveraged in the optimization. This highly structured matrix is also the asymptotic Hessian of our maximum likelihood objective function at the optimal solution point. This tight coupling in our estimation framework will lead to a very efficient, modified Newton’s method for computing the wavelet coefficients. We maintain all the desirable properties that inherently come with estimating , while circumventing the need to establish the previously mentioned substitute. The focus of this paper is to carefully establish these connections and discuss the resulting algorithms for estimating both one and 2-D densities.
As researchers in the field of image analysis, we have apenchant toward image processing oriented applications. To this end, we provide proof-of-concept demonstrations through information-theoretic shape registration and mutual information (MI) based affine registration of medical imagery [7]. Given a pair of images, one image, designated the source, is considered as similar under affine transformations to a prespecified target image. MI-based image alignment tries to maximize the mutual information between the image pair, which hopefully occurs when they are optimally aligned. The algorithm requires a density estimation step that computes the joint density between the image pairs. This density is then used to calculate the mutual information. For shape registration, we adopt the correspondence-free approach as presented in [8] but replace the performance criterion with the Hellinger divergence [9]. For both applications, we replace the typical density estimator, usually a 2-D histogram, kernel estimator or mixture model, with our wavelet density estimator and analyze its viability. We also provide anecdotal results of the wavelet density estimator’s ability to model true analytic 1-D and 2-D densities such as Gaussian mixture models.
The rest of this paper is organized in the following manner. In Section II, we briefly recap multiresolution wavelet analysis and provide a more in depth discussion concerning wavelet density estimation. Section III discusses our maximum likelihood framework for estimating the wavelet coefficients for . It goes on to detail the modified Newton’s method used to efficiently compute the coefficients. In Section IV, our method is validated in the application setting of image and shape registration and we showcase the capability to model known densities and compare performance against fixed and variable bandwidth kernel density estimators.
II. WAVELET THEORY AND ITS APPLICATION TO DENSITY ESTIMATION
We now provide a basic introduction to the ideas of wavelet multiresolution theory and move on to discussing how these concepts are carried out in wavelet-based density estimation. We will be focused throughout on clearly communicating the conceptual aspects of the theory, diverting much of the mathematical machinery to the (hopefully) appropriately cited references.
A. Multiresolution
For any function f ∈ 𝕃2 and a starting resolution level j0, representation in the wavelet basis is given by
| (3) |
where
| (4) |
are scaled and translated version of the father ϕ(x) and mother ψ(x) wavelets. The key idea behind multiresolution theory is a sequence of nested subspaces Vj, j ∈ ℤ such that
| (5) |
and which satisfy the properties (completeness). The resolution increases as j → ∞ and decreases as j → −∞ (some references show this order reversed due to the fact they invert the scale [10]). At any particular level j + 1, we have the following relationship:
| (6) |
where Wj is a space orthogonal to Vj, i.e., Vj ⋂ Wj = {0}. The father wavelet ϕ(x) and its integer translations form a basis for V0. The mother wavelet ψ(x) and its integer translates span W0. These spaces decompose the function into its smooth and detail parts; this is akin to viewing the function at different scales and at each scale having a low pass and high pass version of the function.
We will assume ϕ(x), ψ(x), and their scaled and translated versions form orthogonal bases for their respective spaces. Under these assumptions, the standard way to calculate the coefficients for (3) is by using the inner product of the space, e.g., the coefficient αj0,k is obtained by
| (7) |
where we have used the 𝕃2 inner product. Most of the existing wavelet-based density estimation techniques exploit this projection paradigm to estimate the coefficients. Replacing the general function f by a density p, the coefficients for (1) can be calculated as
| (8) |
where ℰ is the expectation operator. Given N samples, this is approximated as the sample average
| (9) |
Many density estimation techniques, including ours, require evaluating ϕ(x) and ψ(x) at various domain points in their support region. However, most father and mother wavelets do not have an analytic closed-form expression. The strategy is to use the close coupling between the scaling function and wavelet—find ϕ(x) by numerically solving the dilation equation and then directly obtain ψ(x) by solving the wavelet equation. The dilation equation is given by
| (10) |
where l(k) are the low-pass filter coefficients associated with a particular scaling function family. This can be numerically solved using iterative procedures such as the cascade algorithm [11]. Upon solving for ϕ(x), one can immediately get the associated wavelet function by using the high pass filter coefficients h(k) and solving the wavelet equation
| (11) |
The numerical versions of ϕ(x) and ψ(x) will have values at domain points that are integer multiples of 1/2M (where M controls the discretization level). If an x value lands in between these grid points, it is a straightforward process to interpolate and get the desired value. We have adopted a cubic spline interpolation strategy to obtain the intermediate values.
B. Wavelet Density Estimation
A practical consideration of using wavelets for density estimation requires careful consideration of several issues. First, one must decide the family of wavelets that will be used as the basis. Though this issue has received considerably less attention in theoretical literature, a pragmatic solution suggests that the choice is closely tied to problem domain. However, one almost always assumes that the bases satisfy the desirable orthonormality property and are compactly supported. Also, multiresolution analysis for function approximation requires the use of bases that have a coupled scaling and wavelet function relationship. This restricts our choice of basis functions to families such as Haar, Daubechies, Coiflets, and Symlets. It is worth mentioning that these basis functions are not perfectly symmetric (except for Haar); in fact, in classical wavelet analysis, symmetry is a detriment to perfect reconstruction of the signal [10]. Exact symmetry is not a critical requirement for density estimation and families like Symlets exhibit characteristics close to symmetry for higher order vanishing moments. Throughout this paper, we assume that our bases are orthogonal, compactly supported and have both a scaling and wavelet function.
Second, and perhaps most importantly, we must address efficient computation of the basis coefficients and the impact they have on the properties of the estimated density. The main contribution of this paper is to further advance the theoretical framework for the estimation of these coefficients, while maintaining the important properties of a bona fide density—non-negative in its support and integrating to one.
Finally, it is necessary to consider the practical issue of selecting the basis truncation parameters. Notice that in (2), convergence of the wavelet representation to the true function assumes a starting resolution level j0 and uses an infinite number of detail resolution levels. In practical computation, it is necessary to develop a principled way of choosing j0 and also a stopping level j1 as we cannot have an infinite expansion. These issues are necessarily addressed by model selection methods such as cross validation. Model selection is not the focus of our current work. It is possible to adopt any of the existing model selection methods, as summarized in [12], to appropriately choose these parameters and incorporate it into our framework.
