Abstract
Proteomic analysis of flagella from the green alga Chlamydomonas reinhardtii has identified over 600 putative flagella proteins. The genes encoding nine of these not previously characterized plus the previously described PACRG protein were cloned, inserted into a vector adding a triple-HA tag to the C-terminus of the gene product, and transformed into C. reinhardtii. Expression was confirmed by western blotting. Indirect immunofluorescence located all ten fusion proteins in the flagellum; PACRG was localized to a subset of outer doublet microtubules. For some proteins, additional signal was observed in the cell body. Among the latter was FAP232-HA, which showed a spotted distribution along the flagella and an accumulation at the basal bodies. This pattern is characteristic for intraflagellar transport (IFT) proteins. FAP232-HA co-localized with the IFT protein IFT46 and co-sedimented with IFT particles in sucrose gradients. Furthermore, it co-immunoprecipitated with IFT complex B protein IFT46, but not with IFT complex A protein IFT139. We conclude that FAP232 is a novel component of IFT complex B and rename it IFT25. Homologues of IFT25 are encoded in the genomes of a subset of organisms that assemble cilia or flagella; C. reinhardtii IFT25 is 37% identical to the corresponding human protein. Genes encoding IFT25 homologues are absent from the genomes of organisms that lack cilia and flagella and, interestingly, also from those of Drosophila melanogaster and Caenorhabditis elegans, suggesting that IFT25 has a specialized role in IFT that is not required for the assembly of cilia or flagella in the worm and fly.
Keywords: flagellar proteome, FAP134, cilia, placental protein pp25, small heat-shock-like protein beta-11 (HSPB11), FAP173, PACRG
INTRODUCTION
Cilia and flagella are cell organelles supported by a microtubular scaffold, the axoneme, and surrounded by a membrane that is continuous with the cell's plasma membrane. They protrude from the surface of many eukaryotic cells and have various functions ranging from cell locomotion to the sensing of environmental stimuli. Defects in cilia have been implicated in several human disorders, including primary ciliary dyskinesia and, more recently, polycystic kidney disease, hydrocephalus, juvenile myoclonic epilepsy, blindness, and syndromes such as Bardet-Biedl syndrome, Senior-Loken syndrome, Joubert syndrome, and Meckel syndrome [Adams et al., 2008]. This has increased the interest in the molecular composition of cilia and flagella as a prerequisite for understanding their function, assembly, and role in disease.
Protists such as the green alga Chlamydomonas reinhardtii allow the isolation of flagella for biochemical analysis [Witman et al., 1972]. In a recent proteomic study of flagella isolated from C. reinhardtii, more than 600 proteins were identified by two or more peptides; however, only about 100 of these have been characterized at the molecular level [Pazour et al., 2005]. Thus, there is a need for tools and approaches for efficiently analyzing the many uncharacterized proteins in order to confirm that they are indeed associated with the flagellum, to determine their specific locations there, and to predict their functions.
To this end, we cloned the C. reinhardtii gene encoding the previously described PACRG protein [Ikeda et al., 2007] and nine genes encoding uncharacterized C. reinhardtii flagellar proteins, inserted each gene into a novel vector that adds a C-terminal triple-HA tag to the gene product, and then transformed wild-type C. reinhardtii with the vector. Transformants expressing the tagged proteins were identified and the cellular locations of the tagged proteins determined by immunofluorescence microscopy using an anti-HA antibody. All of the proteins were localized to the flagella, although they differed in their specific localization patterns. Thus, this approach has great promise for the rapid initial characterization of putative flagellar proteins in C. reinhardtii, and provides a basis for selecting those with particularly interesting distributions for further study, which itself is facilitated by the HA tag. For example, we readily determined that PACRG-HA is localized to a subset of outer doublet microtubules, a pattern similar to the distribution of some axonemal substructures but not previously described for any protein.
One of the tagged proteins, FAP232-HA, exhibited a localization pattern similar to that of known intraflagellar transport (IFT) proteins. IFT is the movement of large non-membrane-bound protein complexes, termed IFT particles, from the base to the tip of the flagellum, and then back to the base, where there is a large pool of IFT particles in close proximity to the basal body [Rosenbaum and Witman, 2002]. The particles, which are composed of at least 18 different proteins organized into two discrete assemblies termed complex A and complex B [Cole et al., 1998], carry axonemal and membrane proteins into the flagellum and out to the flagellar tip where axonemal assembly occurs. In C. reinhardtii, the particles are moved from base to tip – i.e., in the anterograde direction – by the microtubule motor kinesin-2; movement in the opposite direction is powered by cytoplasmic dynein 1b. IFT is essential for the assembly of most cilia and flagella, and defects in IFT result in disease in humans [Beales et al., 2007].
To determine the relationship of FAP232 to IFT, we carried out a detailed analysis of the tagged protein. FAP232-HA was found to co-localize, co-sediment, and co-immunoprecipitate with known components of IFT-particle complex B, but not complex A, indicating that it is a novel IFT complex B component. We have renamed this protein IFT25. IFT25 is highly conserved among many organisms with cilia, including mammals but not Caenorhabditis elegans or Drosophila melanogaster, suggesting that it has a specialized function not needed for assembly of some cilia and flagella. The accompanying paper by Follit et al. [2009] describes the localization of IFT25 in mammalian primary cilia and its association with IFT complex B there.
