Abstract
Herpesviruses, which cause many incurable diseases, infect cells by fusing viral and cellular membranes. While most other enveloped viruses use a single viral catalyst, called a fusogen, herpesviruses, inexplicably, require two conserved fusion-machinery components, gB and the heterodimer gH–gL, plus other non-conserved components. gB is a class III viral fusogen but, unlike other members of its class, does not function alone. We determined the crystal structure of the gH ectodomain bound to gL, from herpes simplex virus 2. gH–gL is an unusually tight complex of a novel architecture that, unexpectedly, does not resemble any known viral fusogen. Instead, we propose that gH–gL activates gB for fusion, possibly through direct binding. Formation of a gB–gH–gL complex is critical for fusion and is inhibited by a neutralizing antibody, making gB–gH–gL interface a promising antiviral target.
The Herpesviridae family contains eight important human pathogens including herpes simplex viruses 1 and 2 (HSV-1 and HSV-2), varicella-zoster virus, cytomegalovirus (CMV), Epstein-Barr virus (EBV), and Kaposi’s Sarcoma virus. These enveloped viruses enter cells by fusing their envelopes with host cell membranes. This event delivers the icosahedral capsid containing the dsDNA viral genome into the cell and initiates infection. Unlike most other enveloped viruses, which use a single fusogen, all herpesviruses use the conserved core fusion machinery that consists of glycoproteins gB and the gH–gL heterodimer. Some herpesviruses employ additional receptor-binding glycoproteins (e.g., HSV gD and EBV gp42)1,2, and others require further gH–gL-associated proteins, e.g. UL128-131 of CMV3. Thus, the fusion machinery of herpesviruses is clearly more complex than that of most enveloped viruses and is, perhaps, reminiscent of the fusion machinery involved in cellular fusion processes, e.g., neurotransmitter release4, in that it also engages multiple proteins.
Previously, we determined the crystal structure of the gB ectodomain from HSV-15. gB is a class III viral fusion protein or fusogen6, presumably directly involved in bringing the viral and the cellular membranes together, but unlike other members of this class, glycoprotein G of vesicular stomatitis virus7 and baculovirus gp648, it cannot function on its own. Less is known about the role of gH–gL in fusion. It is highly conserved among herpesviruses and a major target of virus-neutralizing antibodies9, emphasizing its importance for virus infection. Several reports have previously suggested that gH may have inherent fusogenic properties. For example, when cells are transfected with expression plasmids for gH–gL from HCMV, VZV, or KSHV, cell fusion is observed in the absence of any other viral proteins10–12. Also, in HSV-1, gH–gL can cause hemifusion in the absence of gB13. Nevertheless, both gB and gH–gL are required for efficient viral entry and cell fusion in all herpesviruses, and in HSV, gB and gH–gL are thought to interact in response to receptor binding by glycoprotein D14,15.
HSV-2 gH is an 838-residue protein with a signal peptide and a single C-terminal transmembrane region; gL is a 224-residue protein with a signal peptide, but no transmembrane region. In HSV-infected cells and on mature virions, gH and gL are always found together, in a stable 1:1 complex9. Here, we report the crystal structure of the gH ectodomain bound to full-length gL from HSV-2, determined to 3.0-Å resolution. The structure reveals an unusually extensive interaction between gH and gL such that the two proteins clearly need each other to fold properly. Unexpectedly and contrary to previous ideas, the complex revealed by the crystal structure does not resemble any known viral fusogen. We propose that, instead of acting as a fusogen, gH–gL activates the fusogenic potential of gB by binding it directly. A potent anti-gH–gL neutralizing antibody inhibits formation of the gB–gH–gL complex, suggesting that the gB-binding site in gH–gL could be located in the vicinity of its epitope. The gB-binding site is an attractive target for antiviral design, and we propose its possible location. Moreover, the structure of gH–gL suggests a new paradigm for how viral fusion with cell membranes is accomplished.
RESULTS
Crystal structure of the gH–gL complex
The expressed HSV-2 gH–gL complex contains residues Gly48 to Pro803 of gH, followed by a C-terminal His6 tag, and residues Gly20 to Asn224 of gL. Removal of residues His19 to Thr47 of gH from the expression construct was necessary to obtain diffraction quality crystals. These missing N-terminal residues could be located at the top of the molecule (Supplementary Fig. 1). Removal of these residues does not affect cell-cell fusion or viral entry16. Thus, the structure is a good representation of the native HSV-2 gH–gL.
The crystal structure was determined using single anomalous dispersion and a selenomethionine derivative (Table 1 and Supplementary Fig. 2). The final model contains residues Arg49 to Pro797 of gH, except for three disordered loops Gly116 to Pro136, Thr328 to Asp331, and Arg720 to Arg724, and residues Thr24 to Asn203 of gL, except for two disordered loops Phe112 to Ala114 and Leu166 to Pro196 (Fig. 1a).
Table 1.
Data collection and refinement statistics
| Native | SeMet | |
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Cell dimensions | ||
| a, b, c (Å) | 88.26, 88.26, 333.41 |
88.34, 88.34, 332.84 |
| α,β,γ (°) | 90, 90, 90 | 90, 90, 90 |
|
Peak |
||
| Wavelength (Å) | 0.9795 | 0.9789 |
| Resolution (Å) | 39.20-3.00 (3.05-3.00)* |
40.00-3.30 (3.36-3.30) |
| Rsym or Rmerge | 0.131 (0.873) | 0.188 (0.648) |
| I / σI | 31.0 (1.94) | 19.3 (5.2) |
| Completeness (%) | 99.8 (100) | 100 (100) |
| Redundancy | 5.9 (6.0) | 16.3 (16.4) |
| Refinement | ||
| Resolution (Å) | 39.20-3.00 | |
| No. reflections | 25,991 (2,475) | |
| Rwork / Rfree | 0.1701/0.2425 | |
| No. atoms | ||
| Protein | 6567 | |
| Ligand/ion | 66 | |
| Water | 52 | |
| B-factors | ||
| Protein | 53.78 | |
| Ligand/ion | 98.74 | |
| Water | 33.78 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.217 |
A single crystal was used to collect the Native and the Semet SAD datasets, each.
Values in parentheses are for highest-resolution shell.
