Abstract
Mitochondria continuously undergo fusion and fission, the relative rates of which define their morphology. Large mitochondria produce energy more efficiently, whereas small mitochondria translocate better to subcellular sites where local production of ATP is acutely required. Mitochondrial fusion is currently assayed by fusing together cells expressing GFP or RFP in their mitochondria and then scoring the frequency of cells with yellow mitochondria (representing fused green and red mitochondria). However, this assay is labor-intensive and only semi-quantitative. We describe here a reporter system consisting of split fragments of Renilla luciferase and YFP fused to mitochondrial matrix-targeting sequences and to leucine zippers to trigger dimerization. The assay enables fusion to be quantitated both visually for individual cells and on a population level using chemiluminescence, laying the foundation for high throughput small molecule and RNAi screens for modulators of mitochondrial fusion. We use the assay to examine cytoskeletal roles in fusion progression.
Keywords: Mitochondria, Fusion, Split-fluorescence, Split-chemiluminesence, Vimentin
1. Introduction
Regulating the balance of mitochondrial fusion and fission is fundamentally important (Chan, 2006), since disturbances result in mitochondrial dysfunction that causes neurodegenerative disease (Chen et al., 2007; Zuchner et al., 2004) or death of the individual (Chen et al., 2003; Waterham et al., 2007). Cells with predominant fission have small mitochondria, whereas cells with predominant fusion have large, tubulated mitochondria. The relative rates of fusion and fission are regulated by many types of signaling mechanisms (McBride et al., 2006). Metabolic demands, such as signaled by insulin receptor activation, lead to a generalized increase in mitochondrial size and tubulation (Pawlikowska et al., 2007), potentially because larger mitochondria are more efficient producers of energy (Chen et al., 2003). On the other hand, increased needs for energy production at specific subcellular sites, for example at the rear of migrating cells where myosin activity is located (Campello et al., 2006), or at axonal sites of nerve growth factor signaling (Chada and Hollenbeck, 2004), cause mitochondria to fragment and translocate to those sites. As well, mitochondria undergo fragmentation during apoptosis (Suen et al., 2008), and undergo extensive fission and then fusion during the cell cycle (Margineantu et al., 2002).
Several proteins mediating mitochondrial fusion and fission have been identified, although many aspects of how they function remain unknown. In the context of signaling-regulated changes in mitochondrial morphology, the best understood component is dynamin-related protein 1 (Drp1), which mediates fission (Chan, 2006). Drp1 translocation to mitochondria and the ensuing fission is regulated by signaling-activated phosphorylation (Han et al., 2008) and/or dephosphorylation (Cereghetti et al., 2008), SUMOylation (Braschi et al., 2009), and S-nitrosylation (Cho et al., 2009). In contrast, little is known about the signaling mechanisms that regulate activity of Mitofusin1 (Mfn1), Mitofusin 2 (Mfn2), and optic atrophy type 1 (Opa1), the proteins that mediate mitochondrial fusion. As a consequence, although mutation of key sites in Drp1 that respond to signaling cues have suggested that controlling fission rate is an important factor in dynamically controlling mitochondrial morphology, relatively little is known about whether rates of fusion are also controlled by signaling processes or other cues. Moreover, there is a lack of consensus about the role of the cytoskeleton in facilitating mitochondrial movement and morphology, with reports variously suggesting dominant roles for F-actin, for microtubules, or for intermediate filaments (Anesti and Scorrano, 2006).
Mechanistic studies of mitochondrial fusion have relied heavily on a mitochondrial fusion assay reported in 2002 (Legros et al., 2002). In this assay, fluorescent proteins are targeted to the mitochondrial matrix; one line of cells expresses mitochondrially-targeted green fluorescent protein (EGFP), and another, mitochondrially-targeted red fluorescent protein (RFP). The cell lines are co-cultured on a coverslip until confluent, and then fused using polyethylene glycol, following which mitochondrial fusion events lead to matrix mixing and ensuing yellow mitochondria. The assay is generally allowed to proceed for 7–16 h, and the frequency of cells with yellow mitochondria then recorded as a measure of fusion. However, this approach requires manually scanning each experimental sample using confocal microscopy to find and score several hundred fusion events, and thus is labor-intensive and not easily quantifiable or capable of gathering information about kinetics of the reaction. More recently, a quantitative mitochondrial fusion assay has been developed using photoactivatable GFP. Live cell confocal microscopy is used to monitor mitochondrial fusion via the matrix-targeted photoactivated GFP in combination with matrix-targeted red fluorescence protein after a defined region of interest is irradiated (Karbowski et al., 2004; Twig et al., 2006). Although this method allows real time and quantitative analysis of mitochondrial fusion, it can only track limited regions of the mitochondria in one a cell at a time, making it challenging to gather information on a population level, and the approach is labor-intensive for quantification.