MATERIALS AND METHODS
Strains and Culture Conditions
C. reinhardtii strain CC3395 (arg7–8, cwd) was grown in TAP medium [Gorman and Levine, 1965] supplemented with 0.02% arginine in non-aerated liquid cultures at 22 °C and a light/dark cycle of 14/10 hours. Transformants were selected on TAP plates without arginine and grown in TAP or Medium I of Sager and Granick [1953] modified by the addition of 3X the original amount of PO4 (M) [Witman, 1986]. Wild-type strain 137c was grown in M.
Vector Design, Cloning, and Transformation
A PCR fragment encoding a triple-HA tag (3xHA) was generated using plasmid p3xHA [Silflow et al., 2001; Silflow and Rosenbaum, 1981] as a template and primers HAfor and HArev (see Supplementary Table S1). After restriction with BamHI and BglII, the PCR fragment was ligated into pCr-CenAS [Koblenz et al., 2003] digested with BamHI. The resulting plasmid (pKL1) places the 3xHA downstream of the HSP70A promoter and upstream of the Rubisco small subunit (RbcS) 3’ UTR. To amplify target genes from genomic DNA we used GoTaq DNA polymerase (Promega) and touchdown PCR [1 min 95 °C, 20 s 96 °C, 15 s 97 °C, 19–20x (22 s 96 °C, 50–70 s 68–72 °C reduced by 0.4 °C /cycle, 2 min 30 s-6 min 45 s 72 °C) 13–14x (22 s 96 °C, 50–70 s 58–63 °C, 2 min 30 s-6 min 45 s 72 °C), 10 min 72 °C]. The gene-specific primers are listed in Supplementary Table S1. PCR products were restricted using the appropriate enzymes, purified by agarose electrophoresis, and cloned into dephosphorylated pKL3 linearized with XbaI. Linearized plasmids (EcoRI) were co-transformed with pARG7 into CC3395.
To increase expression levels of PACRG-HA, an XbaI/EcoRI fragment of pKL1-PACRG was cloned into pCB740 [Schroda et al., 2000] digested with NheI and EcoRI; this placed the PACRG∷HA∷RbcS fragment downstream of the HSP70A/RbcS fusion promoter and on the same plasmid as the ARG7 gene (Supplementary Figure S1). Expression of PACRG-HA was only slightly improved in transformants (e.g. strain 2.3) carrying this construct (not shown). In an attempt to analyze the function of PACRG, we made an inverted repeat construct by cloning PACRG genomic DNA and the inverted PACRG cDNA into the NheI site of the FLA14 expression vector described in Lechtreck and Witman [2007]. Analysis of ~400 transformants in CC3395, however, did not reveal an obvious phenotype caused by PACRG ablation. To test the efficiency of the construct to suppress PACRG expression, it was co-transformed together with the ble gene as a selectable marker into Strain 2.3, which allows easy monitoring of PACRG-HA levels due to the availability of a specific antibody. Transformants were screened by immunofluorescence using anti-HA, and strain 4.3 was selected for further characterization.
Western Blotting and Indirect Immunofluorescence
Western blotting and immunofluorescence staining were carried out as previously described [Lechtreck and Witman, 2007]. For simultaneous permeabilization and fixation, cells were resuspended in MT buffer (30 mM HEPES, 25 mM KCl, 5 mM MgSO4, 5 mM EGTA) and mixed 1:1 with 1% Nonidet P-40, 6% formaldehyde in the same buffer. Cytoskeletons were allowed to settle onto poly-L-lysine-coated multiwell slides for 15–20 min. For methanol fixation, cells in culture medium or MT buffer were immobilized on poly-L-lysine-coated multiwell slides, fixed in −20 °C methanol for 6–10 min, and air-dried. Wells were washed with PBS, blocked, and processed as previously described. The following antibodies were used in this study: rat monoclonal anti-HA (Boehringer), anti-alpha-tubulin [Silflow and Rosenbaum, 1981], anti-centrin [Geimer et al., 1997], anti-IFT46 [Hou et al., 2007], anti-IFT139 and anti-IFT172 [Cole et al., 1998], and anti-IFT20 [George Witman, unpublished]. Secondary antibodies were obtained from Molecular Probes and Pierce.
Flagellar Preparation
Flagella were isolated, extracted with 1% Nonidet P-40, and the soluble phase ("membrane + matrix") was collected; proteins of the membrane + matrix were analyzed by sucrose density gradient centrifugation, all as previously described [Hou et al., 2007]. The extrusion of central pair microtubules from the axoneme also was as previously described [Lechtreck and Witman, 2007]. To split the axonemes into groups of doublets, flagella were isolated from strain 4.3 (PACRG-HA) by homogenization (glass Potter-Elvehjem homogenizer, 25 strokes on ice). Cell bodies were removed by centrifugation at 2000 × g for 10 min, and the flagella were demembranated with 1% Nonidet P-40, treated with 40 mM ATP for 10 min, and fixed with 3% formaldehyde/0.25% glutaraldehyde for 20 min. During the fixation, axonemes were allowed to adhere to poly-L-lysine-coated multiwell slides. After washes and blocking, specimens were stained with antibodies as indicated in the figure legends.
