Abstract
In response to stress p53 initiates the transcriptional regulation of selected target genes and various cellular responses, including cell cycle arrest, apoptosis and senescence. Recent studies revealed two additional functions of p53 in regulation of IGF-1/AKT/mTOR pathways and energy metabolism, which contribute to p53's role as a tumor suppressor. Oncogenic processes give rise to metabolic pathways focused upon the use of aerobic glycolysis (the Warburg effect) and the pentose shunt providing higher levels of reducing activities. p53 shuts down these pathways and refocuses cells to utilize mitochondrial oxidative phosphorylation maximizing efficient ATP production and minimizing the synthesis of substrates for cell division. The use of these alternative metabolic pathways is an integral part of both normal and oncogenic phenotypes.
The p53 pathway
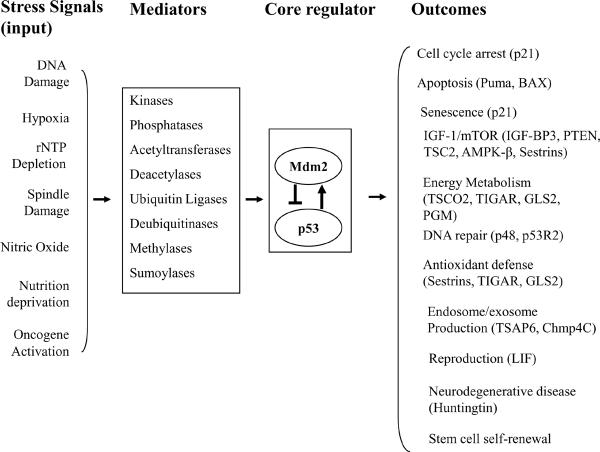
The p53 tumor suppressor gene plays a crucial role in maintaining genomic stability and preventing tumor formation1–3. As a transcription factor, the p53 protein mainly exerts its functions through the transcriptional regulation of its target genes. The p53 protein responds to a wide variety of intrinsic and extrinsic stress signals (Figure 1). These stress signals all interfere with the cellular homeostatic mechanisms that monitor and control the fidelity of DNA replication, chromosome segregation and cell division. Stress signals are detected and communicated to the p53 protein via various enzymes that modify the p53 protein and its regulators (e.g. MDM2), leading to an increase of the half-life of the p53 protein and therefore its accumulation in cells. Once activated, the p53 protein selectively transcribes a set of p53 target genes to initiate various cellular responses. Depending upon the cell type, the transformed state of a cell and the type or degree of stress placed upon a cell, the p53 protein induces either cell cycle arrest, apoptosis or senescence to prevent the propagation of these damaged or mutant cells that could potentially become cancerous (Figure 1).
Figure 1. p53 and its signaling pathway.
Various kinds of stress signals are detected by cells and communicated to the p53 protein and its core constituents by the mediators. Stress signals result in the degradation of the MDM2 protein and the increase in the levels and activity of the p53 protein. Once activated, p53 selectively transcribes a group of its target genes (shown are representative genes) and initiates cellular responses to exert various functions.
While cell cycle arrest, apoptosis and senescence are traditionally thought of as the major outputs of the p53 pathway recent studies are beginning to define additional functions of the p53 pathway (Figure 1). p53 regulates some proteins that aid in DNA repair and the prevention of DNA damage4, 5. These functions act constitutively and are induced after DNA damage. They aid in repair processes that permit the cell to re-enter into the cell cycle without incurring errors that result in a high mutation rate6–9. In addition a stress signal will activate p53 resulting in the transcription of genes that enhance the endosomal compartment and this results in the removal of growth receptors from the cell surface and promotes exosome secretion so as to communicate a cellular stress event to the immune system10, 11. Notably the activated p53 protein enhances the transcription of several genes (Figure 1) that inhibit IGF-1/AKT and mTOR pathways and prevent cell growth and division after a stress signal12–14. p53 also regulates energy metabolism in cells through the transcription regulation of the SCO2, TIGAR and GLS2 genes7, 15, 16, as well as through the p53R2 gene which is important in maintaining mitochrondrial DNA17, 18. In addition to those p53 functions which impact upon energy metabolism and cell growth and division the p53 protein also functions in a number of important developmental regulatory events. The p53 protein is required for the implantation of the embryo into the uterus through the regulation of transcription of the LIF gene19, 20. p53 function was also identified as an important checkpoint during the multifactor reprogramming process in which induced pluripotent stem (iPS) cells are derived from differentiated adult cells21–23. The absence of a functional p53 protein increases the kinetics of formation and enhances the yield of iPS cells, suggesting that p53 may be a major gatekeeper in the reformation of stem cells from more differentiated cell types. Furthermore, the p53 protein plays a role in the central nervous system. The Huntingtin gene is regulated by the p53 protein in response to stress and as such it may be involved in the regulation of at least one neurodegenerative disease24, 25. Clearly the p53 response to stress is a very integrated one in both cell autonomous and cell non-autonomous mechanisms for mobilizing the sources of energy metabolism, modulating down signals for cell growth and division, controlling the formation of stem cells, responding to cell damage and even communicating with the innate and the adaptive immune response. This review explores just how this is accomplished at the cellular and molecular level (Figure 1).