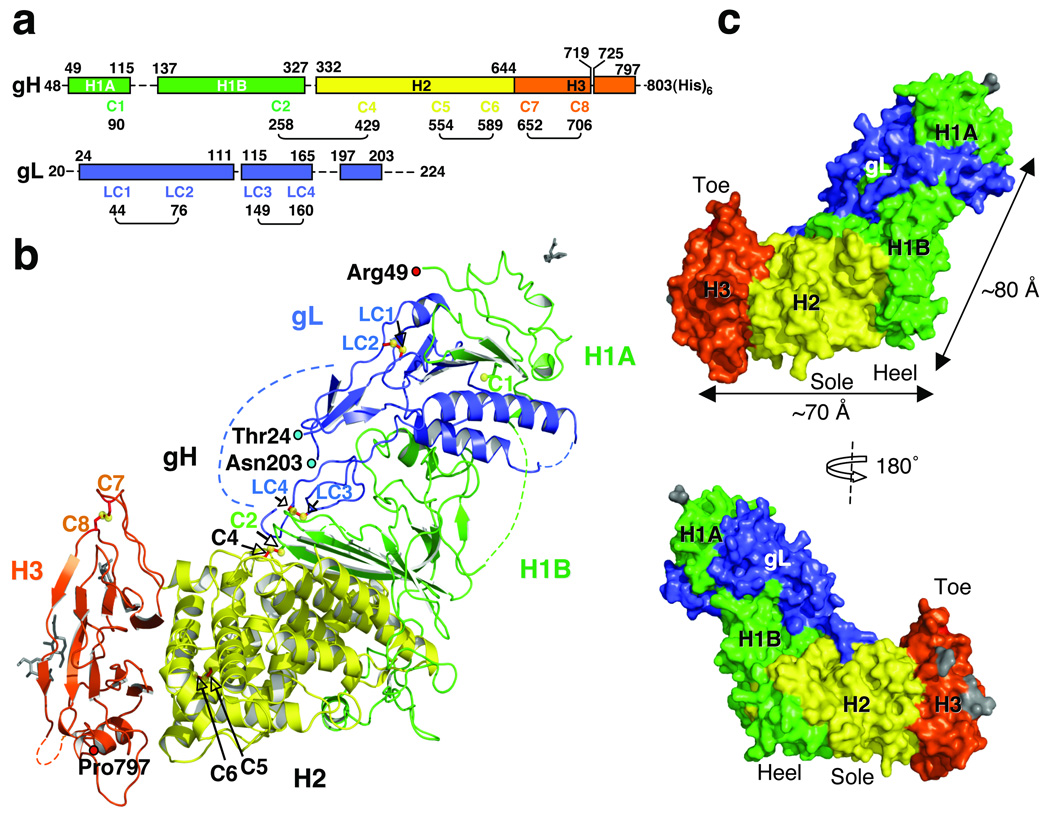
Fig. 1. Structure of HSV-2 gH–gL complex.
(a) Domain arrangement. Regions present in the expression construct but missing from the final model are shown as dashed lines. gH domains are in green (H1), yellow (H2) and red (H3); gL is in blue. The numbering scheme for gH cysteines is based on HSV-1 gH, which has eight cysteines. HSV-2 gH has seven cysteines, lacking C3. (b) Side view of gH–gL structure showing disordered segments (dotted lines), disulfides (yellow spheres and red sticks), and sugars (grey). (c) gH–gL in molecular surface representation. View in the top panel is the same as in (b). View in the bottom panel was generated by 180° rotation around the vertical axis.
The gH–gL heterodimer has the overall shape of a “boot”, approximately 80-Å high and 70-Å long (Fig. 1b,c). The C terminus of the gH ectodomain, which would normally lead into its transmembrane region, is located near the “sole” side of the “toe”. Using cryoelectron tomography of the HSV-1 virions, Grünewald and colleagues observed 15–20 nm glycoprotein spikes, which emerge from the envelope at an angle and often appear curved17. Although the identity of these spikes was not determined, their unusual shape is highly reminiscent of the gH–gL structure. Different projections of the gH–gL spike would yield curved or straight images, depending on the orientation. So, on the virion surface each gH–gL molecule is probably oriented such as to appear standing on its “toe” and leaning to the side (Supplementary Fig. 3a). The gH–gL heterodimer is smaller than gB, but larger than gD (Supplementary Fig. 3b).
Architecture of gH
gH has three distinct domains – the N-terminal domain that binds gL (domain H1), the central helical domain (domain H2), and the C-terminal β-sandwich domain (domain H3) (Figs. 1, 2 and Supplementary Fig. 4). None of the domains have any previously described structural homologues18.
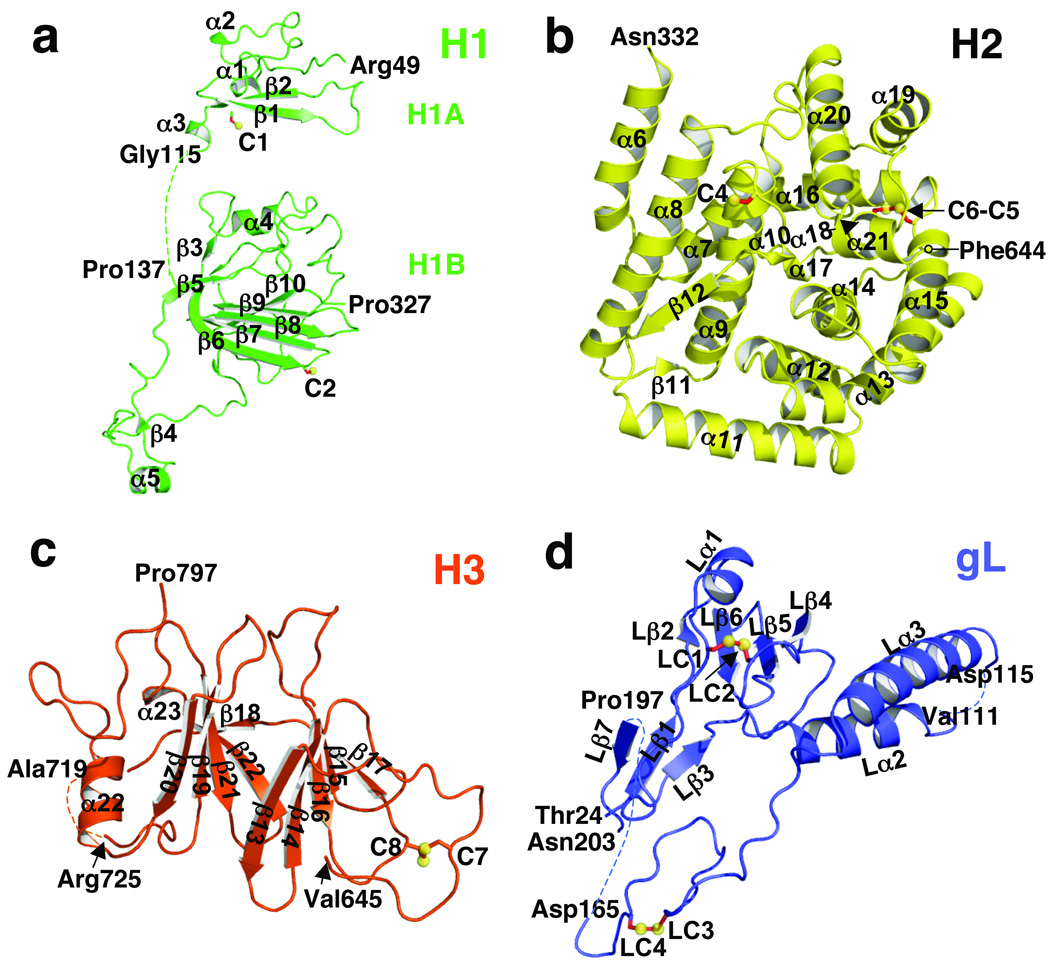
Fig. 2. Domains of gH and gL.