Immunoprecipitation
Membrane + matrix or fractions from sucrose density gradients of membrane + matrix obtained from the flagella of a strain expressing FAP232-HA were mixed with protein G-agarose (Roche) preloaded with anti-HA or with Protein A Sepharose 4 Fast Flow beads (Amersham) preloaded with anti-IFT46. Wild-type membrane + matrix or beads without antibody were used as controls. Suspensions were incubated overnight at 4 °C with agitation. After repeated washes, SDS-sample buffer was added to the beads, and the eluates were analyzed by western blotting.
Quantitative PCR
The relative amount of cDNA representing FAP232 and PACRG messages in samples was determined by real-time PCR as previously described [Lechtreck and Witman, 2007].
RESULTS
HA-tagging Confirms Flagellar Localization of Nine Conserved Uncharacterized Proteins from the Flagellar Proteome
We selected the parkin co-regulated gene protein (PACRG protein) and nine uncharacterized conserved proteins from the flagellar proteome of C. reinhardtii for HA-tagging (Table I). The PACRG protein has been reported to be located along the entire length of the doublet and triplet microtubules of the axoneme and basal body, respectively [Ikeda et al., 2007]. The other proteins ranged in predicted mass from ~ 21,000 – 81,000 Da and were represented by five to 31 unique peptides in the flagellar proteome (Table II). They varied in terms of the flagellar fraction (Nonidet-soluble membrane + matrix, KCl extract, extracted axonemes) in which they were found in the flagellar proteome (Table II). Peptides of FAP232 and FAP173 were present primarily in the membrane + matrix fraction. FAP146, FAP73, and FAP295 were found exclusively or primarily in the KCl-extract fraction, suggesting that they are readily solubilized axonemal components. During the course of this work, FAP146 was shown in an unrelated study to be a subunit of an inner dynein arm [Yamamoto et al., 2008]. FAP134, FAP122, FAP207, and FAP52 were found exclusively or primarily in the KCl-extracted axoneme fraction, suggesting that they are integral components of the axoneme. Sequence comparison identified various motifs in these proteins. For example, FAP295 appears to be a catalytic subunit of a cyclic nucleotide regulated protein kinase (Table II). FAP173 is predicted to have a Band 7 domain characteristic of the flotillin/stomatin/prohibitin family of membrane-associated proteins, and has homologues in both ciliated and non-ciliated organisms. All other proteins are highly conserved in ciliated organisms.
Table I.
Genes selected for HA-tagging
| gene (v2.0) |
protein ID | protein name |
size of PCR product (bp) |
upstream of start codon (bp) |
start codon - pen- ultimate codon (bp) |
human homologue |
BLAST E value |
|---|---|---|---|---|---|---|---|
| C_20334 | EDP07596 | PACRG | 3120 | 1483 | 1637 | NP_689623 | 8e-66 |
| C_30068 | EDP04330 | FAP146 | 4267 | 1628 | 2639 | NP_115633 | 3e-16 |
| C_310134 | EDO99184 | FAP173 | 4384 | 883 | 3501 | NP_038470* | 3e-11 |
| C_320063 | EDP08000 | FAP232 | 2508 | 1522 | 986 | NP_057210 | 3e-19 |
| C_380055 | EDP03601 | FAP134 | 5155 | 1666 | 3489 | NP_112584 | 1e-56 |
| C_410061 | EDP09736 | FAP122 | 3795 | 1469 | 2326 | BAB15699 | 8e-16 |
| C_450085 | EDP05720 | FAP207 | 2802 | 819 | 1983 | AAH57760 | 3e-30 |
| C_630058 | EDP05376 | FAP52 | 5062 | 1567 | 3495 | AAH25392 | 1e-161 |
| C_650031 | EDO98890 | FAP73 | 3842 | 1720 | 2122 | NP_653282 | 4e-17 |
| C_60149 | EDO99770 | FAP295 | 4254 | 340 | 3914 | NP_006250 | 5e-90 |
The gene numbers in version 2.0 of the C. reinhardtii genome (JGI), the protein names, details of the PCR products, putative human homologues, and the E values obtained in BLAST searches between the human and C. reinhardtii gene products are shown. The size of the PCR products does not include the overhangs and restriction sites added to the primers for cloning of the PCR product.
Although NP_038470 is the closest match to C_310134 in humans, C_310134 is much more closely related to band 7 proteins in Leishmania and plants.
Table II.