Returning to the second point, classical wavelet density estimation [13], [14] does not try to explicitly ensure that the density is non-negative and usually suffers from negative values in the tails of the density. For example, in the work of Donoho et al. [13], this artifact is introduced by the necessity to threshold the coefficients. Nonlinear thresholding obtains better convergence and achieves the optimal minimax rate under the global integrated mean squared error (IMSE) ℰ [‖p̂ − p‖2] measure, where ℰ is the expectation operator. Though these are favorable properties, it is still somewhat unsettling to have a density with negative values. Also, thresholding leads to the problem of having to renormalize the coefficients to maintain the integrable property of the estimated density. Under the usual nonlinear estimation process, this is not a straightforward procedure and may require further integration to work out the normalizing constant. Next, we will show how it is possible to incorporate the benefits of thresholding the coefficients while maintaining the integrity of the estimated density.
The preferred way to maintain these properties is to estimate the square root of the density rather than p. Estimating has several advantages: i) non-negativity is guaranteed by the fact , ii) integrability to one is easy to maintain even in the presence of thresholding, and iii) the square root is a variance stabilizing transform [15]. The present work also falls into this category of techniques that estimate . To our knowledge, there are only two previous works [5], [6] that estimate the square root of the density using a wavelet basis expansion.
We begin by representing the square root of the density using
| (12) |
Imposing our integration condition, , implies that
| (13) |
Notice that if (13) is not one but some arbitrary constant D, such as when a thresholding scheme changes the weights, it is possible to perform a straightforward renormalization by merely dividing the coefficients by .
In the previous two works [5], [6], estimation of αj0,k and βj,k is motivated by applying the previously discussed projection method. We are now working with , however, which changes (8) to
| (14) |
The βj,k coefficients are defined by analogy. In [5], the authors propose a suitable substitute to the empirical estimator , but the coefficient estimation is sensitive to the preestimator of p(x). In the work by Penev and Dechevsky [6], the coefficient computation is based on order statistics of the sample data. As we illustrate in Section III, the method we present avoids these issues by casting the density estimation problem in a maximum likelihood setting. The maximum likelihood model also ensures the asymptotic consistency of our estimated coefficients. To complete the spectrum of non-negative density estimation techniques, it is worth mentioning that Walter [16] presents an alternative using a clever construction of non-negative wavelets; exploration of this method, however, is beyond the present scope.
III. MAXIMUM LIKELIHOOD FOR WAVELET DENSITY ESTIMATION
We now discuss how to cast wavelet density estimation in a maximum likelihood framework. Often, maximum likelihood is designated a parametric technique and reserved for situations where we are able to assume a functional form for the density. Thus, current research typically categorizes wavelet density estimation as a nonparametric estimation problem. Treating the coefficients as the parameters we wish to estimate, however, allows us to move the problem into the parametric realm. This interpretation is also possible for other formulations such as kernel density estimation, where maximum likelihood is applied to estimate the kernel bandwidth parameter. For estimating the wavelet coefficients, adopting the maximum likelihood procedures will lead to a constrained optimization problem. We then investigate the connections between estimating and the Fisher information of the density. Exploring this connection will allow us to make simplifying assumptions about the optimization problem, resulting in an efficient modified Newton’s method with good convergence properties. We will present derivations for both 1-D and 2-D density estimation, extensions to higher dimensions would follow a similar path.
A. One-Dimensional Constrained Maximum Likelihood
Let , xi ∈ ℝ represent N i.i.d. samples from which we will estimate the parameters of the density. As is often customary, we will choose to minimize the negative log likelihood rather than maximize the log likelihood. The negative log likelihood objective is given by (15), shown at the bottom of the page. Trying to minimize (15) directly w.r.t. αj0,k and βj,k would result in a density estimator which does not integrate to one. To enforce this condition, we require the following equality constraint:
| (15) |
| (16) |
This constraint can be incorporated via a Lagrange parameter λ to obtain the following constrained objective function:
| (17) |
The constraint (16) dictates that the solution to (17) lives on a unit hypersphere; it can be solved using standard constrained optimization techniques. Before presenting our particular solution, we explore the properties of the Fisher information matrix associated with this problem.
B. Many Faces of Fisher Information
The classic form of the Fisher information matrix is given by
| (18) |
where the (u, υ) index pair denotes the row, column entry of the matrix and consequently the appropriate parameter pair. Intuitively, one can think of the Fisher information as a measure of the amount of information present in the data about a parameter θ; for wavelet density estimation θ = {αj0,k, βj,k} and the (u, υ)-indexing is adjusted to be associated with the appropriate level, translation index pair, i.e., {u = (j, k), υ = (l, m)} where j and l are the level indices and k and m are the translation indices. For the current setting, where we are estimating , the Fisher information has a more pertinent form
| (19) |
Hence, g̃uυ computed using the square root of the density differs only by a constant factor from the true Fisher information. Using this algebraic relationship, the Fisher information of the wavelet density estimator can be calculated by substituting the wavelet expansion of given in (2). This gives
| (20) |
where we have leveraged the orthogonal property of the wavelet basis functions. This is an identity matrix and the Fisher information of p(x; θ)can be written as guυ = 4I.
There is another algebraic manipulation that allows us to compute the Fisher information using the Hessian of the log likelihood, specifically
| (21) |
where ∇ is the gradient operator w.r.t. the parameters and H is the Hessian matrix of the multiparameter negative log likelihood. Recalling that (15) is the negative log likelihood, we can immediately make the connection that (17)’s asymptotic Hessian should be Hℒ = guυ = 4I. To verify this, let
| (22) |
where Hnll and Hh are the Hessian of the negative log likelihood (15) and constraint (16), respectively. Equation (22) is the Hessian of the Lagrangian which is typical of constrained minimization problems. We illustrate the computation of the asymptotic Hnll by providing results for a particular coefficient β; other coefficients are calculated in a similar manner. The second partial derivative of (15) is
| (23) |
and taking its expected value, we get
| (24) |
(Note: For readability, we have let p ≡ p(X; {αj0,k, βj,k})). The Hessian of (16) is constant and is consequently equal to its expected value, i.e.,
| (25) |
Both (24) and (25) are 2I matrices. Hence, referring back to (22), in order for Hℒ = 4I, we require λ = 1 at the optimal solution point; the proof of which is obtained through algebraic manipulation of the Lagrangian’s, (17), first-order necessary conditions. Intuitively, what we have shown is that under an orthonormal expansion of the square root of density, the Fisher information matrix essentially specifies a hypersphere [17].