To address these limitations, we have employed a split-protein complementation system to develop a quantitative, less labor-intensive, and population-based mitochondrial fusion assay. The system consists of two inactive fragments split from a reporter protein that exhibit function only upon association. The association is promoted through fusion of the reporter protein fragments to a pair of strongly-interacting proteins (matched synthetic leucine zippers), and targeting to the mitochondrial matrix is achieved using a mitochondrial localization sequence (MLS). We validate the assay by demonstrating a loss of signal in the absence of the pro-fusion protein Mfn or mitochondrial potential or glycolytic energy sources. Finally, we examined the role of the cytoskeleton, and in particular, intermediate filaments, in mitochondrial fusion, a relatively unexplored and unsettled topic, as a proof-of-concept.
2. Materials and methods
2.1. Materials
Cell culture media, DMEM and MEMα were purchased from Invitrogen (Carlsbad, CA). Oligomycin, 2-deoxyglucose, valinomycin, CCCP, taxol, nocodazole, cytochalasin D, acrylamide and 2,5-hexanedione were purchased from Sigma (St. Louis, MO). Jasplakinolide was purchased from Calbiochem (La Jolla, CA).
2.2. Plasmid construction
To construct the plasmid N-MitoVZL, DNA sequences encoding amino acid residues 1–173 of mVenus (monomeric Venus Fluorescent Protein) (Rizzo et al., 2006), a linker GGSGSGSS, the synthetic leucine zipper peptide (Z1) ALKKELQANKKELAQLKWELQALKKE-LAQ (Magliery et al., 2005), another GGSGSGSS linker (L), an HA epitope peptide, and finally residues 1–91 of hRLuc, were fused in tandem (see Fig. 1B). C-MitoLZV was generated by fusing DNA sequences encoding residues 92–311 of hRLuc, a FLAG epitope peptide, a linker GGSGSGSS, the complementary synthetic antiparallel leucine zipper peptide (Z2) ASEQLEKKLQALEKKLAQ-LEWKNQALEKKLAQ, another GGSGSGSS linker, and finally residues 155–238 of mVenus. The chimeric coding regions were cloned into a pQCXIP retroviral vector (Clontech, Mountain View, CA) containing the mitochondrial localization sequence (MLS) SVLTPLLLRG LTGSARRLPVPRAKIHSL at the N-terminus of the final protein product. Vector sequences available upon request.
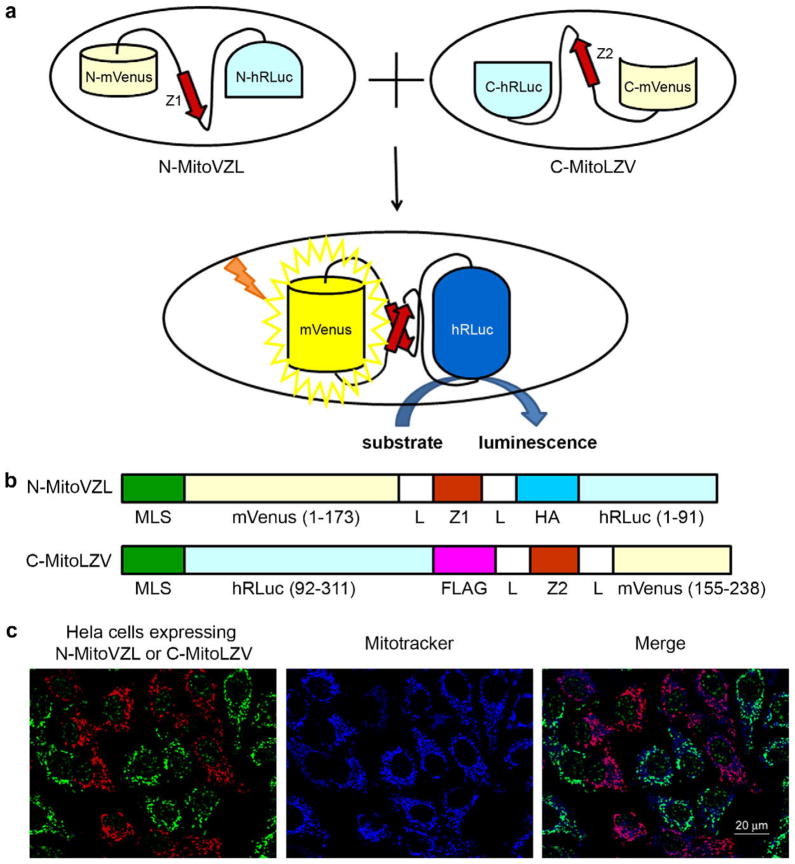
Fig. 1.