Proteins tagged by HA in this study
| protein | size without 3xHA (aa) |
predicted mass incl. 3xHA/ apparent mass in SDS-PAGE (Da) |
localization | transformants (positive/tested) |
flagellar proteome unique peptides/fraction |
predicted domains /motifs |
|---|---|---|---|---|---|---|
| PACRG | 308 | 38913/44000 | fla/c | 21/57 | 24/axoneme | PACRG superfamily |
| FAP146 | 381 | 45719/50000 | fla/bb | 9/24 | 9/KCl extract | - |
| FAP173 | 303 | 37435/42000 | fla/bb/c | 5/24 | 7/membrane + matrix and KCl extract | Band 7 superfamily |
| FAP232 | 190 | 25156/35000 | fla/bb | 26/48 | 5/membrane + matrix | Galactose-binding domain-like |
| FAP134 | 525 | 65205/70000 | fla/c | 3/40 | 10/axoneme | Leucine-rich repeats |
| FAP122 | 374 | 47414/54000 | fla/c | 7/36 | 11/axoneme and tergitol insoluble | - |
| FAP207 | 257 | 33174/39000 | fla | 20/24 | 6/axoneme and tergitol insoluble | MORN ? |
| FAP52 | 634 | 73271/79000 | fla/c | 4/12 | 30/axoneme | WD40 repeats |
| FAP73 | 309 | 40410/49000 | fla/bb | 4/38 | 18/KCl extract | coiled-coil |
| FAP295 | 719 | 85471/90000 | fla/c | 4/48 | 31/KCl extract | cGMP-dependent protein kinase |
The protein names, sizes, predicted molecular mass of the HA-tagged products, molecular mass observed on western blots, localization in this study, number of tested and positive transformants, number of unique peptides and predominate fraction in which they were found in the proteomic analysis of flagella, and predicted domains of the gene products are shown. aa, amino acids; fla, flagella; c, cell body; bb, near the basal bodies.
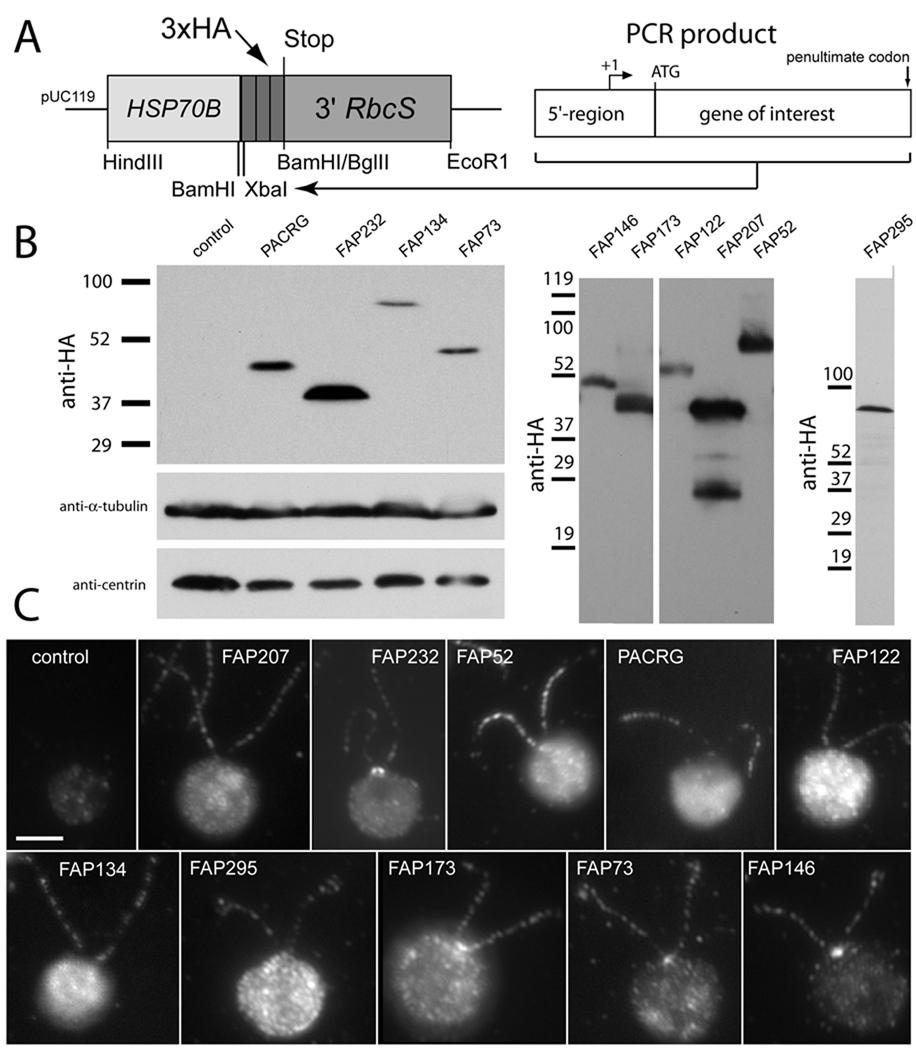
The 10 target genes, including parts of their 5'-upstream regions, were amplified from genomic DNA by means of gene-specific primer pairs (Supplementary Table SI) and a touchdown PCR protocol. The PCR products, ranging from 2508 to 5155 bp, were cloned into pKLl (Fig. 1A), a novel vector consisting of the HSP70B promoter region as an enhancer element, and a triple HA-tag followed by the RbcS terminator sequence. After co-transformation into C. reinhardtii strain CC3395, randomly selected transformants were screened by indirect immunofluorescence of methanol-fixed cells using a commercially available rat monoclonal antibody to the HA epitope. Positive strains were identified for each construct and expression of the fusion proteins was confirmed by western blotting of whole-cell extracts (Fig. 1B). Typically one or two closely spaced bands were detected by the anti-HA antibody; western blotting of the strain expressing FAP207-HA showed several minor bands below the major band of 39,000 Da that we assume were caused by proteolytic processing of FAP207-HA. Table II shows that the apparent mass in SDS-PAGE of the reactive bands generally exceeded the theoretical mass of the target protein plus the triple-HA tag by 4,000 to 10,000 Da, suggesting that the HA tag reduced the mobility of the proteins in SDS-PAGE more than expected based on the tag's molecular mass of 4796 Da.