C. Efficient Minimization Using a Modified Newton’s Method
In light of the discussion in the previous section, we proceed to design an efficient optimization method to iteratively solve for the coefficients. A Newton’s method solution to (17) would result in the following update equations at iteration τ:
| (26) |
where . When the Hessian is positive definite throughout the feasible solution space Ω it is possible to directly solve for xτ and λτ [18]. For (17), it is certainly true that xT Hℒx > 0 over Ω. In order to avoid the computationally taxing Hℒ update at each iteration, we adopt a modified Newton’s method [18]. Modified Newton techniques replace Hℒ by B, where B is a suitable approximation to Hℒ. Here, we can take advantage of the fact we know Hℒ at the optimal solution point; hence, we let . In practice, this method is implemented by solving the system
| (27) |
where C = (Aτ)T B−1Aτ, F = (Aτ)T B−1 and the coefficient updates are given by xτ+1 = xτ + dτ. Notice that these equations can be simplified even further by taking advantage of the simple structure of B to avoid explicit matrix inverses and making it very efficient for implementation, i.e., set . This method depends on having a unit step size and has convergence properties comparable to the standard Newton’s method [18]. It was also shown in [19] and [20] that using the known Hessian at the optimal solution points has the effect of doubling the convergence area, thus making it robust to various initializations.
D. Two-Dimensional Density Estimation
Extensions to bivariate, wavelet density estimation are made possible by using the tensor product method to construct 2-D wavelet basis functions from their 1-D counterparts [10]. The notation becomes noticeably more complicated and requires careful attention during implementation. Let (x1, x2) = x ∈ ℝ2 and now the expansion is given by
| (28) |
where (k1, k2) = k ∈ ℤ2 is a multi-index. The tensor products are
| (29) |
Again, our goal is to estimate the set of coefficients . As in the univariate case, we repeat the necessary steps by first creating the objective function that incorporates the negative log likelihood and the Lagrange parameter term to handle the equality constraint. Then, the minimization procedure follows according to Section III-C. The resulting equations are exactly the same form with straightforward adjustments during implementation of (27) to incorporate the 2-D nature of the indices and wavelet basis. The algorithm to perform one or 2-D wavelet density estimation using our modified Newton’s method is presented in Algorithm 1.
Algorithm 1.
Wavelet density estimation using modified Newton’s method.
|
IV. EXPERIMENTAL RESULTS
As our work is a general density estimation technique, it is applicable to a whole host of applications that rely on estimating densities from observational data. We are still in the early stages of identifying and exploring the best-of-class applications that are well suited to take advantage of the underlying flexibility that comes with wavelet-based analysis. The present experimental evaluation of the proposed methods was conducted on both synthetic and real data. We measure performance by validation against true, analytical densities (both 1-D and 2-D) and illustrate proof-of-concept scenarios for real data applications that require density estimation as part of their solution. Specifically, the two applications we showcase here are shape alignment and mutual information-based image registration. Though our method has some advantages over contemporary wavelet density estimators, there are still practical considerations that all, including our, wavelet-based solutions bump against. These considerations are peppered throughout our analysis of the experimental results.
A. One- and Two-Dimensional Density Approximation
The approximation power of the present method was validated against the class of known densities as presented in [21] and [22], where the authors provide constructions of several densities (which are all generated as appropriate mixtures of Gaussians) that analytically exercise representative properties of real densities, such as spatial variability, asymmetry and concentrated peaks. Most other wavelet density estimators have also showcased their results on a small subset of these densities. In order to provide comprehensive, robust analysis of our method, we selected the following 13, 1-D densities to analyze approximation capabilities: Gaussian, skewed unimodal, strongly skewed unimodal, kurtotic unimodal, outlier, bimodal, separated bimodal, skewed bimodal, trimodal, claw, double claw, asymmetric claw, and asymmetric double claw. The reader is referred to [21] for a visual depiction of all 13 densities. During preliminary empirical evaluation, we noticed a trend that best results were observed when using a single-level, scaling function representation of the density. This was further confirmed via private communication with G. G. Walter and also discussed in [23]. We performed the estimation over a range of scale values, i.e., j0 in (12), from j0 = −1 to 5. (Note: We initially used a cross validation method, see [12], to automatically select j0 but opted to test over a range to be more thorough). We also used three different families of wavelet basis, with multiple orders within a family, to approximate each of the densities—Daubechies of order 1–10, Symlets of order 4–10, and Coiflets of order 1–5. The analyses across families provide some guidance as to the approximation performance capabilities of different bases. All densities were estimated using 2000 samples drawn from the known analytic densities. The approximation error of each test was measured using the integrated squared error (ISE) between the known p and estimated p̂ densities, i.e., ∫ℝ (p − p̂)2 dx. This was computed by discretizing the 1-D support of the density at equally spaced points and then summing area measures. The best wavelet basis, order and starting level j0 were selected based on this error measure. We executed 15 iterations of our modified Newton’s method with many densities converging in fewer (8 to 10) iterations—15 iterations on 2000 samples takes approximately 30 s with a Matlab implementation. We compared the wavelet reconstructed densities with kernel density estimators (KDE) [2]. All experiments were conducted using a Gaussian kernel, a reasonable choice since the true densities are mixtures of Gaussians, with the smoothing parameter (bandwidth of kernel) selected two ways: 1) automatically as described in [2, Ch. 3] and 2) a nearest-neighbor variable bandwidth (see [2, Ch. 5]). The freely available KDE Toolbox for Matlab [24] by Iheler provides fast, robust implementation of these kernel methods. We refer to the automatically selected bandwidth as fixed since it is the same for all kernels. The 1-D results are summarized in Table I.
TABLE I.