The mTOR pathway
The mTOR and IGF-1/AKT pathways are two evolutionarily conserved pathways that play critical roles in regulation of cell proliferation, survival and energy metabolism. They sense the environmental or external signals demonstrating the availability of adequate glucose and amino acid levels as well as the presence of mitogens that signal for cell maintenance and/or division. mTOR is a conserved Serine/Threonine kinase. It forms two complexes in cells, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which have distinctive physical structures and physiological functions. The mTORC1 is composed of mTOR, Raptor, AKTS1 and mLST8 proteins, and is sensitive to rapamycin inhibition26–28, whereas the mTORC2 is composed of mTOR, Rictor, Sin1, and mLST1, and is insensitive to rapamycin inhibition29–32. The mTORC1 functions to regulate cell growth and energy metabolism, and the mTORC2 is mainly involved in cytoskeleton reorganization and cell survival (Figure 2). The mTOR pathway responds to various signals, including nutrients (glucose and amino acids), energy (ATP and AMP), hypoxia, and growth factors (via IGF-1/AKT pathway). LKB1 and AMPK are two main upstream kinases of the mTOR pathway which monitor the levels of glucose and ATP. AMPK exists as a heterotrimeric complex comprising catalytic α-subunits and regulatory β- and γ-subunits.33, 34. In response to glucose starvation and low intracellular ATP levels, LKB1 phosphorylates α-subunits of AMPK on Thr172 and this in conjunction with AMP which binds to γ-subunits activates AMPK35. Activated AMPK then phosphorylates and activates the TSC2 protein36, a GTPase negatively regulating GTPbinding protein Rheb which activates mTORC1 (Figure 2). In addition to LKB1 and AMPK, hVPS34 lipid kinase, a class III PI3K, can signal the availability of amino acids to mTORC1 independently of TSC1-TSC237. Under the condition of hypoxia, HIF-1 induces the expression of REDD1, which inhibits mTORC1 activity by releasing TSC2 from the association with inhibitory 14-3-3 proteins38.
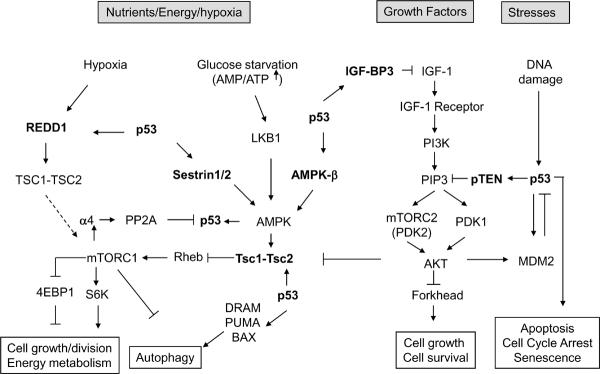
Figure 2. The coordinated regulation of p53, mTOR and IGF-1/AKT pathways.
p53 inhibits IGF-1/AKT and mTOR pathways thorough its induction of IGF-BP3, PTEN, TSC2, AMPK β1, Sestrin1/2 and REDD1 in response to stress. mTORC1 activates the α-4 subunit of the PP2A phosphatase to dephosphorylate p53 on Ser15 to inactivate p53, while AMPK phosphorylates p53 on Ser15 to activate p53. AKT-MDM2-p53 forms a negative feedback loop to negatively regulate p53, while p53-PTEN-AKT-MDM2 forms a positive loop to positively regulate p53. Furthermore, p53 activates autophagy through its regulation of mTOR pathway and p53 target genes, including DRAM, PUMA and Bax. These extensive communication and coordination between p53 and IGF-1/AKT/mTOR pathways slows down cell growth and division to prevent the accumulations of errors in response to stress and restores cellular homeostasis after stress is resolved.