Fig. 1.
HA-tagging of flagellar proteins. (A) Vector and cloning strategy used for HA tagging of flagellar proteins. HSP70B, HSP70B promoter region; 3xHA, sequence encoding triple-HA tag; 3' RbcS, Rubisco small subunit terminator sequence; +1, first base of predicted 5'-untranslated region of the target gene. (B) Western blots of whole-cell extracts from the indicated transformants and a wild-type control probed with antibodies to HA (top and right panels), α-tubulin, or centrin. Numbers at left of each panel indicate molecular weights of protein standards. (C) Immunofluorescence microscopy of transformants and wild-type control probed with anti-HA. Bar = 5 µm.
In indirect immunofluorescence, each of the 10 HA-tagged proteins was detected as a more or less punctuate staining pattern along the entire length of the flagella (Fig. 1C). Exceptions were PACRG-HA, which was localized to the distal region of the flagellum more strongly than the proximal region (but only under some conditions; see Discussion), and FAP52-HA, which was largely absent from the distal part of the flagellum. Additionally, the region near the basal bodies was decorated in cells expressing FAP232-HA, FAP73-HA, FAP146-HA, and often FAP173-HA; interestingly, FAP73-HA was localized to a single spot of variable size that appeared to be located between the two basal bodies (Figs. 1C and 2A). A generalized labeling of the cell body was observed for most of the constructs and strains analyzed; some of the HA-tagged proteins, e.g. FAP207-HA, tended to accumulate in the cell body when over-expressed (not shown).
Fig. 2.
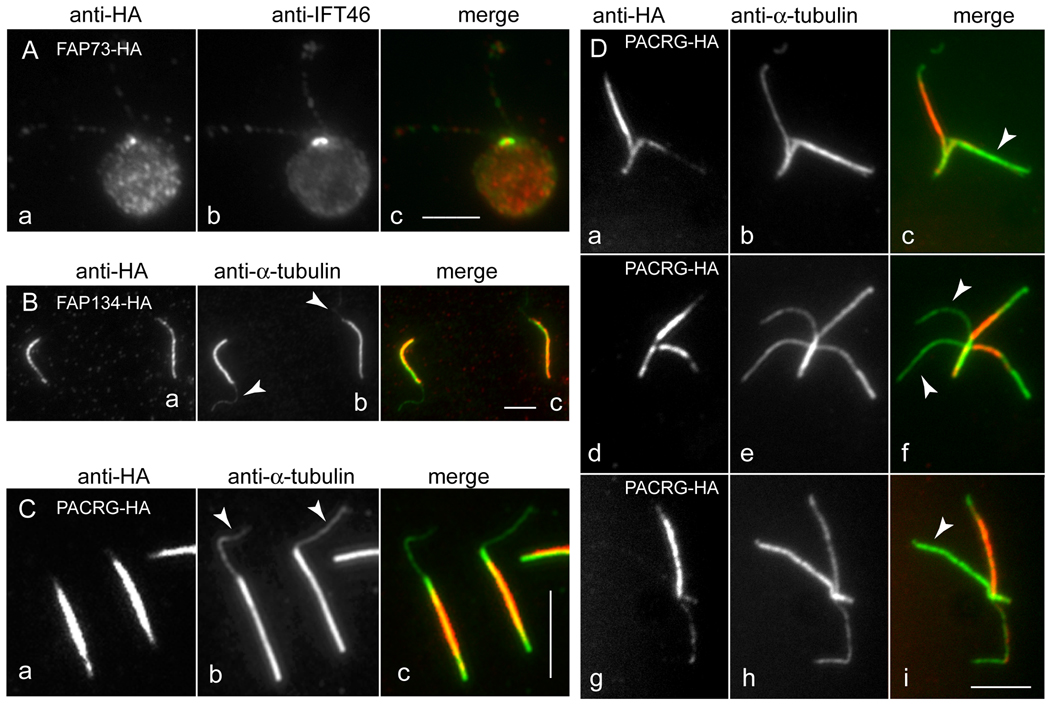
Co-localization studies of FAP73-HA, FAP134-HA, and PACRG-HA.