(a) domain H1, (b) domain H2, (c) domain H3, (d) gL. The coloring scheme is the same as in Fig. 1. Disulfides are shown as yellow spheres and red sticks and labeled. All secondary structure elements are labeled. Disordered segments are shown as dotted lines. Labeled residues indicate the limits of individual domains and the disordered loops.
Domain H1 is located in the upper part of the gH–gL “boot” and consists of subdomains H1A and H1B that are connected by a linker, residues Gly116 to Pro136, missing from the structure. Subdomain H1A, residues Arg49 to Leu115, contains a β-hairpin that forms a six-stranded mixed β-sheet with four strands coming from gL (β2-β1-Lβ4–Lβ5–Lβ6-Lβ2), plus three short helices, α1–α3. Subdomain H1B, residues Ala137 to Pro327, contains a six-stranded antiparallel β-sheet (β12-β6–β7–β8–β9–β10), in which five strands come from domain H1 and one, β12, from domain H2. This β-sheet curves around the upper boundary of the central helical domain and has the appearance of a “diaphragm” that separates the bulk of domain H1 and almost all of gL from the helical domain H2. Subdomain H1B also includes a three-stranded antiparallel β-sheet (β3–β5–β6), a short strand β4 that makes a small two-stranded antiparallel β-sheet (β4–β11), and two short helices, α4 and α5. Residues Arg176 to Thr230 have almost no contacts with the rest of the H1 domain. They form a long, sling-like extension that loops around the “heel” of the gH–gL boot, tethering domain H1 to domain H2. Domain H1 does not have a folded core, with the possible exception of the six-stranded β-sheet, which likely accounts for the misfolding of gH in the absence of gL16,19.
The central domain H2, residues Asn332 to Phe644, is globular and mostly helical. It contains 13 α-helices and three short 310 helices, α6 through α21. In addition to helices, domain H2 has a strand β12, which participates in the six-stranded β-sheet within the subdomain H1B, and a short strand β11 that makes a small antiparallel β-sheet with strand β4 of the subdomain H1B.
The C-terminal domain H3, residues Val645 to Pro797, is located at the “toe” end of the “boot”. It is a 10-stranded β-sandwich where each side is composed of a five-stranded β-sheet. The outward-facing sides of the sandwich are decorated with many extended loops giving the domain an elongated appearance. One side has an extension that leads to the transmembrane region. The other side has three loops, two of which are connected with a disulfide (C7–C8).
Although gH proteins are conserved among different herpesviruses, the sequence conservation is uneven across the protein (Supplementary Fig. 5a). Domain H1 is the most divergent, which is unsurprising given that only about 30% of its residues adopt regular secondary structure. Domain H2 and especially domain H3 are more conserved and probably have the same fold in different gH proteins. Domain H3 is the most highly conserved (Supplementary Fig. 5a) and thus is likely to be functionally important. Indeed, several non-functional mutations map to this domain20.
Six of the seven cysteines in HSV-2 gH form three disulfide bonds. Their pairwise assignment (C2–C4, C5–C6, and C7–C8) in our structure agrees with that previously surmised from genetic studies21. Unpaired C1 is present only in HSV-1 and HSV-2 (Fig. 1 and Supplementary Fig. 5a), but gH proteins from other herpesviruses also contain non-conserved unpaired cysteines. C2 and C4 are conserved in most alphaherpesviruses. C5 and C6 are completely conserved; mutation of either residue causes HSV gH–gL to misfold21. While C8 is completely conserved, C7 is positionally conserved in all herpesviruses except pseudorabies virus (PRV), which uses another nearby cysteine as C7 (Supplementary Fig. 5a). Several non-conserved cysteines in other herpesviruses, when mapped onto our structure, closely approach each other and would require only minor conformational changes to make disulfide bonds. This provides further evidence that gH proteins from different herpesviruses share a conserved fold, especially domains H2 and H3.
Architecture of gL and its role in the gH–gL complex
Like domain H1 of gH that it binds, gL does not have a stable core. Only ~30% of gL residues adopt regular secondary structure, which include three helices Lα1–Lα3 and two β-sheets (Figs. 1, 2 and Supplementary Fig. 4). One four-stranded antiparallel β-sheet forms a six-stranded β-sheet with two strands from gH. Another is a short three-stranded mixed β-sheet (Lβ3-Lβ1-Lβ7). Four cysteines in gL form two disulfide bonds (LC1–LC2 and LC3–LC4), which are conserved in HSV-1 (Supplementary Fig. 5b), and all are critical for proper processing and activity of gH–gL21. In the structure, the disulfides clearly maintain the proper fold of gL. Most residues within the Leu166 to Asn224 region of gL are missing from the structure, except for a 7-amino-acid peptide. Its sequence has been tentatively assigned to residues Pro197 to Asn203. Removal of residues Gly162 to Asn224 from either HSV-2 gL (Supplementary Fig. 6) or HSV-1 gL22 reduces the cell surface expression of the gH–gL complex but not its activity in cell fusion, because cell fusion decreased in proportion to the cell surface expression. Therefore, residues Gly162 to Asn224 do not appear critical for fusion in either HSV-1 or HSV-2. Further, these residues are absent from gL proteins in closely related alphaherpesviruses PRV, bovine herpesvirus (BHV), and VZV (Supplementary Fig. 5b). Thus, the missing C-terminal region of gL is probably of limited functional importance.