The mTORC1 has two major substrates: the 4EBP1 protein (eIF4E binding protein) and the S6K (p70 ribosomal protein S6 kinase). Upon stimulation of growth signals, mTORC1 binds to eIF3 (eukaryotic initiation factor 3) and phosphorylates S6K and 4EBP139–41. Phosphorylation of S6K results in the enhanced translation of mRNAs involved in ribosomal biogenesis, mitochondrial biogenesis, and oxygen consumption. Phosphorylation of 4EBP1 leads to the release of eIF4E (the eukaryotic initiation factor 4E), which is essential for cap-dependent initiation of translation and promoting cell growth. Thus, activated mTORC1 promotes protein translation, increases cell mass and promotes cell growth. The mTORC1 also regulates autophagy, which is an important biological process of sequestration and subsequent digestion of cytoplasmic components. Autophagy provides a survival mechanism in cells by providing nutrients for cells through a catabolic breakdown of cellular components under the conditions of nutrient starvation42. At the same time, autophagy plays an essential role in maintaining genomic stability by destroying damaged, potentially harmful cytoplasmic organelles (e.g. defective mitochondria). It has been shown that loss of one allele of either of the two haploinsufficient autophagy genes Beclin 1 or UVRAG, which is a Beclin 1-binding protein, is sufficient to promote carcinogenesis43, 44. Therefore, autophagy is both a survival and a fidelity mechanism in cells. The mTORC1 kinase negatively regulates autophagy. It has been shown that the activation of Akt and PI3K in IGF-1/AKT pathway, which can activate mTORC1, results in the inhibition of autophagy, whereas the activation of LKB1, TSC1, or TSC2, which are negative regulators for mTORC1 activity, can all result in the activation of autophagy45, 46.
The IGF-1/AKT pathway
The mTOR pathway is co-regulated by IGF-1/AKT pathway to ensure both a reasonable level of nutrients and a positive signal for cell growth and division from mitogen stimulation. The binding of IGF-1 to its tyrosine kinase receptor (IGF-1R) results in the recruitment of the PI3 kinase (PI3K) to the plasma membrane and its activation, which in turn phosphorylates the phosphoinositides, increasing the concentration of PIP3 at the plasma membrane. Increased PIP3 activates PDK1 and mTORC2 (PDK2). The resultant phosphorylation of AKT on Thr308 by PDK1 and on Ser473 by mTORC2, respectively, leads to the full activation of AKT47–49. In turn, AKT phosphorylates several cellular proteins, including FOXO transcription factors, BAD, MDM2, and GSK3α/β to facilitate cell survival and cell cycle entry50–52. IGF-1/AKT pathway regulates mTORC1 through AKT phoshporylation of TSC2 on multiple sites, which inhibits the TSC2 GTPase activity resulting in the activation of Rheb and mTORC153. This coordination of IGF-1/AKT and mTOR pathways enables the activation of mTOR, which in turn promotes cell growth in response to the stimulation of growth factors, including insulin and IGF-1 (Figure 2).
The frequent mutation of signaling components in the mTOR and IGF-1/AKT pathways in tumors have suggested the critical role of these two pathways in tumorigenesis54, 55. PI3K, AKT, and Rheb are mutated or highly expressed in a variety of tumors, which can all promote the activities of these two pathways. LKB1, PTEN, TSC1, TSC2 are tumor suppressors, and their mutations result in a loss of function which has been frequently observed in various tumors. These findings suggest that altered activation of these two pathways regulating cell growth and division, cell survival, and energy metabolism play critical roles in tumor initiation and/or progression.