(A) Methanol-fixed cell expressing FAP73-HA stained with anti-HA (a) and anti-IFT46 (b). The merged image is shown in c. (B) Isolated flagella with protruding central pair (arrowheads in b) from a strain lacking endogenous FAP134 and expressing FAP134-HA, stained with anti-HA (a) and anti-tubulin (b). The merged image is shown in c. (C) Same as in B but from strain 4.3 expressing PACRG-HA. (D) Split axonemes of strain 4.3 expressing PACRG-HA, stained with anti-HA (a, d, g) and anti-tubulin (b, e, h). The merged images (c, f, i) show that staining representing PACRG-HA is strongly reduced or absent from some outer doublet microtubules. Strain 4.3 used for the images in C and D carries a PACRG RNAi construct in addition to the PACRG-HA vector. See Supplementary Fig. 1 for a more detailed characterization of this strain. Bars = 5 µm.
The HA tag also facilitated more detailed localization of the proteins within the flagellum. For example, immunofluorescence microscopy indicated that PACRG-HA and FAP134-HA remained with the axoneme after demembranation of whole cells (not shown). This is in good agreement with the distribution of peptides from these two proteins in fractions of the C. reinhardtii flagellar proteome [Pazour et al., 2005]: eight peptides from FAP134 were found in the KCl-extracted axoneme fraction, but none in the membrane + matrix or KCl fractions, and 19 peptides from PACRG were found in the KCl-extracted axoneme fraction and 6 in the KCl extract fraction, but none in the membrane + matrix fraction. The immunofluorescence microscopy and proteomic results together indicate that these two proteins are likely to be axonemal components. To determine if FAP134 and PACRG are associated with the outer doublet or central microtubules, flagella were isolated and demembranated in the presence of trypsin and ATP to induce extrusion of the central pair of microtubules [Lechtreck and Witman, 2007]. The axonemes with extruded central microtubules were then double labeled with an anti-tubulin antibody and an anti-HA-antibody and examined by immunofluorescence microscopy. In both cases, the anti-HA antibody labeled the outer doublets but not the central pair of microtubules (Figs. 2B and C), indicating that these proteins are associated with the former but not the latter. Surprisingly, immunofluorescence microscopy of splayed axonemes revealed that PACRG-HA was localized to a subset of doublet microtubules (Fig. 2D). To reduce the amount of endogenous wild-type protein that competed with the HA-tagged protein for assembly into the axoneme, these experiments were carried out using a PACRG RNAi strain expressing PACRG-HA (see Supplementary Fig. 1) or a mutant that lacks part of the FAP134 gene and expresses no wild-type FAP134 in the flagellum (Awata and Witman, unpublished results).
In summary, HA-tagged derivatives of PACRG and nine uncharacterized conserved proteins identified by the flagellar proteome were localized to the flagellum; variations in the labeling patterns and simple fractionation experiments indicated that several of the proteins represented structures or compartments of special interest.
FAP232 Co-Localizes with IFT46
Flagellar staining in strains expressing FAP173-HA and FAP232-HA was abolished by detergent extraction, indicating that these are soluble components of the flagellum (not shown). Consistent with this, in the analysis of the flagellar proteome [Pazour et al., 2005], peptides representing FAP232 were found exclusively in the membrane + matrix fraction, and peptides representing FAP173 were found primarily in the membrane + matrix fraction (6 membrane + matrix, 4 KCl extract). The membrane + matrix fraction also includes proteins involved in IFT. To determine if there is a relationship between either of these proteins and IFT, double immunofluorescence was used to test whether FAP173-HA or FAP232-HA co-localized with IFT46, an established IFT complex-B component. FAP173-HA, which also was present in the cell body and near the basal bodies, showed a punctate distribution along the flagellum, but only some of these foci overlapped with IFT particles as revealed by staining with IFT46 (Figs. 3A and B). Similar results were obtained regardless of whether cells were fixed with methanol or simultaneously lysed and fixed by addition of 0.1% Triton X-100 and 3% formaldehyde in a microtubule-stabilizing buffer. The different distribution of FAP173-HA and IFT46 was especially apparent during flagellar regeneration, when IFT components typically accumulate at the tip of growing flagella; in contrast, FAP173-HA accumulated in the middle of the elongating flagellum (Figs. 3C and D). We conclude that FAP173-HA is not part of the established IFT machinery but represents a novel compartment in the flagellum.
Fig. 3.
FAP173-HA does not co-localize with IFT46. (A) and (B) Detached flagella from a transformant expressing FAP173-HA were double labeled with antibodies to HA (a, red in c) and IFT46 (b, green in c). The right panels show a merger of the left and center panels. Numerous foci appeared to contain FAP173-HA only (open arrowheads) or IFT46 only (arrowsheads). (C) and (D) Cells expressing FAP173-HA were deflagellated and allowed to regenerate flagella for 20 min, then fixed and labeled with anti-HA (red) and anti-IFT46 (green) antibodies. IFT46 is concentrated at the flagellar tips (arrowheads) whereas FAP173-HA is concentrated in the middle region of the flagella (arrows). Bars = 5 µm.