Mitochondrial fusion quantification via split-Renilla luciferase complementation. (A) Principle of split-Renilla luciferase complementation to quantitate mitochondrial fusion. Upon fusion of two mitochondria individually expressing a hybrid protein encoding half of the split-Renilla luciferase, the antiparallel leucine zipper pair (red) pulls the constructs together and the luciferase fragments reconstitute and generate luminescence in the presence of substrate. The split-Venus also reconstitutes and generates yellow fluorescence. (B) Schematic representations of domain structures of split-Renilla luciferase. Both constructs have a mitochondrial localization sequence (MLS) in their N-termini. The N-terminal monomeric (m) Venus fragment is fused to the N-terminal hRLuc fragment with a leucine zipper peptide (Z1) in the middle flanked by two linkers (L) and an HA peptide. The C-terminal hRLuc fragment is fused with the C-terminal Venus fragment and the corresponding antiparallel leucine zipper peptide (Z2) flanked by the two same linkers (L) and a FLAG peptide. (C) Mitochondrial localization of split-Renilla luciferase constructs. HeLa cells expressing each construct were mixed and stained for the HA (green) and FLAG (red) epitopes built into the fusion proteins. Note that the anti-HA antibody non-specifically detects a speckled nuclear protein present in all cells. The mitochondria were stained with MitoTracker far red 633 (blue). Scale bar, 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Viral infection and stable population selection
The split constructs, together with pVSVG (Clontech, Mountain View, CA), were transfected separately into GP2-293 cells using Fu-Gene HD transfection reagent (Roche Applied Science, Basel, Switzerland). The viruses were collected 48 h later, filtered, and used to infect cells in medium containing 6 μg/ml polybrene (Sigma, St. Louis, MO). Stably-transfected cell populations were selected 2–3 days after infection using puromycin (InvivoGen, San Diego, CA) at 5 μg/ml for HeLa cells, 10 μg/ml for COS-7 cells, and 2 μg/ml for MEF cells for 2 weeks.
2.4. Cell culture and mitochondrial fusion assay
GP2–293, HeLa and COS-7 cells were maintained in DMEM supplemented with 10% calf serum (Hyclone, Logan, UT). Mfn 1/2−/− MEF and wild-type MEF cells were maintained in DMEM supplemented with 10% fetal bovine serum (Hyclone, Logan, UT).
For the mitochondrial fusion assay, 4 × 105 cells were cultured per well in 12-well plates 18 h before cell fusion. Cycloheximide (100 μg/ml (HeLa), 50 μg/ml (COS-7), 50 μg/ml (wild-type MEFs) or 400 μg/ml (Mfn1/2−/− MEFs)), optimal concentrations predetermined by fusing each cell type in increasing concentrations of cyclohexamide until baseline levels of luciferase activity were attained, as depicted in Fig. 2A for HeLa cells and in Supplemental Fig. 4A–C for other cells) was added 30 min before fusion and kept in the media thereafter. The cells were incubated with 50% PEG (polyethylene glycol) 1500 (Roche Applied Science, Basel, Switzerland) for 60 s, washed four times with complete media, cultured for the indicated times, and harvested in 5 mM EDTA in PBS. Cell pellets were stored frozen until all samples were ready for the Renilla luciferase activity assay.
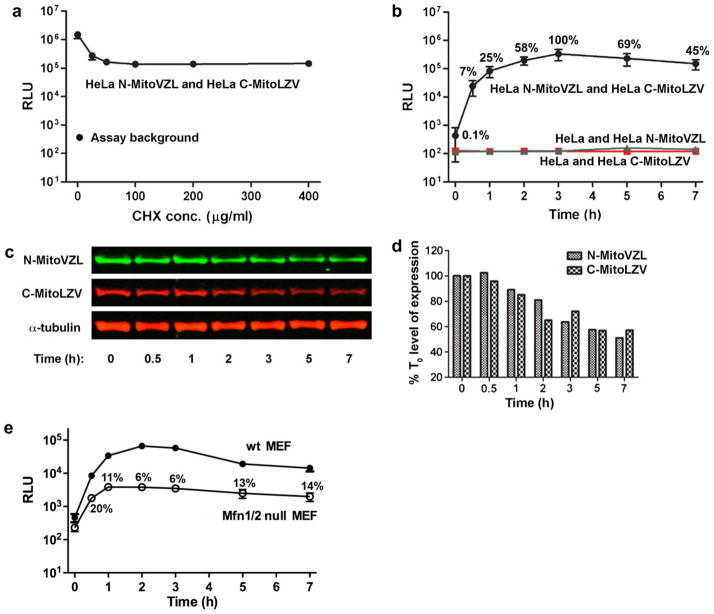
Fig. 2.