The mTOR/IGF-1/AKT pathways signal for cell growth and division in response to high levels of glucose and amino acids to support that growth. The p53 pathway responds to stress signals that will introduce errors or mutations into the process of cell division. To prevent such a high mutation rate from occurring in a dividing cell a signal that activates p53 inactivates the mTOR/AKT pathways. Just how this is accomplished is discussed in the next section.
p53 regulation of IGF-1/AKT/mTOR pathways
The IGF-1/mTOR pathways are monitored by the p53 pathway because stress makes the process of cell growth and cell division subject to high error rates. Thus in responding to stress the p53 pathway shuts down the IGF-1/AKT and mTOR pathways so as to limit the error frequency during cell growth and division12–14, 56. This increases the fidelity of these processes over the life time of an organism. p53 induces the expression of a list of p53 target genes in the IGF-1/AKT and mTOR pathways, including IGF-BP3, PTEN, TSC2, AMPK β1, Sestrins 1 and 2, and REDD112–14, 57, 58. All these gene products negatively regulate the IGF-1/AKT and mTOR pathways in response to stress signals (Figure 2). IGF-BP3 protein binds to IGF-1 and prevents IGF-1 binding to the IGF-1 receptors to shut down IGF-1/AKT signaling. p53 induces PTEN, a PIP3 phosphotase which degrades PIP3 to PIP2 which no longer activates PDK-1 and mTORC2, and thus decreases AKT activity. Therefore, the regulation of IGF-BP3 and PTEN by p53 negatively regulates IGF-1/AKT signaling, which further leads to the negative regulation of mTORC1 activity through the direct phosphorylation of TSC2 by AKT. p53 also induces TSC2 and β-subunits of AMPK12, 13. p53 induces the expression of Sestrin 1 and Sestrin 2, which interact with the α-subunits of AMPK and result in the phosphorylation of AMPK on Thr172. This leads to the activation of AMPK and TSC2, which inhibits mTORC1 activity14. Furthermore, p53 can regulate the expression of REDD1, which can negatively regulate mTORC1 activity in response to hypoxia38, 58 (Figure 2). Thus through the transcription of seven different target genes, p53 negatively regulates the IGF-1/AKT and mTOR pathways creating an inter-pathway network that permits cells under stress to shut down cell growth and division to avoid the introduction of errors during these processes. In this way p53 monitors the activity of cell growth and division.
The balance between a response to cellular stress or a commitment to cell growth and division is modulated by several regulatory loops between p53 and the IGF-1/AKT/mTOR pathways. The activation of p53 in response to stress leads to the phosphorylation of AMPK on Thr172 and its activation12, 59, in which Sestrins 1 and 2 may be involved14. mTORC1 phosphorylates and activates the alpha-4 subunit of the PP2A phosphatase which then dephosphorylates p53 on Ser1560. On the other hand, AMPK can directly or indirectly phosphorylate p53 on Ser15 in response to nutrient starvation24, 61, 62. It has been shown that the phosphorylation of p53 on Ser15 through AMPK induced by glucose starvation leads to cell cycle arrest and cell survival in normal MEF cells61, while it leads to the p53-mediated cell apoptosis in oncogene (E1A) transformed MEF cells13. Thus, mTORC1 and AMPK obtain signals from both the environment (glucose and amino acid levels) and the transformed state of a cell (an abnormal signal transduction pathway) and can then coordinately regulate the responses of p53. PTEN connects another feedback loop between p53 and the IGF-1/AKT pathway. p53 activation induces the expression of PTEN in selective cells and tissues13, 57, which in turn inhibits the activity of AKT. The AKT can phosphorylate MDM2 on ser166/186, which activates MDM2 to decrease p53 levels and activity, and thus forms a negative feedback loop with p5351. This p53-PTEN-AKT-MDM2 loop positively regulates p53 activity after stress and this positive feedback will kill a cell with a PTEN mutation (in a tissue specific fashion). Interestingly, it appears that the mTOR pathway is able to positively regulate p53 activity and function under certain circumstances. Loss of TSC1 or TSC2, the negative regulators of mTORC1, results in constitutive activation of mTORC1, which leads to the accumulation of p53 protein and increased apoptosis in response to stress conditions (e.g. glucose starvation, and DNA damage)63. Consistently, p53 protein levels are elevated in TSC tumors (hamartoma), which rarely become malignant. This coordinated relationship between mTOR and p53 during cellular stress provides a possible explanation for the benign nature of hamartomas. It is then clear that communication flows both between p53 and the mTOR/AKT pathway and back from mTOR/AKT to the p53 pathway.