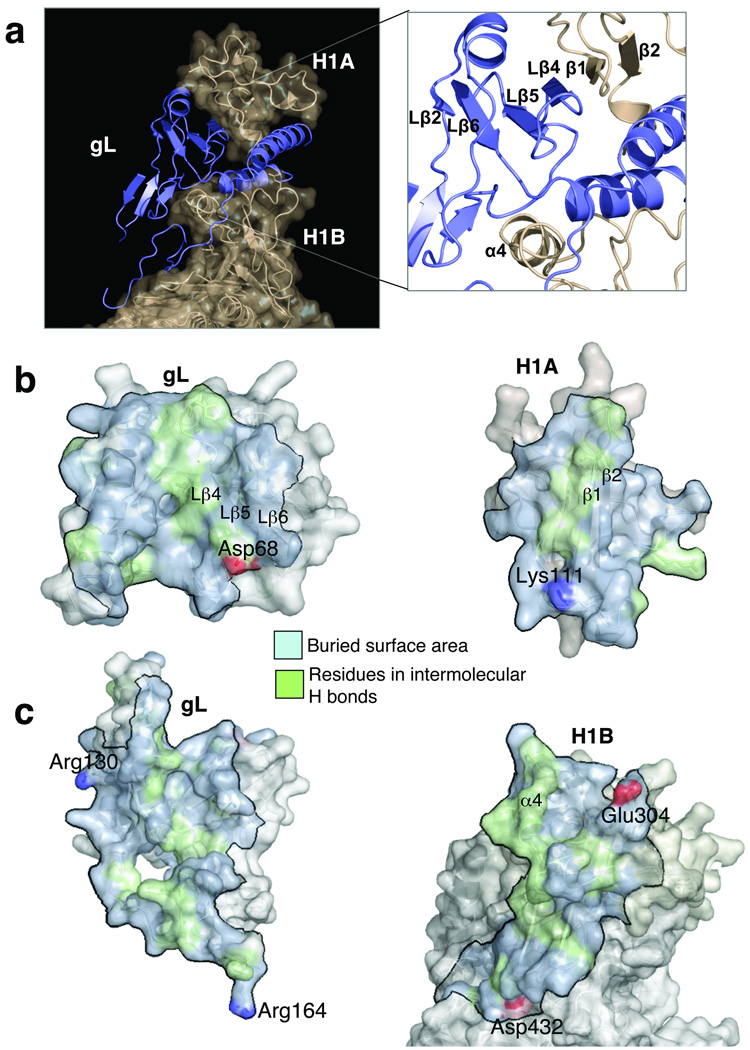
gL is required for correct folding and trafficking of gH19,23 but is not a true chaperone. Chaperones assist the non-covalent folding/unfolding and the assembly/disassembly of other macromolecules but do not remain bound to them when the latter perform their normal biological functions24. By contrast, the gH–gL complex is very stable, so gL is more of a scaffolding protein for gH, perhaps, with some additional function. In the structure, gL interacts almost exclusively with the H1 domain of gH, consistent with our previous data9. Subdomains H1A and H1B clamp gL like tongs (Fig. 3a) and make extensive contacts. Formation of the gH–gL complex buries a large accessible surface area, 7326.8 Å2 25, most of which is hydrophobic (Fig. 3b,c). Subdomain H1A packs very well into the V-shaped groove on top of the gL molecule (Fig. 3a) and shares a six-stranded mixed β-sheet with gL, in which two strands are contributed by gH and four are contributed by gL. The interacting surfaces are highly complementary; thus, the two proteins need each other to fold correctly.
Fig. 3. gH–gL interface.
(a) The gH “clamp”. The molecular surface of gH is shown in beige; gL is rendered as a blue ribbon. Inset: up-close view of the H1/gL interface. Secondary structure at the H1/gL interface is labeled. (b) “Open-book” view of the H1A/gL interface viewed from the “heel” of the gH–gL “boot”. (c) “Open-book” view of the H1B/gL interface viewed from the top of the gH–gL “boot”. Intermolecular interactions in (b) and (c) are color-coded as follows: buried hydrophobic surface (light blue), buried hydrophilic surface (green), and negatively charged (red) and positively charged (blue) residues, in salt bridges.
gH–gL does not resemble a viral fusogen
Previously, several reports suggested that gH may have inherent fusogenic properties10–13. Moreover, some have proposed that gH may be a viral fusogen, based on studies carried out with peptides corresponding to predicted heptad repeats and fusion peptides26–33. The overall architecture of the gH–gL complex revealed by the crystal structure does not resemble any known viral fusogens. Currently, 3 classes of viral fusogens are recognized34. These proteins have different architecture and form trimers (class I and III) or dimers (class II) in their prefusion form. Despite differences in architecture, all form trimeric rod-like structures in their postfusion form.
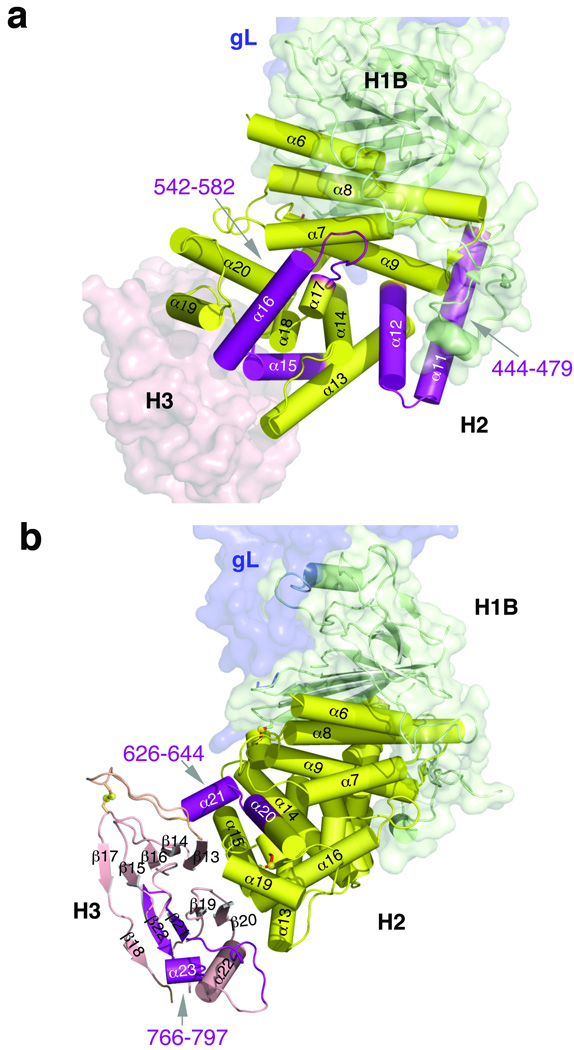
We analyzed the locations of predicted heptad repeats and fusion peptides in the structure of gH–gL. Figure 4a highlights two predicted heptad-repeat peptides, residues 444–479 and 542–582, that inhibit cell fusion30. Both peptides form helical hairpins and appear important for the stability of the helical core of domain H2. We want to emphasize, however, that heptad repeats are commonly found in helices that form coiled coils and helical bundles. Although many helices within domain H2 of gH form helical bundles, the trimeric hairpin bundle that is characteristic of the postfusion form of class I and class III fusogens34 is absent from the gH–gL structure.