Mitochondrial fusion kinetics measured by split-Renilla luciferase complementation. (A) Luminescence of HeLa cells stably expressing the N-MitoVZL and C-MitoLZV split-Renilla luciferase constructs 7 h after PEG-1500 treatment to initiate cellular fusion, followed by mitochondrial fusion, in the indicated CHX concentrations. Assay background indicates luminescence of stably-transfected cells without PEG-1500 treatment. (B) Luminescence of HeLa cells stably expressing N-MitoVZL and C-MitoLZV (black), N-MitoVZL only (green), and C-MitoLZV only (red) at the indicated times after PEG-1500 treatment in 100 μg/ml CHX. Time 0 represents the point of PEG-1500 addition; samples at time 0 were not treated with PEG-1500. % values indicate the RLU normalized to the signal at the 3 h fusion time point. (C) Western blot analysis of HeLa cell lysates harvested at indicated times after PEG-1500 treatment. N-MitoVZL was visualized using anti-HA antibody, and C-MitoLZV using anti-FLAG antibody. (D) Quantification of protein levels in Fig. 2c normalized to α-tubulin. (E) Luminescence of Mfn1/2−/− MEF cells stably expressing the split-Renilla luciferase constructs at indicated times after PEG-1500 treatment, in comparison to the levels of fusion observed for wild-type MEF cells.% values indicate the RLU normalized to that measured in wild-type MEF cells at the corresponding time-point. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For drug treatments, the mitochondrial inhibitors were added to the media after PEG treatment to avoid impairing the cell fusion efficiency. Drugs that disrupted cytoskeleton components were added 30 min before PEG treatment, except for jasplakinolide and cytochalasin D, which shrank the cells promptly after treatment and thus dramatically decreased the extent of cell fusion. Instead, jasplakinolide and cytochalasin D were added to the media 1 h after PEG treatment, by which point the cells are thought to have completed the plasma membrane fusion process.
Vimentin Stealth RNAi™ siRNA and control siRNA were purchased from Invitrogen (Carlsbad, CA) and transfected into cells using Lipofectamine™ RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA). The cells were harvested 72 h later, plated, and cultured overnight before being used in the fusion assay.
2.5. Renilla luciferase activity assay
Renilla luciferase activities were measured using Renilla luciferase assay system (Promega, Madison, WI). Briefly, cells collected at the indicated time points were lysed in 1× Renilla luciferase assay buffer, mixed with Renilla luciferase assay substrate diluted in Renilla luciferase assay buffer, and measured for luminescence using a 20/20 single-tube luminometer (Turner) with integration time of 5 s.
2.6. Western blotting
Cell lysates were subjected to 10% SDS–PAGE, transferred to nitrocellulose membranes, and probed with primary antibodies recognizing the HA epitope (3F10; Roche Applied Science, Basel, Switzerland), FLAG epitope (M2; Sigma, St. Louis, MO), α-tubulin (B-5-1-2; Sigma, St. Louis, MO), vimentin (clone V9; Sigma, St. Louis, MO), or β-actin (Sigma, St. Louis, MO), followed by secondary antibodies conjugated with Alexa 680 (Molecular Probes, Carlsbad, CA) or IRDye 800 (Rockland Immunochemicals, Gilbertsville, PA). Fluorescent signals were detected with an Odyssey infrared imaging system (LICOR Biosciences, Lincoln, NB).
2.7. Immunofluorescence microscopy
HeLa cells stably expressing the split constructs were mixed and plated onto coated coverslips. Twenty-four hours later, they were stained with 500 nM MitoTracker Deep Red 633 (Molecular Probes, Carlsbad, CA) for 30 min, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 5% normal goat serum. The cells were then immunostained using primary antibodies against the HA (Rockland Immunochemicals, Gilbertsville, PA) and FLAG (M2; Sigma, St. Louis, MO) epitopes followed by fluorescent dye-conjugated secondary antibodies. Stained cells were visualized using a Leica TCS SP2 confocal microscope. Images were processed using Adobe Photoshop.
2.8. Statistics
Experiments to determine appropriate concentrations of cyclohexamide to use were repeated three times. Kinetic experiments were repeated at least five times unless otherwise stated. Error bars on graphs display the standard deviation.
3. Results
3.1. A quantitative mitochondrial fusion reporter system usable for high through-put analysis
We set out to develop a high-throughput plate-reader assay using the split-YFP system (Hu and Kerppola, 2003) by adding a mitochondrial matrix-targeting sequence to the amino-termini of fragments of the Venus YFP in combination with a pair of complementary synthetic antiparallel leucine zipper peptides (Magliery et al., 2005) to trigger assembly of the fluorescent protein fragments upon mitochondrial fusion. However, the yellow fluorescent signal produced as a result of mitochondrial fusion, which was readily visible using confocal microscopy (Supplemental Fig. 1A), was not easily or sensitively detected using standard plate readers. Therefore, we further added split fragments of Renilla luciferase (RLuc) (Kaihara et al., 2003) to amplify the fusion signal and enable quantitative measurement via chemiluminescence (Fig. 1A and B). A number of approaches to generating the constructs yielded unstable reporter proteins (data not shown), potentially due to the unpaired leucine zippers or reporter fragments. However, the reporter components N-MitoVZL and C-MitoLZV exhibited stability as individually expressed proteins (Fig. 1C and western blot analysis, not shown, but see also Fig. 2C and Supplemental Fig. 4D), and localized to mitochondria as designed (Fig. 1C). Luciferase activity was observed when the reporter constructs were co-expressed (1420,000 RLU, Supplemental Fig. 1B), but not when they were expressed individually (<50 RLU above negative, non-transfected control), and not to a substantial extent when expressed in different populations of cells that were then lysed and assayed together (<700 RLU above negative control), indicating that <0.1% of the non-complexed components reconstitute into active protein during the lysis and assay steps.