In some organisms mutations which reduce the activity of the IGF-1/AKT pathway result in longer longevity. There is growing evidence that p53 levels in mice can affect the longevity of these organisms, connecting these two pathways with a different phenotype.
The activation of p53 enhances autophagy in cells. It has been shown that p53 activation in response to stress induces much more autophagosomes in p53 wild type MEF (mouse embryo fibroblast) cells than p53 null MEF cells12. This occurs through the inhibition of mTOR pathway by p53, and/or through the regulation of some p53 target genes directly involved in autophagy. p53 has been shown to enhances autophagy through its induction of DRAM (damage-regulated autophagy modulator), a lysosomal protein64. PUMA and Bax, two well-known p53 target genes which positively regulate apoptosis, have also been recently shown to induce autophagy65. Interestingly, it has been shown that the mutant p53 protein accumulated in the cytoplasm of tumor cells can inhibit autophagy66. Although the mechanism by which mutant cytoplasmic p53 protein inhibits autophagy remains unclear, it supports the conception the activation of autophagy by p53 contributes to p53's function as a tumor suppressor.
Energy metabolism and cancer
Metabolic changes have been regarded as a hallmark of tumor cells67, 68. The great majority of tumor cells primarily utilize aerobic glycolysis rather than the much more efficient aerobic mitochondrial respiration for their energy needs, a switch known as the Warburg effect69. Because glycolysis produces ATP much less efficiently (2 ATP per glucose molecule) than aerobic respiration (36 ATP per glucose molecule), tumor cells compensate by having a much higher rate of glucose uptake and utilization than normal cells. Based on the Warburg effect and its high utilization of glucose, Positron Emission Tomography (PET) has been established and widely used for tumor detection. The tumor cells take more of the glucose analog18 flurodeoxyglucose than normal cells. Recently, metabolic changes in tumors have been identified as a possible key contributor to malignant progression68, 70. Metabolic changes in tumor cells can confer tumor cells advantages of proliferation and survival. For instance, altered metabolism may provide biosynthetic substrates (e.g. nucleotides, fatty acids and proteins) for tumor cell proliferation, may avoid apoptosis of tumor cells and confer their resistance to therapy. Emerging evidence has shown that these changes could be targeted for the development of new cancer therapies. Lactate dehydrogenase A (LDH-A) and pyruvate kinase M2 are two enzymes that regulate the rate-limiting final steps of glycolysis, and are highly expressed in many tumors. It has been shown that knockdown of LDH-A levels diminished the ability of tumor cells to metabolize pyruvate to lactate and resulted in the stimulation of mitochondrial respiration and severely compromised the tumorigenicity of cancer cells71. Similarly, knockdown of pyruvate kinase M2 expression in tumor cells was able to reverse the Warburg effect and reduce the tumorigenicity of cells72. These results suggest that reversing the Warburg effect in tumor cells greatly compromised tumorigenecity of cancer cells, and targeting the metabolic changes could be an effective strategy for cancer treatment.
The molecular mechanism underlying the Warburg effect is not well-understood67, 68, 73. The activation of several oncogenes in cancer cells have been shown to contribute to the Warburg effect, including Myc and Akt, and hypoxia inducible factor 1 (HIF-1). Myc transcriptionally activates many of the glycolytic enzymes, and activation of Akt increases both glucose uptake and metabolism74, 75. Activation of HIF-1 is also involved in mediating the switch to aerobic glycolysis through its ability to increase the expression of genes encoding glucose transporters and glycolytic enzymes76, 77. Furthermore, HIF-1 induces pyruvate dehydrogenase kinase 1, which phosphorylates and inactivates pyruvate dehydrogenase, and thus suppresses the TCA cycle and aerobic respiration78, 79.