One dimensional density estimation. optimal start level was selected by taking lowest ISE for j0 ∈ [−1,5]
| WDE | KDE | |||||
|---|---|---|---|---|---|---|
| Best Basis | ISE | FixedBW† ISE | Var.BW† ISE | |||
| Gaussian | SYM10 | −1 | 3.472E-04 | 3.189E-04 | 5.241E-03 | |
| Skewed Uni. | DB7 | 1 | 3.417E-04 | 1.970E-04 | 7.551E-03 | |
| Str. Skewed Uni. | SYM7 | 3 | 2.995E-03 | 6.947E-02 | 7.610E-03 | |
| Kurtotic Uni. | COIF2 | 2 | 2.399E-03 | 2.869E-02 | 1.388E-02 | |
| Outlier | SYM10 | 2 | 1.593E-03 | 3.911E-03 | 3.962E-02 | |
| Bimodal | COIF5 | 0 | 5.973E-04 | 2.084E-04 | 4.223E-03 | |
| Sep. Bimodal | SYM7 | 1 | 5.354E-04 | 6.237E-03 | 5.419E-03 | |
| Skewed Biomodal | DB10 | 1 | 8.559E-04 | 1.461E-03 | 3.885E-03 | |
| Trimodal | COIF3 | 1 | 9.811E-04 | 1.439E-03 | 3.787E-03 | |
| Claw | SYM10 | 2 | 1.511E-03 | 3.692E-02 | 7.014E-03 | |
| Dbl. Claw | COIF1 | 2 | 2.092E-03 | 1.795E-03 | 5.283E-03 | |
| Asym. Claw | DB3 | 3 | 2.383E-03 | 1.373E-02 | 6.490E-03 | |
| Asym. Dbl. Claw | COIF1 | 2 | 2.250E-03 | 4.759E-03 | 4.940E-03 | |
For the 2-D evaluation, we selected the following five difficult densities, i.e., ones that exhibited more variations or closely grouped Gaussians, from [22]: bimodal IV, trimodal III, kurtotic, quadrimodal, and skewed. The experimental procedure was similar to that of the 1-D densities and results are summarized in Table II. Again, our modified Newton’s method was able to converge with fewer than 15 iterations. In all 2-D test cases, our wavelet density estimator was able to outperform both the fixed and variable bandwidth kernel density estimators. Overall, in both the 1-D and 2-D cases the wavelet bases were able to accurately represent the true densities. Of the families tested, there was no clear-cut winner as to which basis was better than another. The performance of a particular basis depended on the shape of the true density. Some applications may prefer Symlets or Coiflets as they are “more symmetric” than other bases. In comparison to the kernel estimators, our method provided better results on the more difficult densities and performed only slightly worse on slowly varying ones. Also, there were instances where variable bandwidth KDE provided a lower ISE but visually exhibited more peaks than the true density. In the future, we plan to do further analysis to better evaluate this bias-variance tradeoff when selecting the best density approximation. Examples of estimated densities and some of their properties are illustrated in Figs. 1 and 6.
TABLE II.
Two–dimensional density estimation. Optimal start level was selected by taking lowest ISE for j0 ∈ [−1,3].
| WDE | KDE | |||||
|---|---|---|---|---|---|---|
| Best Basis | ISE | Fixed BW† ISE | Var. Bw† ISE | |||
| Bimodal IV | SYM7 | 1 | 6.773E-03 | 1.752E-02 | 8.114E-03 | |
| Trimodal III | COIF2 | 1 | 6.439E-03 | 6.621E-03 | 1.037E-02 | |
| Kurtotic | COIF4 | 0 | 6.739E-03 | 8.050E-03 | 7.470E-03 | |
| Quadrimodal | COIF5 | 0 | 3.977E-04 | 1.516E-03 | 3.098E-03 | |
| Skewed | SYM10 | 0 | 4.561E-03 | 8.166E-03 | 5.102E-03 | |
BW is bandwitdth
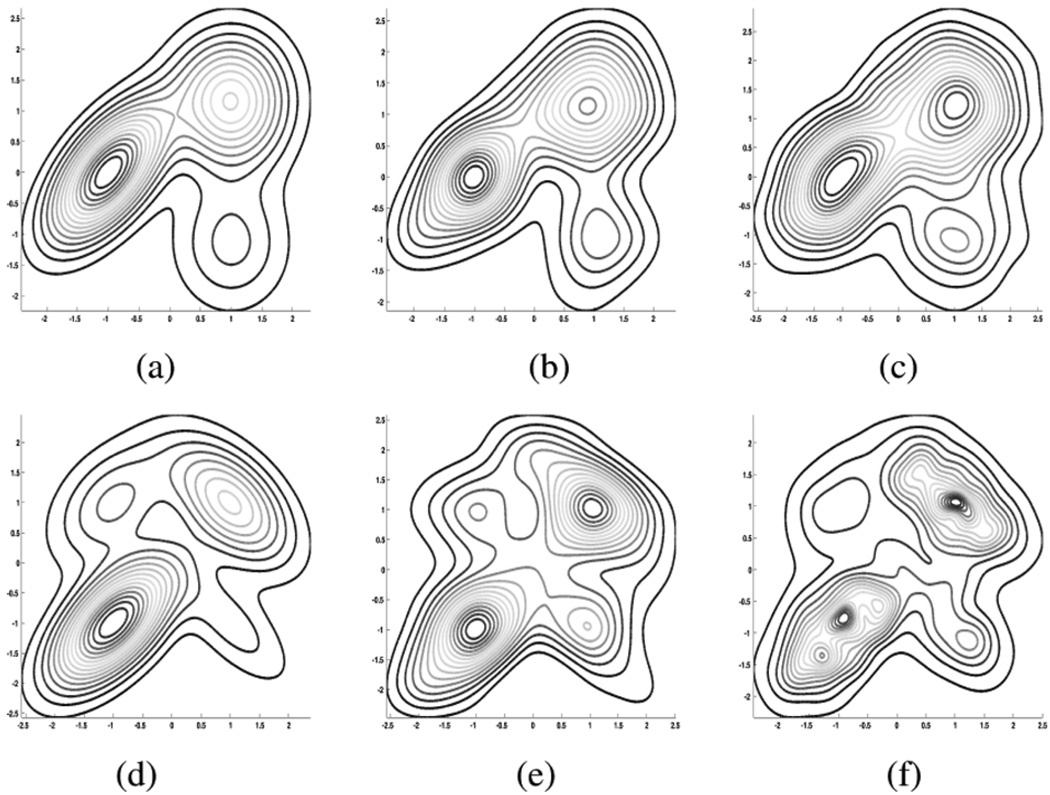
Fig. 1.
Two–dimensional density estimation comparison: WDE versus KDE contours. See Table II for WDE estimation parameters. (a)True trimodal III. (b) WDE of (a). (c) Fixed BW KDE of (a). (d) True quadrimodal (e). WDE of (d). (f) Variable BW KDE of (d).