3.2. Validation of the Renilla luciferase reporter system for mitochondrial fusion
A key issue for the mitochondrial fusion assay is to ensure that protein synthesis ceases before cell fusion, since otherwise both of the reporter components would co-translocate to all mitochondria as newly-synthesized proteins and thus generate a false-positive signal. Cycloheximide (CHX) was used to block protein synthesis in co-cultures of HeLa cells stably expressing N-MitoVZL (HeLa N-MitoVZL) or C-MitoLZV (HeLa C-MitoLZV) and the cells then fused using PEG-1500 treatment (Fig. 2A). Measurement of luciferase activity in the cell lysates collected 7 h after cell fusion showed that the luminescence decreased more than 90% as the concentration of CHX increased, reaching baseline at 100 μg/ml. The resulting output (138,000 RLU, n = 3), which was approximately 1000-fold above the assay background (143 RLU, representing the signal obtained from lysis of the co-cultures without PEG-1500-mediated fusion), accordingly came from reconstitution of the pre-existing split reporter components subsequent to the cell fusion event, rather than from association of co-translocated newly-synthesized reporter proteins.
We next followed the increase in luciferase activity as function of time subsequent to cellular fusion. In the classic assay, mitochondrial fusion is optimally scored at 7–16 h post-cellular fusion. To our surprise, luciferase activity (7% of the peak activity) could be detected as early as 30 min after PEG-1500-induced cell fusion (in some experiments, activity could be detected within 10 min of the assay start point, data not shown), peaked at 3 h at 1000-fold over background levels of activity, and then subsequently declined 2-fold (Fig. 2B). The decline in signal was partially due to the gradual degradation of the reporter proteins (Fig. 2C and D), which appeared to be less stable than α-tubulin, and death of a small fraction of the cells as triggered by the cell fusion and/or CHX treatment. This result indicated 3 h after the cell fusion step to be an optimal assay time point for HeLa cells, rather than 7–16 h as in the classic assay. As expected, no increase in signal was observed over time in HeLa cells expressing each fragment alone, since the individual reporter constructs do not exhibit luciferase activity.
Mfn1 and 2 are key mediators of the mitochondrial fusion process (Chan, 2006); in the absence of both isoforms, no fusion is though to occur. To validate our fusion assay, we performed it in Mfn1/2−/− mouse embryo fibroblast (MEF) cells. Monitoring the luciferase signal over time revealed a small rise in chemiluminescence that peaked at 2 h at 6% of the peak signal observed during fusion of control, wild-type MEF cells (Fig. 2E). The source of this low level of reporter reconstitution and signal in the fused Mfn1/2−/− cells is uncertain, but could reflect localization and reconstitution of a small fraction of the reporter proteins in the cytoplasm, release of the reporter proteins from mitochondria into the cytoplasm during apoptotic events, reconstitution during mitochondrial autophagy, or low levels of mitochondrial fusion independent of Mfn. Although visualization of the reporter constructs confirmed that they predominantly localized to mitochondria (as shown in Fig. 1C for HeLa cells and data not shown), low levels of mis-targeting to the cytoplasm can not be ruled out.
Taken together, these findings show that the fusion assay is quantifiable and robust, with a 18:1 signal-to-noise ratio at 2 h after cell fusion (in comparison to Mfn1/2−/− cells), and that it is capable of detecting fusion events within 30 min of the beginning of the assay period.
3.3. Mitochondrial fusion is selectively inhibited by mitochondrial inhibitors to different extents
The classic mitochondrial fusion assay has been used to demonstrate that glycolysis, but not ATP synthesis, is required in mitochondrial fusion (Malka et al., 2005). We thus examined effects of the glycolysis inhibitor, 2-deoxyglucose, and the ATP synthase inhibitor, oligomycin. Consistent with the prior report, oligomycin had no effect on mitochondrial fusion (Fig. 3A), whereas 2-deoxy-glucose decreased the rate of fusion. It should be noted though that the pattern of luciferase activity over time is also informative. Although the rate of fusion was inhibited in cells cultured in 2-deoxyglucose (20% of the untreated control value at 1 h, and 21% at 3 h), the luciferase signal continued to rise between 1 and 3 h, suggesting that fusion was occurring, but 5-fold slower than in cells with normal glycolysis.