p53 and energy metabolism
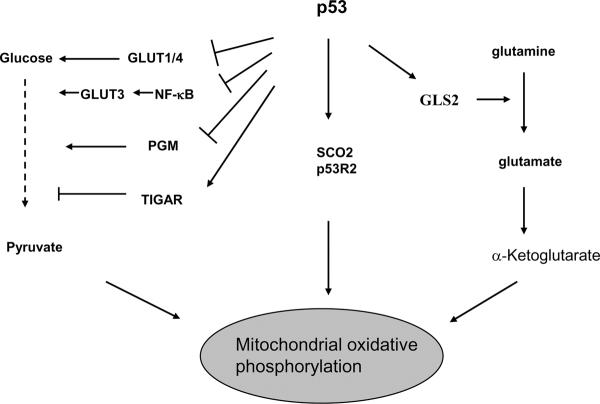
Recent studies revealed a new function for p53 in the regulation of energy metabolism. Loss of p53 has been shown to result in decreased oxygen consumption and impaired mitochondrial respiration, and promote a switch to high glucose utilization in aerobic glycolysis in cells15. In mice, p53 loss results in reduced endurance during physical exercise, suggesting a crucial role for p53 in ensuring efficient ATP production by aerobic respiration for prolonged exercise15. SCO2 (Synthesis of cytochrome c oxidase 2) and TIGAR (TP53-induced glycolysis and apoptosis regulator) were identified as two p53 target genes involved in the regulation of energy metabolism7, 15. The SCO2 gene is a key regulator of the cytochrome c oxidase complex that is essential for mitochondrial respiration. p53 induces the expression of SCO2 to ensure the maintenance of the cytochrome c oxidase complex, and thus enhances mitochondrial respiration15. TIGAR functions to lower the intracellular levels of fructose-2, 6,-bisphosphate, thereby slowing glycolysis and directing glucose to an alternative pathway, the pentose phosphate pathway (PPP) which produces more NADPH. p53 induces the expression of TIGAR, and thus slows glycolysis7. In addition to the SCO2 and TIGAR, p53 modulates glycolysis through the transcriptional regulation of some other key enzymes along the glycolytic pathway. The p53 protein reduces the transcriptional expression of phosphoglycerate mutase (PGM); loss of p53 results in the increased PGM expression, which can enhance glycolysis and thus contribute to the Warburg effect80. The p53 protein represses the transcriptional expression of glucose transporters 1 and 4 (GLUT1 and GLUT4)81, and also reduces the expression of glucose transporter 3 (GLUT3) through its negative regulation of the NF-κB pahtway82. p53 loss can lead to the activation of NF-κB, which causes an increase in the rate of aerobic glycolysis and upregulation of GLUT3 expression, while p53 expression inhibits the activation of IKK-NF-κB and decreases GLUT3 expression and therefore inhibits the high utilization of glucose in glycolysis82. Furthermore, p53 may regulate mitochondrial respiration through its regulation of the expression of ribonucleotide reductase subunit p53R2, which plays an important role in the maintenance of mitochondrial DNA17. Loss of p53R2 has been reported to result in decreased mitochondrial DNA and mitochondrial function in cells18. We recently identified GLS2 (glutaminase 2) as a novel p53-regulated gene16. GLS2 encodes a mitochondrial glutaminase which catalyzes the hydrolysis of glutamine to glutamate. p53 increases the GLS2 expression under both non-stressed and stressed conditions. GLS2 regulates cellular energy metabolism by increasing production of glutamate and α-ketoglutarate, which in turn results in enhanced mitochondrial respiration and ATP generation. Furthermore, GLS2 regulates anti-oxidant defense function in cells by increasing reduced glutathione (GSH) levels and decreasing ROS levels, which in turn protects cells from oxidative stress (e.g. H2O2) -induced apoptosis16. These findings together link the p53 protein with energy metabolism, and provide a novel mechanism that contributes to the Warburg effect (in the loss of several p53 functions) and also suggests a novel mechanism of p53 in tumor suppression (via metabolic regulation). Considering the importance of p53 in tumor suppression and the high mutation rate of p53 (> 50%) in human tumors, these findings suggest that the mutation of the p53 gene and the resultant loss of function of the p53 protein in tumors could be an important genetic change contributing to the Warburg effect.