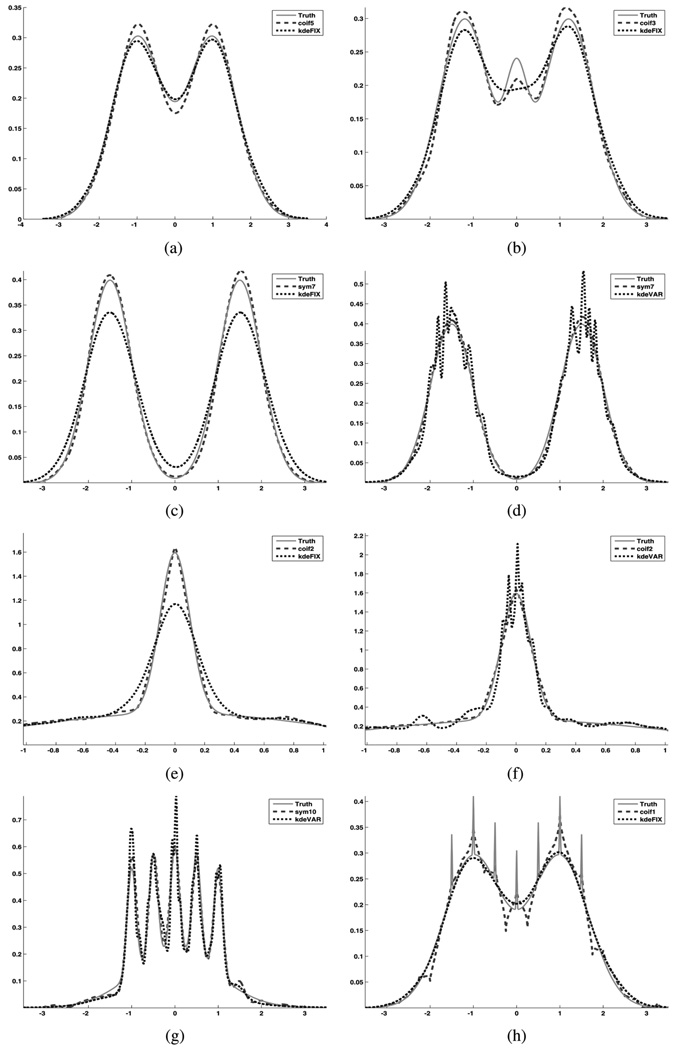
Fig. 6.
One-dimensional density estimation comparison: WDE versus KDE. True analytic density is solid line, WDE is dashed line and KDE is dotted line. See Table I for WDE estimation parameters. (a) Fixed BW KDE performs slightly better than our WDE on this bimodal density. (b) The WDE captures the middle peak of this trimodal density. The fixed BW KDE misses it. (c) Fixed BW KDE under estimates peaks of this separated bimodal density. (d) Variable BW KDE has lower ISE than fixed BW KDE in (c), but incorrectly gives several peaks. (e) WDE captures the main peak area of this kurtotic density; fixed BW KDE fails. (Zoomed around peak) (f) Variable BW KDE also fails on kurtotic density estimating several peaks. (Zoomed around peak) (g) The WDE captures all peaks of this claw density while the variable BW KDE over shoots peaks 1, 3, and 4. (h) The fixed BW KDE has a lower ISE than WDE on this double claw density, but it misses all the sharp peaks.
B. WDE for Registration and Shape Alignment
Information-theoretic approaches have been applied to a variety of image analysis and machine learning problems [25]–[27]. These techniques typically require estimating the underlying density from which the given data are generated. We applied our new density estimation procedure to two information-theoretic methods that utilize the Shannon entropy and the Hellinger divergence. We begin with the well established image registration method based on mutual information [7]. This multimodal registration method iteratively optimizes the mutual information (MI) between a pair of images over the assumed parameter space of their differing transformation. We next implement an adaptation of a more contemporary method which minimizes the Jensen–Shannon divergence in order to align two point-set representation of shapes [8]. Both of these methods require estimating 2-D densities.
1) Registration Using Mutual Information
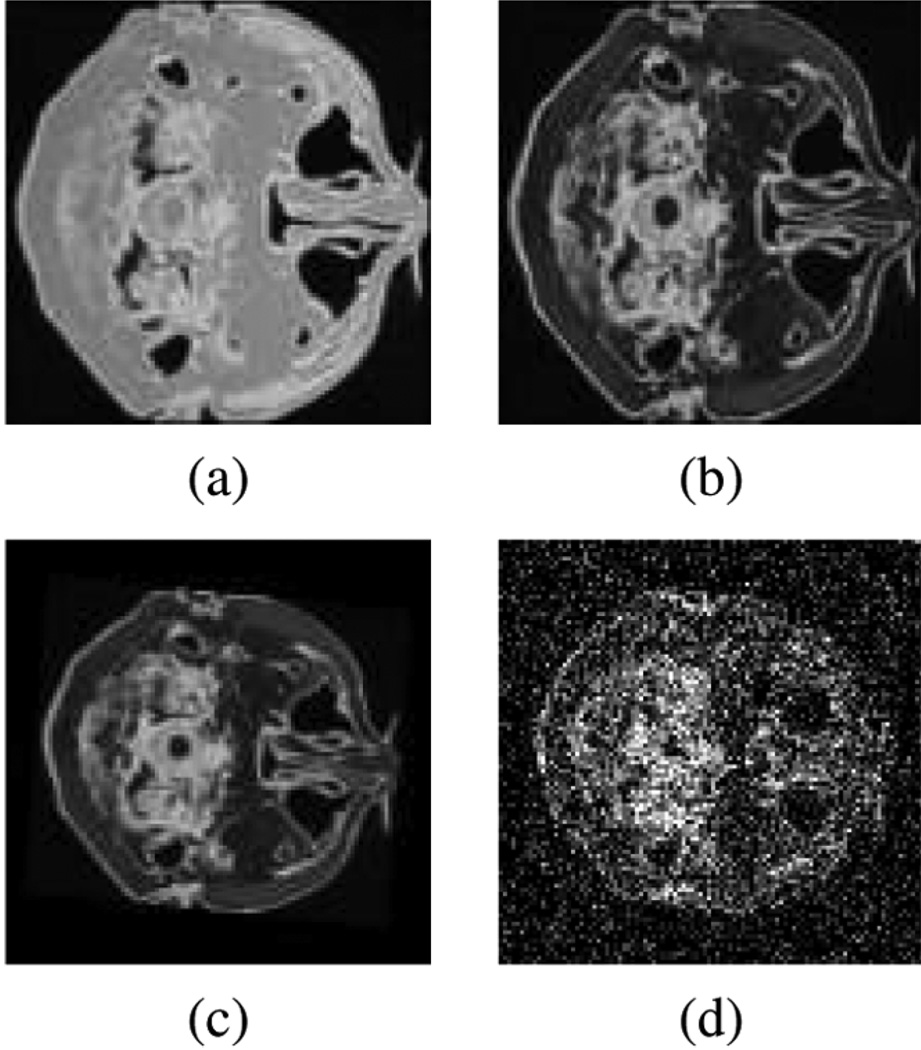
For the MI registration experiments, we used slices from the Brainweb simulated MRI volumes for a normal brain [28]. The goal was to recover a global affine warp between an image pair. (Note: To expedite experiments, we did not include translations in the affine warp). We follow the affine warp decomposition used in [29], which results in a four parameter search space (θ, ϕ, s, t). In order to minimize experimental variability, we manually imposed a known affine transformation between the source and target image. The optimal parameter search was conducted using a coarse-to-fine search strategy over a bounded, discretized range of the parameter space. Calculating the mutual information performance criterion between image pairs requires estimating the joint density between them. This is typically accomplished using a simple 2-D histogram or Parzen window estimator (i.e., the kernel estimators evaluated in Section IV-A). We replace these methods with our wavelet-based density estimator, leaving the rest of the algorithm unchanged. Fig. 2 shows the source and target images used in the trials and the results are listed in Table III. In the absence of noise, we were able to perfectly recover the transformation parameters using our method whereas KDE (both fixed and variable bandwidth) failed to estimate the optimal parameters. In the noise trial, we were able to correctly recover two, s and t, out of four parameters with the other two, θ and ϕ, values only missing the ground truth by one and two discretization steps, respectively (see Table III). The KDE performed about the same as our method in these noise trials. All experiments were conducted using the Daubechies order 1 (db1) family, with a multiresolution basis starting at level j0 = −3 stopping at j1 = −2. This means that both scaling and wavelet functions were used in the density estimation; see Fig. 3 for an example. It is also possible to use other basis families. Because our optimization does not use a step size parameter, we did encounter some cases where choosing a bad starting level caused convergence issues. This can be remedied by utilizing any standard optimization method that incorporates a line search to control the descent direction. Currently, we are using both qualitative analysis (visual inspection) and a 2-D version of the cross-validation procedure, as referenced above in the approximation experiments, in order to select the start (j0) and stop (j1) levels.