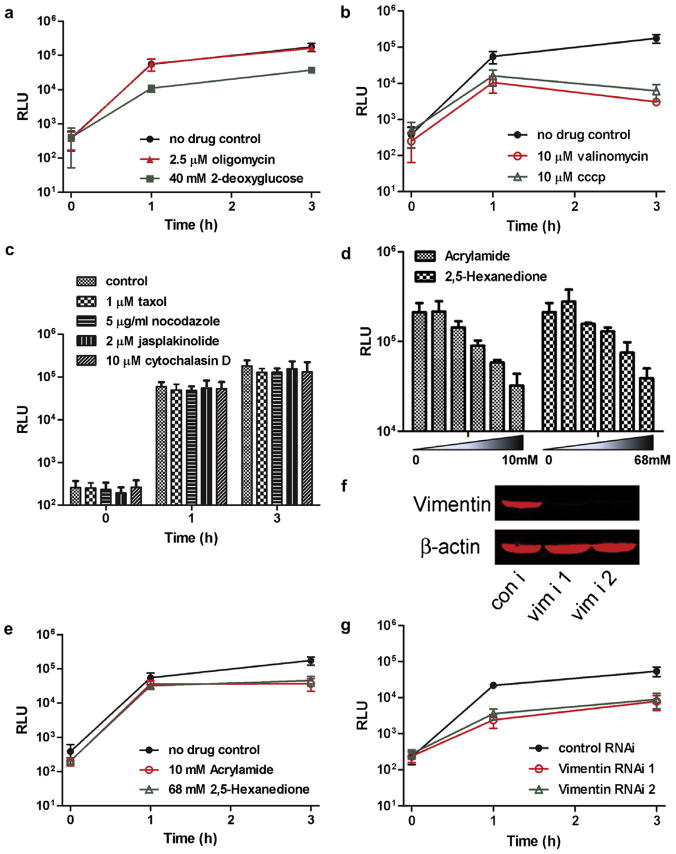
Fig. 3.
Mitochondrial fusion rates are affected by mitochondrial function inhibitors and cytoskeleton integrity. (A) Luminescence at the indicated times after cell fusion with no drug, 2.5 μM oligomycin, or 40 mM 2-deoxyglucose in medium lacking glucose (glucose-lacking medium in itself did not alter rates of fusion in comparison to control medium; data not shown). (B) Luminescence after cell fusion with no drug treatment, 10 μM valinomycin, or 10 μM CCCP. (C) Luminescence after fusion with no drug treatment, 1 μM taxol, 5 μg/ml nocodazole, 2 μM jasplakinolide or 10 μM cytochalasin D. (D) Luminescence 3 h after fusion in the indicated acrylamide or 2,5-hexanedione concentrations. (E) Luminescence after fusion with no drug treatment, 10 mM acrylamide, or 68 mM 2,5-hexanedione. Experiments from A to E were performed in HeLa cells. (F) Western blot analysis of vimentin, 90 h after siRNA transfection of COS-7 cells. (G) Luminescence of COS-7 cells stably expressing the split-Renilla luciferase reporters at the indicated times after cell fusion, 90 h after control siRNA or vimentin siRNA transfection.
The classic mitochondrial fusion assay has also been used to show that dissipation of the mitochondrial membrane potential abolishes mitochondrial fusion (Malka et al., 2005). We further validated our assay by examining the effects of the protonophore, CCCP, and a potassium-specific ionophore, valinomycin, on the fusion reaction. Both inhibitors had a marked impact on fusion (Fig. 3B). An increase in luciferase signal was observed at 1 h in the drug-treated cells, but the signal declined thereafter. In contrast, the control, untreated cells exhibited a 5-fold greater increase in RLU at 1 h, and a 25–50-fold higher activity at 3 h. The luciferase activity in the valinomycin and CCCP-treated cells at 3 h was similar in magnitude to that seen for the Mfn−/− cells (Fig. 2E), suggesting that after an initial modest round of fusion detectable at 1 h, no further fusion occurred.
Taken together, these findings provide evidence that the split-luciferase mitochondrial fusion assay provides an accurate and quantitative method for assessing rates of fusion in mammalian cells.
3.4. Mitochondrial fusion efficiency depends on the integrity of intermediate filaments
Mitochondrial translocation proceeds primarily via microtubules in mammalian cells, and via the actin network in yeast (Boldogh and Pon, 2007). Mitochondrial fragmentation occurs in yeast when cells are treated with actin-depolymerizing drugs such as latrunculin (Drubin et al., 1993), and ablation of Kif5b, a member of the mouse Kinesin-1 family that regulates microtubule-based mitochondrial transport, results in mitochondrial clustering in the perinuclear region (Anesti and Scorrano, 2006). We next examined the effect of taxol/nocodazole and jasplakinolide/cytochalasin D, which stabilize or destabilize microtubules and the actin network, respectively, without substantially affecting mitochondrial localization in HeLa cells (Supplemental Fig. 2A and B). However, no significant changes in luciferase activities were observed (Fig. 3C), indicating that mitochondrial fusion appears to proceed independently of microtubule and actin dynamics in HeLa cells.