Summary
Evolutionary processes have brought about a sophisticated and integrated set of responses to a wide variety of stresses. As cells in a multicellular organism grow and divide each stage in the process is monitored by check points that determine the fidelity of progression through cell division. Stress can result in an increase in the rate of mistakes for premature entry into S-phase, DNA replication, and chromosome segregation, resulting in mutations and the initiation of cancers. The broad responsibility for surveillance of progression through the cell cycle and ensuring the fidelity of these processes falls upon the p53 protein. This review demonstrates just how coordinated this process is in cells. This regulation takes place at both the levels of the IGF-1/AKT/mTOR pathways and the metabolic pathways (which are of course related). In response to stress and the activation of the p53 protein cellular endocytosis is enhanced by removing growth and maintenance receptors from the plasma membrane. Glucose transport into cells is slowed by the p53-mediated repression of glucose transporter genes (GLUT 1, 3 and 4). Glycolysis is slowed while oxidative phosphorylation is enhanced burning available glucose to CO2 and H2O limiting substrates for growth. The IGF/AKT-1 and mTOR pathways are inhibited by the p53 regulated transcription of several genes that encode proteins that negatively regulate these pathways. A series of proteins are synthesized in a p53 regulated fashion that prevent oxidative DNA damage and maintain a reducing environment. This is a coordinated response to stop cell growth and division and minimize damage. p53 also regulates the metabolic switch between aerobic glycolysis, observed in wound healing, embryonic development, the immune response and cancers and mitochondrial oxidative phosphorylation which is employed for the maintenance of steady state normal adult growth and turnover. In situations where abnormal signals for cell division over-rides these inhibitory steps, such as oncogenic transformation of cells, p53 mediated cell death or senescence eliminates these clones of cells resulting in benign tumors. All of these processes contribute to the tumor suppressor phenotype of the p53 gene.
Many questions remain to be answered. Most tumors, but not all cancers, take up glucose in large amounts using aerobic glycolysis to fuel the growth of the cells. Are these largely the tumors with p53 mutations or a compromised p53 functional pathway? We don't really know this. Does every PET-scan positive tumor have a p53 mutation or are other mutations (such as PI3K mutations) also over riding the impact of wild type p53 upon the Warburg effect? Several normal processes in cells (the immune response, developmental stages of growth and wound healing) also utilize the Warburg effect. Is the p53 protein inactivated during those processes? NF-kB which controls the transcriptional responses of the immune system to external stresses (organisms, cytokines) is known to have several mechanisms which inactivate the functions of the p53 protein. So when immune cells undergo clonal expansion using NF-kB and the metabolic Warburg effect p53 is transiently inactivated.
Over the next few years drugs will be developed to inhibit the enzyme activities that favor the Warburg effect in tumor cells. It will be interesting to see if these eliminate cancers. It will also be of interest to determine if the wild type p53 protein in cells treated with these drugs will contribute to the elimination of a cancer through apoptosis or senescence. The p53 protein has the ability to turn off mTOR activity. When mTOR is modulated down autophagy is activated. What is the role of autophagy in tumor cell death (mice and rats defective in autophagy develop tumors) given that it is also thought of as a survival mechanism? Will inhibiting autophagy inhibit a cancerous growth or will it eliminate a mechanism used to kill cancer cells?
It is of some interest that the activation of p53 by a stress signal, such as chemotherapy, results in enhanced exosome production by those cells10, 11. Exosomes can sensitize immune cells to respond by killing cells expressing foreign antigens. Does this happen with cancers that have a wild type p53 protein but not a mutant p53 gene? Could this contribute to tumor regression? These ideas should be tested.
These studies reviewed here bring together several signal transduction pathways that interact in cells to respond to signals for cell growth and division and stress. The next challenge for research in this area is to understand how diverse signal transduction pathways interact, what nodes they share and the impact of signaling in one pathway has upon the other pathway. For example it is already clear that the p53 pathway can interact with the Wnt-pathway, the hedgehog pathway, Myc functions, the NFκB oncogenic pathway and others. Evolutionary processes have finely integrated these pathways and if we truly understand them we can develop novel therapies that alter selectively one or more pathways. The signal transduction pathways discussed here are involved in a number of diseases; cancers, diabetes, metabolic disorders, neurological degenerative disease and normal processes such as; growth and cell division, damage repair, development of the organism, longevity and many others. We are at the beginning of understanding these many of these normal cellular processes and what goes wrong when they fail. Over the next few years we will come to appreciate the common themes and interactions that connect these molecular circuits in cells and there will be a new appreciation for the targets that cause disease and the means to prevent or stop these diseases.