Fig. 4. Locations of several predicted heptad repeats and fusion peptides in gH.
(a) predicted heptad repeat sequences in gH that inhibit cell-cell fusion as synthetic peptides, residues 444–479 and 542–582, are shown in purple and labeled. Domain H3 is shown as a light pink surface. (b) putative fusion peptide sequences of gH, residues 626–644 and 766–797, are shown in purple and labeled. Domain H3 is shown as light pink ribbon. Secondary structure elements within domains H2 and H3 are numbered according to Figs. 2 and 3.
Fusion peptides are hydrophobic stretches within viral fusogens that are capable of binding to membranes. Several fusion peptides have been proposed in gH, some of which have been shown to bind liposomes as synthetic peptides, with modest affinity26,27,31. This is, perhaps, not surprising given the hydrophobicity of these peptides. But in the gH–gL structure, these putative fusion peptides are buried helices or β hairpins that participate in multi-stranded sheets. For example, residues 626–644 compose 2 helices that are mostly buried (Fig. 4b). As another example, residues 766–802 form two strands in the middle of a 5-strand β sheet in conserved domain H3 (Fig. 4b). Obviously, removing these two strands would completely destroy the β sheet. It is difficult to envision how either residues 626–644 or 766–802 be “yanked” out of the hydrophobic core to promote fusion in the context of the entire protein. Further, we have been unable to pinpoint any fusion peptides in the gH–gL structure based on known fusion peptides. For example, N-terminal helical fusion peptides, characteristic of class I fusogens, are generated from a precursor protein by proteolytic cleavage. But currently, there are no data to suggest that any fusion-relevant proteolytic cleavage takes place in gH. Likewise, we have been unable to identify internal fusion peptides of class II and III viral fusogens - loop-like structures with hydrophobic residues at their tips - within the structure of gH–gL.
The mechanism that underlies the ability of some synthetic peptides to inhibit fusion is currently unclear. Some regions of these peptides are exposed on the surface, however (Supplementary Fig. 7), and if they were involved in interactions with other proteins, this could, perhaps, explain the ability of corresponding synthetic peptides to inhibit cell fusion.
Epitopes of anti-gH–gL neutralizing antibodies
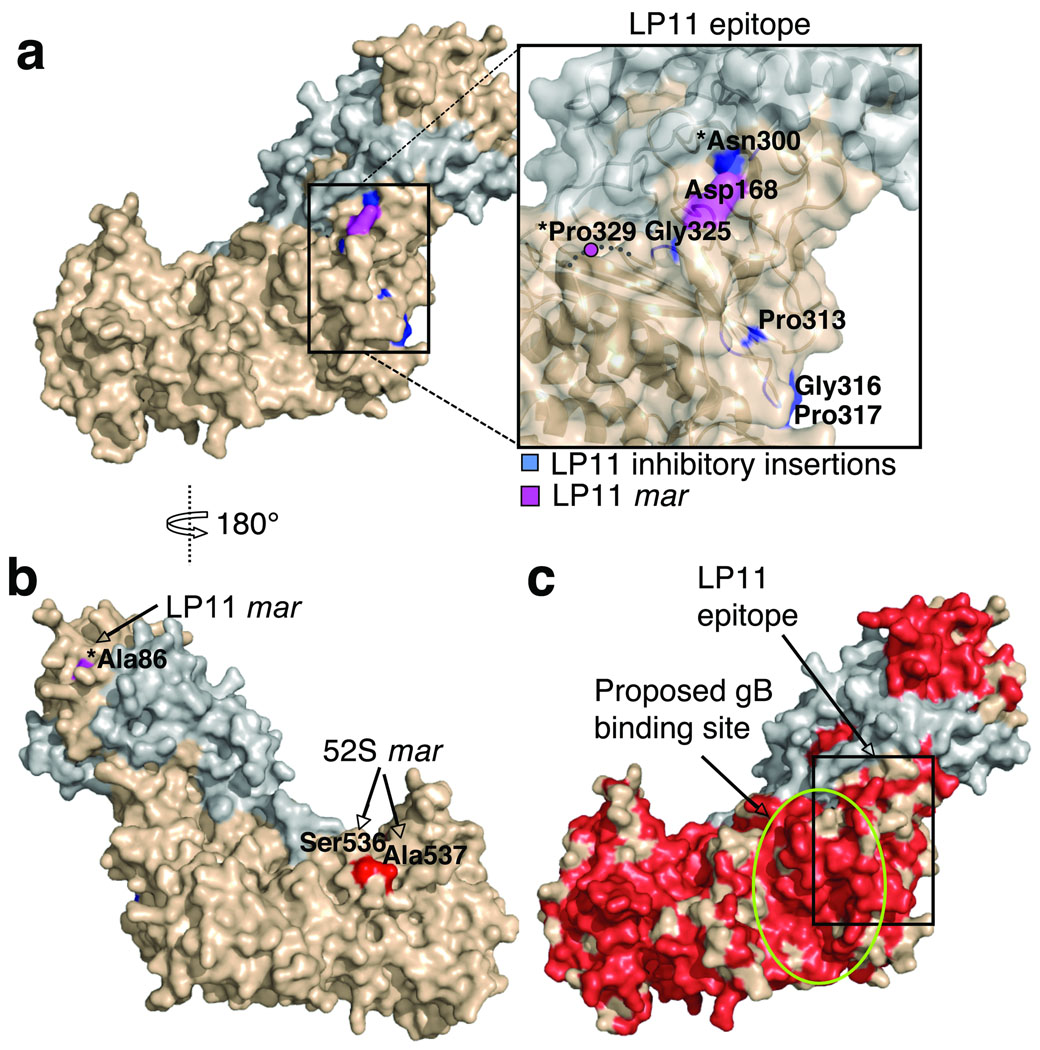
gH–gL is a major target of virus-neutralizing antibodies9. LP11 and 52S are two potent neutralizing antibodies against HSV-1 gH–gL35–37. The high sequence identity between the gH and gL proteins from HSV-1 and HSV-2 (Supplementary Fig. 5c) implies that their structures are very similar and that the epitopes of anti-HSV-1 antibodies can be mapped onto the structure of the HSV-2 gH–gL. The LP11 epitope is present only on the intact HSV-1 gH–gL. Five LP11 mar mutant viruses have been isolated, and each contain on of the following mutations38: E86K, D168G, D168N, R329W, or R329Q. HSV-1 gH residues Glu86, Asp168, and Arg329 correspond to HSV-2 gH residues Ala86, Asp168, and Pro329. In the structure, two of the HSV-2 gH equivalents, residues Asp168 and Pro329, are located near each other, but the third, Ala86, is on the opposite face of the molecule (Fig. 5a,b). Insertions that destroy LP11 binding20 map to the region of the molecular surface that contains residues Asp168 and Pro329 (Fig. 5a). Therefore, the LP11 epitope is likely located near residues Asp168 and Pro329 (Fig. 5a). Resistance to LP11 neutralization in mar mutants D168G, D168N, R329W, and R329Q is probably due to a loss of a charge that affects LP11 affinity for its epitope. It is harder to explain the mechanism of resistance in the E86K mutant because residue 86 is located far from the proposed LP11 epitope (Fig. 5a,b).