Fig. 2.
Registration using mutual information. (a), (b) Registration image pair. (c) Affine warp applied to (b) without noise. Target image (c) with noise.
TABLE III.
Mutual information registration results. The left-most columns are affine parameters (see text). With no noise, σ2 = 0, the parameters were exactly recovered when using WDE whereas KDE estimates were slightly off. With noise added, the recovered values were close to the truth, but there was not a significant advantage using KDE versus our method
| Truth | WDE | Fixed BW KDE | Var. BW KDE | ||||
|---|---|---|---|---|---|---|---|
| σ2 = 0 | σ2 = 0.05 | σ2 = 0 | σ2 = 0.05 | σ2 = 0 | σ2 = 0.05 | ||
| θ | 10 | 10 | 9.8 | 9.8 | 10.2 | 10 | 10.4 |
| ϕ | −5 | −5 | −5.4 | −5 | −4.8 | −4.6 | −5 |
| s | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.2 | 0.3 |
| t | −0.1 | −0.1 | −0.1 | −0.1 | −0.1 | 0 | −0.1 |
Fig. 3.
Example of joint density estimation from two images utilized in registration experiments; scaling and wavelet functions from Haar basis using levels j0 = −3 to j1 = −2.
2) Shape Alignment Under Hellinger Divergence
Next, we applied this density estimation method to shape analysis. The applications of landmark and point set-based shape analysis are often cast in a probabilistic framework which requires a density estimation procedure. The compact, localized nature of the wavelet basis allows one to model a rich class of shapes, with intricate structures and arbitrary topology. We qualitatively illustrate this by estimating the density corresponding to a dog snapper fish shape consisting of 3040 points, Fig. 4(a). The density estimation was carried out using a Coiflet 4 basis with only scaling basis functions starting at level j0 = 3. The estimated density is shown in Fig. 5. Notice how the wavelet basis captures the detailed structures such as the fins and closely hugs the spatial support region of the original points. We use our estimation method in a correspondence-free registration framework, as described in [8] and [30], to recover an affine transformation between two point sets. In [8], their goal was to determine a probabilistic atlas using Jensen–Shannon divergence. Here, we wish to showcase the wavelet density estimator for probabilistic shape matching, as in [30]. However, rather than using the Kullback– Leibler divergence measure as in [30], we elect to use the Hellinger divergence instead [9]
| (30) |
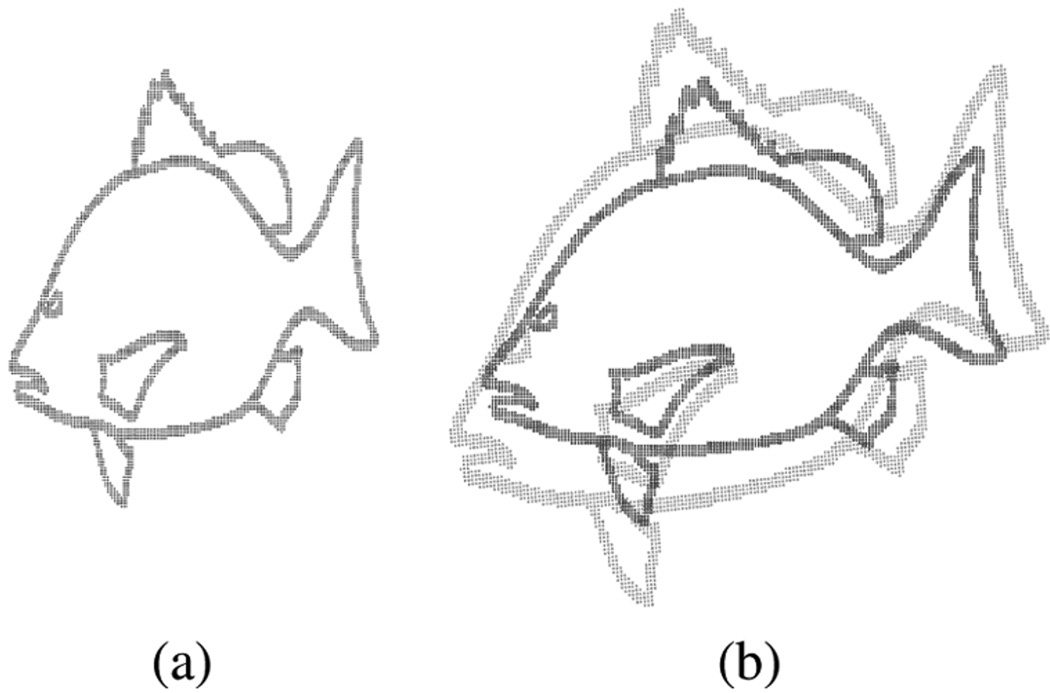
where (α(1), β(1)) and (α(2), β(2)) are the wavelet parameters of p1 and p2, respectively. The Hellinger divergence is also closely related to the geodesic distance on a sphere where each point on the sphere is a wavelet density. The advantage in using the Hellinger divergence (or the geodesic distance) over the Kullback–Leibler divergence is that the divergence is in closed form and does not need to be estimated from the data using a law of large numbers-based approach as in [8] and [30]. In order to control experimental variability, we used a brute force coarse-to-fine search over the affine parameters. The target shape’s density, p1 in (30), is estimated once at the beginning using our method. At each iteration of the affine parameters, the source point set is deformed by the current affine parameters and a new p2 is estimated with the wavelet density estimator using these transformed points. The Hellinger divergence error criterion in (30) is minimized when the two densities are best aligned and this in turn gives the optimal parameters of the affine transformation. Following a strategy similar to those described in the MI experiments, we were able to successfully recover the affine transformation. The ground truth affine parameters were the same as in the MI tests. In these experiments, the KDE—using fixed and variable bandwidths—again failed to estimate all of the parameters correctly (see Table IV). Some of the KDE’s inaccuracies could be attributed to the fact that (30) is available in closed form under our representation but has to be numerically computed for the KDE. Fig. 4(b) illustrates the source and target point set used in this matching experiment.