Figure 3. p53 regulation of energy metabolism.
p53 induces the expression of TIGAR and inhibits the expression of PGM to inhibit glycolysis. p53 induces SCO2 and GLS2 to enhance mitochondrial respiration, and induces p53R2 to maintain mitochondrial DNA. Furthermore, p53 represses the transcriptional expression of GLUT1 and GLUT4, and inhibits the NF-κB pathway to reduce the expression of GLUT3 to reduce glycolysis. Thus, p53 inhibits glycolysis and enhances mitochondrial respiration in cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine AJ, et al. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, et al. P53 is a tumor suppressor gene. Cell. 2004;116:S67–69. doi: 10.1016/s0092-8674(04)00036-4. 61 p following S69. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Hwang BJ, et al. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci U S A. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 6.Budanov AV, et al. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 7.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Tan M, et al. Transcriptional activation of the human glutathione peroxidase promoter by p53. J Biol Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 9.Yoon KA, et al. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–140. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, et al. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 11.Yu X, et al. The regulation of the endosomal compartment by p53 the tumor suppressor gene. FEBS J. 2009;276:2201–2212. doi: 10.1111/j.1742-4658.2009.06949.x. [DOI] [PubMed] [Google Scholar]
- 12.Feng Z, et al. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Z, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 14.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 16.Hu W, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1001006107. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulawiec M, et al. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourdon A, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 19.Hu W, et al. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 20.Kang H, et al. Single nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marion RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Z, et al. p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene. 2006;25:1–7. doi: 10.1038/sj.onc.1209021. [DOI] [PubMed] [Google Scholar]
- 25.Bae BI, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 28.Kim DH, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 29.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 31.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 32.Sarbassov DD, et al. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Kahn BB, et al. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Hardie DG. AMPK and Raptor: matching cell growth to energy supply. Mol Cell. 2008;30:263–265. doi: 10.1016/j.molcel.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 35.Shaw RJ, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Inoki K, et al. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 37.Byfield MP, et al. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 38.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holz MK, et al. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 40.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280:26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- 41.Hannan KM, et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 43.Yue Z, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 45.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheid MP, et al. Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol Cell Biol. 2002;22:6247–6260. doi: 10.1128/MCB.22.17.6247-6260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayascas JR, Alessi DR. Regulation of Akt/PKB Ser473 phosphorylation. Mol Cell. 2005;18:143–145. doi: 10.1016/j.molcel.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 49.Sarbassov DD, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 50.Bader AG, et al. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 51.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 52.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 53.Inoki K, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 54.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 55.Cully M, et al. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 56.Levine AJ, et al. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20:267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 57.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 58.Ellisen LW, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 59.Braunstein S, et al. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29:5645–5656. doi: 10.1128/MCB.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kong M, et al. The PP2A-associated protein alpha4 is an essential inhibitor of apoptosis. Science. 2004;306:695–698. doi: 10.1126/science.1100537. [DOI] [PubMed] [Google Scholar]
- 61.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 62.Imamura K, et al. Cell cycle regulation via p53 phosphorylation by a 5'-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem Biophys Res Commun. 2001;287:562–567. doi: 10.1006/bbrc.2001.5627. [DOI] [PubMed] [Google Scholar]
- 63.Lee CH, et al. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 2007;26:4812–4823. doi: 10.1038/sj.emboj.7601900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 65.Yee KS, et al. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–1145. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morselli E, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 67.Garber K. Energy deregulation: licensing tumors to grow. Science. 2006;312:1158–1159. doi: 10.1126/science.312.5777.1158. [DOI] [PubMed] [Google Scholar]
- 68.Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 70.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 71.Fantin VR, et al. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 72.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 73.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 74.Shim H, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elstrom RL, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 76.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 77.Plas DR, Thompson CB. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- 78.Kim JW, et al. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 79.Papandreou I, et al. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 80.Kondoh H, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 81.Schwartzenberg-Bar-Yoseph F, et al. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 82.Kawauchi K, et al. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]