It has recently been reported, however, that vimentin intermediate filaments interact with mitochondria, and that depletion of vimentin by siRNA causes mitochondrial fragmentation (Tang et al., 2008), which we confirmed using COS-7 cells (Supplemental Fig. 3). It is not known, though, whether the fragmentation of mitochondria after vimentin knock-down is caused by inhibition of mitochondrial fusion, or by stimulation of mitochondrial fission. To address this question, we first treated HeLa cells with two compounds that have been shown to cause central collapse of the vimentin network, acrylamide and 2,5-hexanedione (Sager, 1989), and found that they inhibited mitochondrial fusion in a dose-dependent manner (Fig. 3D). Integrity of the vimentin network appeared to be important, but not vital for mitochondrial fusion, since only partial inhibition was observed (Fig. 3E; compare to Fig. 3B).
Acrylamide and 2,5-hexanedione cause degeneration of the peripheral nervous system, characteristics of patients with mutations in Mfn2, suggesting that their clinical effects might ensue from defect in mitochondrial fusion. However, these compounds presumably cross-link and impede the function of other proteins as well. Accordingly, to explore the importance of the vimentin network on mitochondrial fusion, we knocked down vimentin mRNA in COS-7 cells using two different siRNAs (Fig. 3F, Supplemental Fig. 3), and found that mitochondrial fusion was decreased almost 10-fold in the absence of vimentin-based intermediate filaments in COS-7 cells (Fig. 3G). These results suggest that the vimentin-based intermediate network has a more profound role in promoting mitochondrial fusion in HeLa and COS-7 cells than do those of actin and tubulin.
4. Discussion
We have developed a quantitative and facile mitochondrial fusion assay that can be applied to the study of mechanistic and regulatory aspects of mitochondrial fusion. Microtubules are solidly established as directing long-range mitochondrial distribution in mammalian cells, with actin playing a role over short distances. However, due to the limitations of the widely-used 2-color, manually-scanned mitochondrial fusion assay, the roles of the different cytoskeletal networks in mitochondrial fusion have not been well delineated. A study using mitochondrially-targeted photoactivatable GFP reported previously that microtubules function to transport mitochondria together during mitochondrial fusion (Liu et al., 2009), which differs from the findings we observed here. However, dependence on cytoskeletal components for mitochondria is known to be cell-type specific. In some cells, such as the myoblasts studied in Liu et al., the mitochondria are lined up with microtubules, while in others, such as NIH3T 3 cells (Tang et al., 2008), mitochondria co-localize more with vimentin than with microtubules. In fact, microtubules are better recognized for their role in segregating mitochondria after fission: when mitochondrial fission is inhibited, novel structures consisting of thin mitochondrial extensions are formed, which can be prevented using a micro-tubule disrupting drug (Bowes and Gupta, 2008). We show here that integrity of the vimentin intermediate filament network appears to be much more important to the progression of mitochondrial fusion than integrity of the microtubule or actin cytoskeleton networks, at least in HeLa and COS-7 cells. Vimentin interacts with microtubules, and disruption of microtubules eventually leads to vimentin reorganization (Summerhayes et al., 1983). It is possible that some of the effects on mitochondrial morphology previously defined in the context of manipulation of microtubule networks might be indirect and ensue instead from the consequential effects on the vimentin network. It is also possible that some of the known mitochondrial dysfunctions might well be caused by primary problems in vimentin organization. Intriguingly, tau-like protein, which mediates the interaction of vimentin and tubulin (Capote and Maccioni, 1998), causes neurodegenerative diseases such as Alzheimer’s disease, a component of which we speculate could ensue from disrupting the tau–vimentin interaction, resulting in decreased mitochondrial fusion and hence increased sensitivity to apoptosis.
The assay described here provides a measure of complete mitochondrial fusion, that is, mixing of the matrix contents. However, the inner and outer mitochondrial membranes can fuse independently (Malka et al., 2005), and it would be possible to exploit our assay to examine each step discretely. For example, split Firefly luciferase (FLuc) (Paulmurugan and Gambhir, 2007) could be similarly targeted to the intermembrane space or to the outer membrane. Since Firefly and Renilla luciferase use distinct substrates that luminescence in the presence of different cofactors, both could be measured sequentially in the same lysate sample. Such a dual luciferase activity assay would be more sensitive and convenient than the three-color fluorescence method that has been used for the classic assay approach (Malka et al., 2005).
Fusion is scored in the traditional assay seven or more hours after cell fusion, making it challenging to examine signaling events that take place in a shorter time frame. Our assay robustly detects on-going fusion within 30 min after cell fusion, and in preliminary experiments, as quickly as 10 min after cellular fusion. Employing this new approach will enable many types of studies previously impractical to attempt.
A limitation to real-time measurements using the assay is that the method requires lysing cells to sample the luciferase activity. Live cell substrates exist for Renilla luciferase, such as ViviRen, EnduRen (Promega) and DeepBlue C (Jensen et al., 2002). However, at least in our hands, their sensitivity was modest and insufficiently robust for quantification. Intriguingly, a study on luciferase from Gaussia princeps showed that the signal from the humanized form of G. princeps luciferase (hGLuc) is 100-fold higher than the ones generated by hFLuc and hRLuc, and the substrate for hGLuc is membrane permeable (Remy and Michnick, 2006), making it a very promising candidate for development of a real-time mitochondrial fusion assay in live cells.