Fig. 4.
Fish point sets. (a) Dog snapper represented by 3040 points. (b) Overlay of source and target shape (lighter shade) used in Hellinger divergence-based registration.
Fig. 5.
Example of 2-D density estimated from fish point set using Coiflet 4, only scaling functions at level j0 = 3.
TABLE IV.
Hellinger divergence shape alignment. The WDE recovers all of the transformation parameters exactly
| Truth | WDE | Fixed BW KDE | Var. BW KDE | |
|---|---|---|---|---|
| θ | 10 | 10 | 9.8 | 9.8 |
| ϕ | −5 | −5 | −5.2 | −5.2 |
| s | 0.3 | 0.3 | 0.2 | 0.2 |
| t | −0.1 | −0.1 | −0.1 | −0.1 |
In this section, we have detailed both the approximation power and practical utility of our proposed wavelet density estimator. The estimator is able to accurately represent a large class of parametric and nonparametric densities, a well-known trait of wavelet bases. Our method robustly satisfies the integrability and non-negativity constraints desired from density estimators with the added localization benefits inherent to wavelet expansions. This allowed us to seamlessly plug in our technique into several applications that critically depend on assessing densities from sample data.
V. CONCLUSION
In this paper, we have presented a new technique for non-negative, density estimation using wavelets. The non-negativity and unit integrability properties of bona fide densities are preserved through directly estimating which allows one to obtain the desired density through the simple transformation . In sharp contrast to previous work, our method casts the estimation process in maximum likelihood framework. This overcomes some of the drawbacks of methods that require good preestimators for the density we are trying to find. The maximum likelihood setting consequently resulted in a constrained objective function whose minimization yielded the required basis function coefficients for our wavelet expansion. We were able to develop a efficient modified Newton method to solve the constrained problem by analyzing the relationship to the Fisher information matrix under the wavelet basis representation. We showed that the Hessian matrix at the solution point of the maximum likelihood objective function had a highly structured and simple form, allowing us to avoid matrix inverses typically required in Newton-type optimization. Verification of our proposed method was first empirically demonstrated by testing this method’s capability to accurately reproduce known densities. Success was illustrated across a range of densities and wavelet families and validated against kernel density estimators. We also applied the estimation process to two image analysis problems: mutual information image registration and density estimation for point-set shape alignment. Both illustrated the successful operation of our method.
As a general density estimation procedure, this method could be applied to numerous applications. The compact, localized nature of wavelets and the large class of functions they are capable of representing make them an excellent choice for density estimation. Another property of particular interest to us is the regularity of wavelet families. We hope to explore this in future work, where we can take advantage of the differentiability characteristics to design and optimize information-theoretic objective functions typical in image analysis applications. It is also possible to investigate other optimization techniques to solve the present objective, such as preconditioned conjugate gradient, quasi-Newton or trust region methods. Finally, we plan to leverage the useful property that the Hellinger divergence and the geodesic distance between two wavelet densities (in the same family) are available in closed form in shape matching and indexing applications.
ACKNOWLEDGMENT
The authors would like to thank S. Penev and G. Walter for literature material and helpful suggestions.
This work was supported in part by the National Science Foundation under Grant IIS-0307712 and in part by the National Institutes of Health under Grant R01NS046812. The associate editor coordinating the review of this manuscript and approving it for publication was Dr. Srdjan Stankovic.
Biographies

Adrian M. Peter (S’04) received the B.S. degree (high honors) in computer engineering and the M.S. degree in electrical and computer engineering from the University of Florida, Gainesville, in 1999 and 2003, respectively; he returned there in 2005 to complete a Ph.D. degree in electrical and computer engineering.
After his B.S. degree, he was with Intel Corporation where he last held the position of a technology initiatives program manager responsible for short range wireless products. After his M.S. degree, he was with Harris Corporation developing image analysis algorithms for a variety of remote sensing platforms and was a co-inventor on eight pending patents. His research interests are in machine learning and computer vision with a focus on applying information geometry to shape analysis.

Anand Rangarajan (M’88) received the B.Tech. degree in electronics engineering from the Indian Institute of Technology, Madras, in 1984, and the Ph.D. degree in electrical engineering from the University of Southern California, Los Angeles, in 1991.
From 1990 to 1992, he was a Postdoctoral Associate in the Departments of Diagnostic Radiology and Computer Science, Yale University, Harvard, CT. From 1992 to 1995, he held a joint research faculty position in both departments. From 1995 to 2000, he was an Assistant Professor in the Image Processing and Analysis Group (IPAG), Departments of Diagnostic Radiology and Electrical Engineering, Yale University. He is now an Associate Professor in the Department of Computer and Information Science and Engineering (CISE), University of Florida. His current research interests are best summarized as the application of machine learning to image analysis. He is also interested in the scientific study of consciousness.