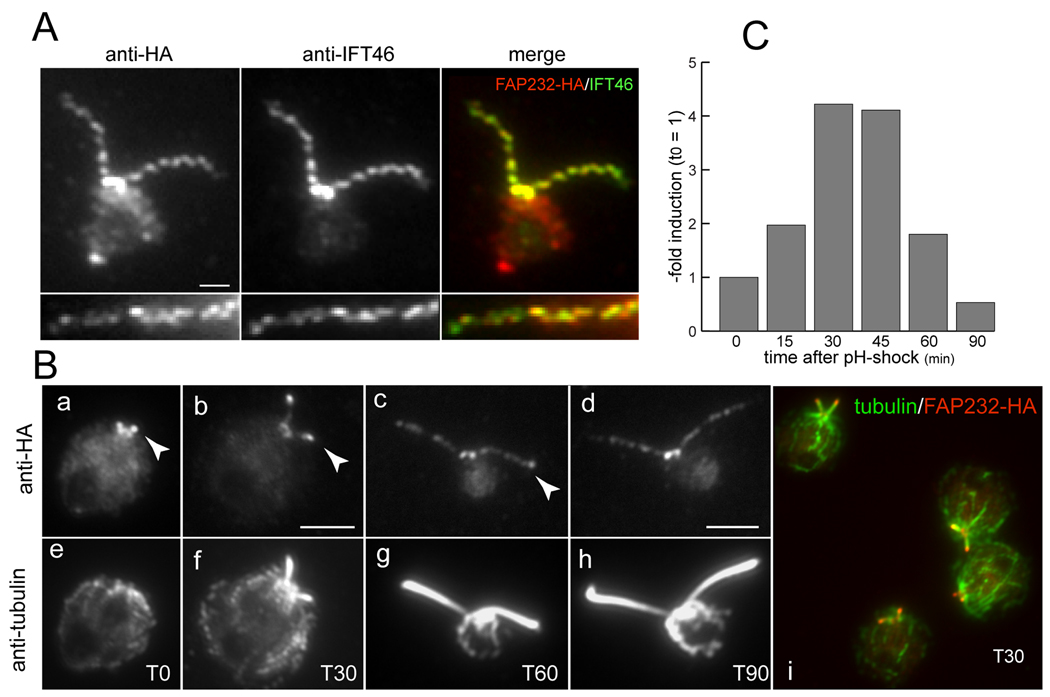
In contrast, FAP232-HA co-localized precisely with IFT46 as revealed by double immunofluorescence of cells (Fig. 4A). The results were identical regardless of whether cells were fixed with methanol or simultaneously lysed and fixed by addition of 0.1% Triton X-100 and 3% formaldehyde in a microtubule-stabilizing buffer. During flagellar regeneration, FAP232-HA accumulated at the tip of the growing flagella (Fig. 4B), as is typical of IFT proteins (cf. IFT46 in Figs. 3C and D). After amputation of flagella from control cells (strain 137c) by pH-shock, the message for FAP232 increased about 4-fold to a peak at 30–45 minutes post-deflagellation, and then decreased (Fig. 4C). This is typical for many mRNAs encoding flagellar components, including IFT proteins. The distribution of FAP232-HA during flagellar regrowth and its co-localization with IFT46 strongly suggest that FAP232 is a component of the IFT machinery.
Fig. 4.
FAP232 co-localizes with IFT46. (A) A transformant expressing FAP232-HA was double labeled with anti-HA (red) and anti-IFT46 (green). The bottom panel shows a detail of another flagellum from the same strain. There is almost complete coincidence between the FAP232-HA and IFT46 foci. Bar = 1 µm. (B) Localization of FAP232-HA during flagellar regeneration. Cells were double labeled with anti-HA and anti-tubulin. The right panel is a merged image of several regenerating cells double-labeled for FAP232-HA (red) and tubulin (green). Arrowheads indicate accumulation of FAP232-HA at the tips of the growing flagella. The time after deflagellation is indicated in minutes (T0, T30, T60, T90). (C) Induction of FAP232 message in wild-type cells after deflagellation by pH shock. Message levels were assayed by quantitative PCR.