Fig. 5. The epitope of the HSV-1 neutralizing antibody LP11 defines the gB-binding site.
(a) gH–gL in surface representation, gH (beige) and gL (grey). The proposed LP11 epitope is boxed and enlarged in the inset. (b) gH–gL is color-coded as in (a). The view in (b) is rotated 180° around the vertical axis relative to (a). Residues with a star are not conserved between HSV-1 and HSV-2. (c) gH–gL is colored by sequence conservation between HSV-1 and HSV-2, with identical residues in red. The view is as in (a). LP11 epitope is boxed. mar is monoclonal antibody resistance mutation.
Two 52S mar mutant viruses have been isolated; they contain mutations S536L and A537V, respectively38. HSV-1 residues Ser536 and Ala537 are the same in HSV-2, and these adjacent residues define the 52S epitope, located on the opposite face of the gH molecule from the LP11 epitope (Fig. 5b). Thus, the epitopes of LP11 and 52S do not overlap (Fig. 5a,b), consistent with previous observations that LP11-resistant viruses are sensitive to 52S and vice versa38.
A neutralizing antibody inhibits interactions between gB and gH–gL
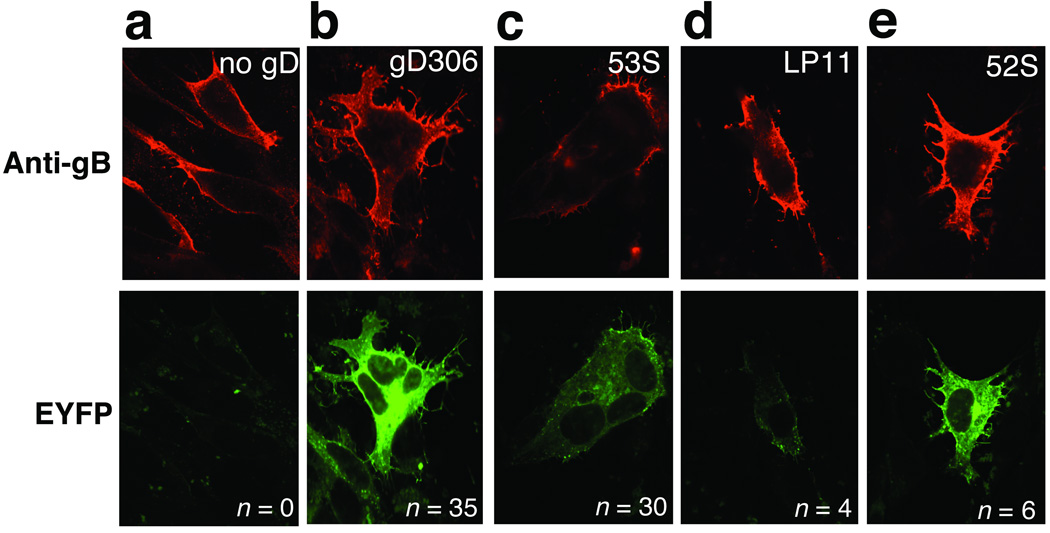
Formation of the gB–gH–gL complex is necessary for fusion39 and is inhibited by certain anti-gB neutralizing antibodies39. Thus, to gain insight into the role of gH–gL in fusion, it is important to know where gB binds gH–gL. An anti-gH or anti-gL neutralizing antibody can possibly inhibit infectivity by preventing gH or gL from binding gB or other entry glycoproteins. To map a gB-binding site on gH–gL, we tested two anti-HSV-1 gH–gL neutralizing antibodies, LP11 and 52S, as well as 6 non-neutralizing antibodies in a bi-molecular fluorescence complementation assay (BiMC) (Supplementary Table 1). In this assay, two inactive halves of enhanced yellow fluorescent protein (EYFP) are fused to the C termini of gB and gH, respectively, and the resulting proteins are transiently expressed in receptor-bearing cells. If gB and gH associate, the two EYFP halves are brought together and fluorescence is restored. Fusion can be monitored in the same experiment by observing the formation of syncytia. This assay was used previously to show that gB and gH–gL interact and that fusion occurs only if gD is also expressed or added as a soluble protein15,39.
In the absence of gD, only background green fluorescence and no syncytia were observed (Fig. 6a). In the presence of soluble gD306, bright EYFP fluorescence was observed, along with syncytia (Fig. 6b), indicating BiMC and thus interactions between gB and gH–gL. Non-neutralizing antibody 53S did not affect the number of syncytia or BiMC (Fig. 6c). The rest of the non-neutralizing antibodies had the same effect as 53S (Supplementary Table 1). Neutralizing antibody LP11 visibly reduced BiMC (Fig. 6d), while neutralizing antibody 52S did not (Fig. 6e). Both LP11 and 52S markedly inhibited fusion, however, reducing the number of syncytia from 35 to 4 and 6, respectively. To summarize, none of the tested non-neutralizing antibodies could inhibit BiMC or formation of syncytia. By contrast, both neutralizing antibodies, LP11 and 52S, inhibited formation of syncytia but only LP11 inhibited BiMC. We conclude that LP11 blocks interactions between gB and gH–gL and prevents fusion, while 52S does not block gB–gH–gL interactions but nonetheless inhibits fusion. Thus, 52S probably blocks a step in the entry process that occurs after the gB–gH–gL interaction.
Fig. 6. Effect of anti-gH antibodies on gB–gH BiMC.
Top panels show gB surface expression (red) detected using immunofluorescence. Bottom panels show EYFP BiMC (bright green fluorescence). (a) no gD, (b) gD306, (c) gD306 plus mAb 53S, (d) gD306 plus mAb LP11, and (e) gD306 plus mAb 52S. n is the number of syncytia from a representative experiment. EYFP is enhanced yellow fluorescent protein.