5. Conclusion
The split-Renilla luciferase system we described here quantitatively measures mitochondrial fusion rates in a high throughput manner, laying the foundation for RNAi and small molecule screens for modulators of mitochondrial fusion.
Supplementary Material
Acknowledgments
We thank Drs. M. Rizzo (U. Maryland) and Lynne Regan (Yale U.) for the kind gifts of plasmids encoding mVenus and the synthetic leucine zippers, respectively, and Dr. H. McBride for critical feedback on the manuscript. Supported by NIH awards GM071520 and GM084251 and the United Mitochondrial Disease Foundation.
Abbreviations
- CHX
cycloheximide
- Drp1
dynamin-related protein
- EGFP
enhanced green fluorescent protein
- hRLuc
Renilla Luciferase
- Mfn
mitofusin
- MLS
mitochondrial localization sequence
- Opa
optic atrophy type 1
- PEG
polyethylene glycol
- RFP
red fluorescent protein
- YFP
yellow fluorescent protein
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.mito.2010.05.003.
References
- Anesti V, Scorrano L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta. 2006;1757:692–699. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Bowes T, Gupta RS. Novel mitochondrial extensions provide evidence for a link between microtubule-directed movement and mitochondrial fission. Biochem Biophys Res Comm. 2008;376:40–45. doi: 10.1016/j.bbrc.2008.08.120. [DOI] [PubMed] [Google Scholar]
- Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello S, Lacalle RA, Bettella M, Manes S, Scorrano L, Viola A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med. 2006;203:2879–2886. doi: 10.1084/jem.20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capote C, Maccioni RB. The association of tau-like proteins with vimentin filaments in cultured cells. Exp Cell Res. 1998;239:202–213. doi: 10.1006/excr.1997.3902. [DOI] [PubMed] [Google Scholar]
- Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chada SR, Hollenbeck PJ. Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr Biol. 2004;14:1272–1276. doi: 10.1016/j.cub.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondrial fusion and fission in mammals. Ann Rev Cell Develop Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, Nairn AC, Takei K, Matsui H, Matsushita M. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nature Biotech. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AA, Hansen JL, Sheikh SP, Brauner-Osborne H. Probing intermolecular protein-protein interactions in the calcium-sensing receptor homodimer using bioluminescence resonance energy transfer (BRET) Eur J Biochem. 2002;269:5076–5087. doi: 10.1046/j.1432-1033.2002.03218.x. [DOI] [PubMed] [Google Scholar]
- Kaihara A, Kawai Y, Sato M, Ozawa T, Umezawa Y. Locating a protein-protein interaction in living cells via split Renilla luciferase complementation. Anal Chem. 2003;75:4176–4181. doi: 10.1021/ac0300800. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen HC, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Lombes A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XG, Weaver D, Shirihai O, Hajnoczky G. Mitochondrial ’kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery TJ, Wilson CGM, Pan WL, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: Scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombes A, Rojo M. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep. 2005;6:853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margineantu DH, Gregory Cox W, Sundell L, Sherwood SW, Beechem JM, Capaldi RA. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochon. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir SS. Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem. 2007;79:2346–2353. doi: 10.1021/ac062053q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska P, Gajkowska B, Orzechowski A. Mitofusin 2 (Mfn2): a key player in insulin-dependent myogenesis in vitro. Cell Tissue Res. 2007;327:571–581. doi: 10.1007/s00441-006-0320-3. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nature Meth. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Springer G, Segawa K, Zipfel WR, Piston DW. Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc Microanal. 2006;12:238–254. doi: 10.1017/S1431927606060235. [DOI] [PubMed] [Google Scholar]
- Sager PR. Cytoskeletal Effects of Acrylamide and 2, 5-Hexanedione-Selective Aggregation of Vimentin Filaments. Tox Appl Pharm. 1989;97:141–155. doi: 10.1016/0041-008x(89)90063-x. [DOI] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Develop. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerhayes IC, Wong D, Chen LB. Effect of Microtubules and Intermediate Filaments on Mitochondrial Distribution. J Cell Sci. 1983;61:87–105. doi: 10.1242/jcs.61.1.87. [DOI] [PubMed] [Google Scholar]
- Tang HL, Lung HL, Wu KC, Le AHP, Tang HM, Fung MC. Vimentin supports mitochondrial morphology and organization. Biochem J. 2008;410:141–146. doi: 10.1042/BJ20071072. [DOI] [PubMed] [Google Scholar]
- Twig G, Graf SA, Wikstrom JD, Mohamed H, Haigh SE, Elorza A, Deutsch M, Zurgil N, Reynolds N, Shirihai OS. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am J Physiol-Cell Physiol. 2006;291:C176–C184. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CWT, Mooyer PAW, Wanders RJA, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. New Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, De Jonghe P, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battologlu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nature Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.