Dr. Rangarajan chaired a Neural Information Processing Systems (NIPS) Workshop entitled “Deterministic Annealing and Combinatorial Optimization” in 1992 and he co-chaired a NIPS Workshop entitled “Statistical and Structural Models in Network Vision” in 1995. He has served on the program committees of Energy Minimization Methods in Computer Vision and Pattern Recognition (EMMCVPR) (1997, 1999, 2001, and 2007), the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2000, 2001, 2003, and 2006), and Information Processing in Medical Imaging (IPMI) (2005 and 2007). He has also co-chaired the IEEE Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA) 2001, EMMCVPR (2003 and 2005), and was an area chair for the IEEE International Conference on Computer Vision (ICCV) 2007. He has also been active in the Toward a Science of Consciousness conferences (1996, 1998, 2000, 2002, and 2008) and the International Congress of Vedanta conferences (2002, 2005, and 2007).
Contributor Information
Adrian M. Peter, Department of Electrical and Computer Engineering, University of Florida, Gainesville, FL 32611 USA.
Anand Rangarajan, Department of Computer and Information Science and Engineering, University of Florida, Gainesville, FL 32611 USA.
REFERENCES
- 1.Scott DW. Multivariate Density Estimation: Theory, Practice, and Visualization. New York: Wiley-Interscience; 2001. [Google Scholar]
- 2.Silverman BW. Density Estimation for Statistics and Data Analysis. Boca Raton, FL: Chapman and Hall/CRC; 1986. [Google Scholar]
- 3.Doukhan P. Formes de Töeplitz associéesà une analyse multiechélle. C. R. Acad. Sci. Paris Sér. A. 1988;vol. 306:663–666. [Google Scholar]
- 4.Vannucci M. Nonparametric density estimation using wavelets. Duke Univ.; ISDS, Tech. Rep. 95-26. 1995 [Online]. Available: http://www.isds.duke.edu.
- 5.Pinheiro A, Vidakovic B. Estimating the square root of a density via compactly supported wavelets. Comput. Statist. Data Anal. 1997;vol. 25(no. 4):399–415. [Google Scholar]
- 6.Penev S, Dechevsky L. On non-negative wavelet-based density estimators. J. Nonparam. Statist. 1997;vol. 7:365–394. [Google Scholar]
- 7.Viola P, Wells WM., III Alignment by maximization of mutual information. Int. J. Comput. Vis. 1997;vol. 24(no. 2):137–154. [Google Scholar]
- 8.Wang F, Vemuri B, Rangarajan A, Schmalfuss I, Eisenschenk S. Simultaneous nonrigid registration of multiple point sets and atlas construction; Eur. Conf. Comput. Vis; 2006. pp. 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beran R. Minimum Hellinger distance estimates for parametric models. Ann. Statist. 1977;vol. 5(no. 3):445–463. [Google Scholar]
- 10.Daubechies I. Ten Lectures on Wavelets. Philadelphia, PA: SIAM; 1992. [Google Scholar]
- 11.Strang G, Nguyen T. Wavelets and Filter Banks. Cambridge, MA: Wellesley-Cambridge; 1997. [Google Scholar]
- 12.Vidakovic B. Statistical Modeling by Wavelets. New York: Wiley; 1999. [Google Scholar]
- 13.Donoho D, Johnstone I, Kerkyacharian G, Picard D. Density estimation by wavelet thresholding. Ann. Statist. 1996;vol. 24(no. 2):508–539. [Google Scholar]
- 14.Hardle W, Kerkyacharian G, Pickard D, Tsybakov A. Wavelets, Approximation, and Statistical Applications. New York: Springer-Verlag; 2000. [Google Scholar]
- 15.Montgomery DC. Design and Analysis of Experiments. New York: Wiley; 2004. [Google Scholar]
- 16.Walter GG. Wavelets and Other Orthogonal Systems with Applications. Boca Raton, FL: CRC; 1994. [Google Scholar]
- 17.Lebanon G. Ph.D. dissertation. Pittsburg, PA: Carnegie Mellon Univ.; 2005. Riemannian geometry and statistical machine learning. [Google Scholar]
- 18.Luenberger D. Linear and Nonlinear Programming. Reading, MA: Addison-Wesley; 1984. [Google Scholar]
- 19.Burkhardt H, Diehl N. Simultaneous estimation of rotation and translation in image sequences; Proc. Eur. Signal Processing Conf; 1986. pp. 821–824. [Google Scholar]
- 20.Vemuri B, Huang S, Sahni S, Leonard CM, Mohr C, Gilmore R, Fitzsimmons J. An efficient motion estimator with application to medical image registration. Med. Image Anal. 1998 Mar.vol. 2(no. 1):79–98. doi: 10.1016/s1361-8415(01)80029-3. [DOI] [PubMed] [Google Scholar]
- 21.Marron SJ, Wand MP. Exact mean integrated squared error. Ann. Statist. 1992;vol. 20(no. 2):712–736. [Google Scholar]
- 22.Wand MP, Jones MC. Comparison of smoothing parameterizations in bivariate kernel density estimation. J. Amer. Statist. Assoc. 1993;vol. 88(no. 422):520–528. [Google Scholar]
- 23.Walter GG, Ghorai JK. Advantages and disadvantages of density estimation with wavelets. Proc. 24th Symp. on the Interface, Computing Science and Statististics; 1992. pp. 234–243. [Google Scholar]
- 24.Ihler A. Kernel Density Estimation Toolbox for Matlab. 2003 [Online]. Available: http://ttic.uchicago.edu/ ihler/code/kde.php. [Google Scholar]
- 25.Peter A, Rangarajan A. Shape matching using the Fisher-Rao Riemannian metric: Unifying shape representation and deformation; Proc. IEEE Int. Symp. Biomedica Imaging; 2006. pp. 1164–1167. [Google Scholar]
- 26.Cootes T, Taylor C. A mixture model for representing shape variation; Proc. Brit. Machine Vision Conf; 1997. pp. 110–119. [Google Scholar]
- 27.Principe J, Xu D, Fisher J. Information theoretic learning. In: Haykin S, editor. Unsupervised Adaptive Filtering. New York: Wiley; 2000. pp. 265–319. [Google Scholar]
- 28.Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans. Med. Imag. 1998 Jun.vol. 17(no. 3):463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Rangarajan A. Affine image registration using a new information metric. Proc. IEEE Conf. Computer Vision Pattern Recognition. 2004:848–855. [Google Scholar]
- 30.Wang Y, Woods K, McClain M. Information-theoretic matching of two point sets. IEEE Trans. Image Process. 2002 Aug.vol. 11(no. 8):868–872. doi: 10.1109/TIP.2002.801120. [DOI] [PubMed] [Google Scholar]