DISCUSSION
Herpesvirus gH–gL is a conserved component of the fusion machinery with a yet unassigned function. Whether gH–gL is a fusogen has been a subject of much speculation26–33. It is also a target of potent neutralizing antibodies. We have determined the crystal structure of the gH–gL complex from HSV-2, which has an unusual boot-shaped architecture. The structure reveals extensive interactions between gL and the N-terminal domain H1 of gH. Both lack stable cores and are clearly required for each other’s folding, which explains why gH and gL are always found in a complex. The sequences of domain H1 and of gL vary substantially among herpesviruses. For example, gL proteins from alpha-herpesviruses cannot be aligned at all with those from either beta- or gamma-herpesviruses. Not surprisingly, gL cannot be functionally swapped between different herpesviruses, with the exception of gL proteins from closely related HSV-1 and HSV-221,40. It appears that the gH–gL pairs in each herpesvirus have co-evolved to form tight complexes, while presenting quite variable surfaces, perhaps, for binding other viral or cellular proteins. By contrast, domains H2 and H3 of gH are independently folded entities. Being more highly conserved, these domains likely have similar folds in gH–gL from different herpesviruses and have conserved functional roles.
What is the function of gH–gL? Most enveloped viruses require only one viral fusogen. Fusion can be triggered by pH or by receptor binding, but typically, the receptor-binding function is performed by the fusogen. In paramyxoviruses, fusogen F is thought to be activated by a homotypic viral attachment protein, e.g., HN41, but HN-independent fusion has been observed for some paramyxovirus F proteins42. In contrast, gB requires the presence of gH–gL for function. Two exceptions to this rule have been reported. First, EBV gB containing truncated cytoplasmic domain has been showed to mediate fusion with epithelial cells in the absence of gH–gL, possibly due to increased surface expression43. Second, either gB or gH–gL can mediate membrane fusion during nuclear egress of HSV-144. But, neither observation has yet been confirmed in gB proteins from other herpesviruses.
Previously, gH–gL has been proposed to be a fusogen in its own right. Given that gH–gL appears to be able to mediate cell-cell fusion in some herpesviruses10–13, even if inefficiently, we cannot exclude the possibility that gH–gL has some intrinsic fusogenic properties. However, the gH–gL complex revealed by the crystal structure does not resemble any known viral fusogen, and based on our analysis of the structure, we conclude that it is not a viral fusogen. Moreover, for efficient membrane fusion during viral entry and cell-cell fusion, both gB and gH–gL are required. We propose that in this biologically more relevant scenario, gH–gL regulates the transition of gB into its fusion-active state. gH–gL could, in principle, function as a negative or a positive regulator of gB. If gH–gL were a negative regulator of gB, it would keep gB in an inactive prefusion state, and gD binding to its receptor would de-repress gB by overcoming the inhibitory effect of gH–gL. If gH–gL were a positive regulator, its role would be to activate gB into a fusion-active state; receptor-bound gD could then be a co-activator. The latter role is more likely because gB and gH–gL interact only when gD and a gD receptor are also present14,15. This implies that gB and gH–gL are not normally associated on the cell surface or in the mature virion, which conflicts with the “negative regulator” model.
Formation of the gB–gH–gL complex is necessary for fusion and is inhibited by certain anti-gB neutralizing antibodies39. Here, we determined that formation of the gB–gH–gL complex is inhibited by a potent anti-gH–gL neutralizing antibody LP11 (Fig. 6d). Moreover, at least one anti-gH–gL neutralizing antibody blocks fusion but not the interaction. These data suggest that formation of the gB–gH–gL complex likely occurs prior to fusion. The fact that non-neutralizing antibodies do not block the interaction supports the idea that formation of a gB–gH–gL complex is a critical step in the fusion mechanism. This makes the interface of the gB–gH–gL complex a promising target for the future antiviral design efforts.
Because LP11 competes with gB for binding to gH–gL, we propose that the gB-binding site on gH–gL likely overlaps the LP11 epitope, unless LP11 interferes with gB binding due to a long-range steric hindrance. Several insertions that inhibit LP11 binding20 map to one margin of the LP11 epitope (Fig. 5a). These insertions do not interfere with gB binding, because they do not affect fusion20. Therefore, we propose that gB binds near the other margin of the LP11 epitope, in a large groove, highly conserved between HSV-1 and HSV-2 gH (Fig. 5c). gB-binding site is expected to be conserved because gB proteins from HSV-1 and -2 can substitute each other in fusion assays40. Future work will map the exact location of the gB-binding site on gH–gL. Whether the gB-binding site overlaps the LP11 epitope or is located nearby remains to be determined. Also, although we are convinced that BiMC reflects a direct interaction between gB and gH–gL, future work is necessary to unequivocally demonstrate that gB and gH–gL interact directly rather than through a partner protein such as gD. We have proposed a possible location of the gB-binding site on gH–gL; future work will map its exact location. Recent studies show that the gH–gL-binding site in gB is likely located in domain I or domain II39, but it also awaits precise mapping.
The structure of the gH–gL complex does not suggest a direct role in fusion but rather that gH–gL may activate gB for fusion through direct binding. Two other class III viral fusogens, VSV G and baculovirus gp64, use proton binding as a trigger for fusogenic conformational changes. gH–gL may fulfill a role that functionally substitutes for proton binding. If indeed the function of gH–gL is to enable the transition of gB into its proper fusogenic state, it will establish a new paradigm for how viral fusion with cell membranes is accomplished. Nevertheless, much remains unknown about the fusion mechanism in herpesviruses. For example, does gH–gL normally dissociate from gB upon activating it or does it remain bound to gB during subsequent steps? Additional questions that beg answers are how fusion is activated in herpesviruses that lack gD or its analog, and how gB is kept inactive prior to fusion.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
Acknowledgements
We thank L. Friedman for making the gHΔ48/gL baculovirus expression construct, M. Shaner for preliminary characterization of the gL(161t) mutant, D. King of the University of California at Berkeley for mass spectrometry, and A. Héroux of the National Synchrotron Radiation Source for collecting x-ray diffraction data on selenomethionine derivative crystals. We also thank H. Lou, C. Whitbeck, and M. Ponce de Leon for their earlier contributions to this project, and S. C. Harrison for helpful discussions and critical reading of the manuscript. This work was funded by the NIH grant 1DP20D001996 and by the Pew Scholar Program in Biomedical Sciences (E.E.H.), and by the NIH grants AI18289 (G.H.C.), AI056045 (R.J.E.), and AI076231 (R.J.E.). This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from NCRR at NIH. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Footnotes
Accession codes: Protein Data Bank: Atomic coordinates and structure factors have been deposited into the RCSB Protein Data Bank under accession number 3M1C.
Note: Supplementary information is available on the Nature Structural and Molecular Biology website.
Author Contributions T.K.C., T.M.C., and D.A. performed the experimental work, T.K.C. and E.E.H. performed computational analysis of data, and T.K.C., T.M.C., D.A., G.H.C., R.J.E., and E.E.H. interpreted the results and wrote the manuscript.
References
- 1.Spear PG, Longnecker R. Herpesvirus entry: an update. J Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci. 2008;65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryckman BJ, et al. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol. 2008;82:60–70. doi: 10.1128/JVI.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 6.Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Curr Opin Struct Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 8.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 9.Peng T, et al. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J Virol. 1998;72:6092–6103. doi: 10.1128/jvi.72.7.6092-6103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinzler ER, Compton T. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J Virol. 2005;79:7827–7837. doi: 10.1128/JVI.79.12.7827-7837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole NL, Grose C. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev Med Virol. 2003;13:207–222. doi: 10.1002/rmv.377. [DOI] [PubMed] [Google Scholar]
- 12.Pertel PE. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J Virol. 2002;76:4390–4400. doi: 10.1128/JVI.76.9.4390-4400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian RP, Geraghty RJ. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc Natl Acad Sci U S A. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avitabile E, Forghieri C, Campadelli-Fiume G. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol. 2007;81:11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atanasiu D, et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A. 2007;104:18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairns TM, et al. N-terminal mutants of herpes simplex virus type 2 gH are transported without gL but require gL for function. J Virol. 2007;81:5102–5111. doi: 10.1128/JVI.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunewald K, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 19.Hutchinson L, et al. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galdiero M, et al. Site-Directed and linker Insertion Mutangenesis of Herpes Simplex Virus Type 1 Glycoprotein H. J. Virol. 1997;71:2163–2170. doi: 10.1128/jvi.71.3.2163-2170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairns TM, Landsburg DJ, Whitbeck JC, Eisenberg RJ, Cohen GH. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology. 2005;332:550–562. doi: 10.1016/j.virol.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Klyachkin YM, Stoops KD, Geraghty RJ. Herpes simplex virus type 1 glycoprotein L mutants that fail to promote trafficking of glycoprotein H and fail to function in fusion can induce binding of glycoprotein L-dependent anti-glycoprotein H antibodies. J Gen Virol. 2006;87:759–767. doi: 10.1099/vir.0.81563-0. [DOI] [PubMed] [Google Scholar]
- 23.Roop C, Hutchinson L, Johnson DC. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 25.Tsodikov OV, Record MT, Jr, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 26.Galdiero S, et al. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J Biol Chem. 2005;280:28632–28643. doi: 10.1074/jbc.M505196200. [DOI] [PubMed] [Google Scholar]
- 27.Galdiero S, et al. Evidence for a role of the membrane-proximal region of herpes simplex virus Type 1 glycoprotein H in membrane fusion and virus inhibition. Chembiochem. 2007;8:885–895. doi: 10.1002/cbic.200700044. [DOI] [PubMed] [Google Scholar]
- 28.Galdiero S, et al. Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H. J Biol Chem. 2008;283:29993–30009. doi: 10.1074/jbc.M803092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopper M, Compton T. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J Virol. 2004;78:8333–8341. doi: 10.1128/JVI.78.15.8333-8341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galdiero S, et al. Analysis of synthetic peptides from heptad-repeat domains of herpes simplex virus type 1 glycoproteins H and B. J Gen Virol. 2006;87:1085–1097. doi: 10.1099/vir.0.81794-0. [DOI] [PubMed] [Google Scholar]
- 31.Gianni T, Martelli PL, Casadio R, Campadelli-Fiume G. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha-helix with attributes of an internal fusion peptide, positionally conserved in the herpesviridae family. J Virol. 2005;79:2931–2940. doi: 10.1128/JVI.79.5.2931-2940.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gianni T, Menotti L, Campadelli-Fiume G. A heptad repeat in herpes simplex virus 1 gH, located downstream of the alpha-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J Virol. 2005;79:7042–7049. doi: 10.1128/JVI.79.11.7042-7049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gianni T, Piccoli A, Bertucci C, Campadelli-Fiume G. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J Virol. 2006;80:2216–2224. doi: 10.1128/JVI.80.5.2216-2224.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gompels U, Minson A. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology. 1986;153:230–247. doi: 10.1016/0042-6822(86)90026-7. [DOI] [PubMed] [Google Scholar]
- 36.Buckmaster EA, Gompels U, Minson A. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 10(3) molecular weight. Virology. 1984;139:408–413. doi: 10.1016/0042-6822(84)90387-8. [DOI] [PubMed] [Google Scholar]
- 37.Showalter SD, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gompels UA, et al. Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J Virol. 1991;65:2393–2401. doi: 10.1128/jvi.65.5.2393-2401.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atanasiu D, et al. Bimolecular complementation defines functional regions of Herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol. 2010;84:3825–3834. doi: 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muggeridge MI. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 2000:2017–2027. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- 41.Lamb RA, Parks GD. Paramyxoviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott, Williams, and Wilkins; 2006. pp. 1449–1496. [Google Scholar]
- 42.Paterson RG, Hiebert SW, Lamb RA. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc Natl Acad Sci U S A. 1985;82:7520–7524. doi: 10.1073/pnas.82.22.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McShane MP, Longnecker R. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc Natl Acad Sci U S A. 2004;101:17474–17479. doi: 10.1073/pnas.0404535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farnsworth A, et al. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc Natl Acad Sci U S A. 2007;104:10187–10192. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannah BP, et al. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J Virol. 2009;83:6825–6836. doi: 10.1128/JVI.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: CCW Jr, M SR, editors. Methods in Enzymology. Vol. 276. New York, NY: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 47.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–222. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 48.Fortelle EdL, Bricogne G. Maximum-likelyhood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. In: CCW Jr, M SR, editors. Methods in Enzymology. Vol. 276. New York, NY: Academic Press; 1997. pp. 472–494. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 51.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Barton GJ. Alscript: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 54.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 56.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 57.Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. Host range of human T-cell bleukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology. 1999;254:235–244. doi: 10.1006/viro.1998.9530. [DOI] [PubMed] [Google Scholar]
- 58.Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001;279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 59.Connolly SA, et al. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA(HVEM) J. Virol. 2002;76:10894–10904. doi: 10.1128/JVI.76.21.10894-10904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.