FAP232 is a Component of IFT Complex B
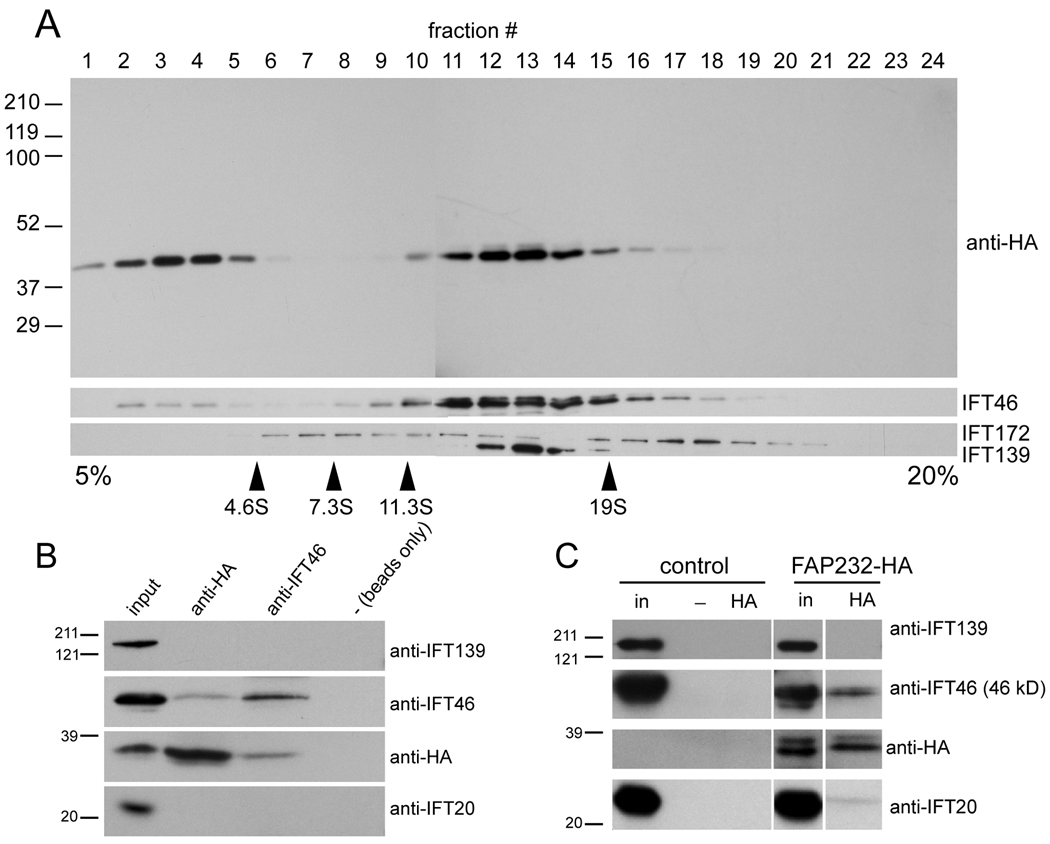
To confirm that FAP232 is part of the IFT machinery, and to determine if it is associated with IFT complex A, complex B, or the motors that move the IFT particles, we carried out a biochemical analysis of FAP232-HA. When proteins from the membrane + matrix fraction of strains expressing FAP232-HA were separated by sucrose density gradient centrifugation, most of the FAP232-HA co-migrated with IFT complex B as represented by IFT46 (Fig. 5A). IFT139, a component of complex A, peaked at a slightly higher density than IFT46 and FAP232-HA. Considerable amounts of FAP232-HA were present in the fractions above 4.6S, indicating that some FAP232-HA dissociated from the IFT complexes under the conditions used; some IFT46 also was present in these fractions. IFT172, a peripheral complex B protein, was almost completely dissociated from complex B and sedimented in two peaks, one above and one below the otherwise intact complex B at ~15S, as previously observed by Hou et al. [2007].
Fig. 5.
FAP232 is a component of IFT complex B. (A) Proteins in the membrane + matrix fraction obtained from flagella of cells expressing FAP232-HA were separated by sucrose density gradient centrifugation and analyzed by western blotting. The positions of marker proteins are indicated. FAP232-HA co-sediments precisely with complex B protein IFT46 but not complex A protein IFT139. (B) Western blot analysis of immunoprecipitates obtained from fractions 12 and 13 of the sucrose gradient shown in A. Protein G beads loaded with anti-HA antibody, protein A beads loaded with anti-IFT46 antibody, and protein G beads alone were used for the immunoprecipitations. The anti-HA antibody co-precipitated FAP232-HA and IFT46 but not IFT139. Similarly, the anti-IFT46 antibody co-precipitated IFT46 and FAP232-HA but not IFT139. The position of the marker proteins is indicated. (C) Western blots of immunoprecipitates obtained from membrane + matrix fractions from flagella of wild-type (control) and a strain expressing FAP232-HA, using anti-HA-loaded or unloaded (−) protein G beads. The anti-HA antibody co-precipitated FAP232-HA and complex B proteins IFT46 and IFT20, but not complex A protein IFT139.
Further evidence that FAP232 is part of IFT complex B was obtained by immunoprecipitation studies. Rat monoclonal anti-HA co-immunoprecipitated FAP232-HA and IFT46 but not IFT139 from fractions 12 and 13 of the above sucrose gradient, and anti-IFT46 co-immunoprecipitated IFT46 and FAP232-HA but not IFT139 (Fig. 5B). Similarly, protein G-sepharose beads loaded with the anti-HA antibody co-immunoprecipitated FAP232, IFT46, and some IFT20, but no IFT139, from the membrane + matrix fraction collected from isolated flagella of a strain expressing FAP232-HA (Fig. 5C). None of these IFT proteins were immunoprecipitated with anti-HA from untransformed control cells. These data provide further independent evidence that FAP232 is a component of IFT complex B.
FAP232 Is Highly Conserved in a Subset of Ciliated Organisms
BLAST searches of nucleotide databases at NCBI and the Joint Genome Institute (JGI) revealed that FAP232 is conserved in many organisms with the ability to assemble cilia or flagella. These include vertebrates such as Danio rerio (XM_692944, BLAST E = 5e-24), Mus musculus (MN_028394, BLAST E = 6e-18), and Homo sapiens (NP_057210, BLAST E = 1e-21) (Fig. 6); the chordate Branchiostoma floridae (EEA72558, BLAST E = 1e-21), the sea urchin Strongylocentrotus purpuratus (XP_001201498, BLAST E = 2e-13), the polychaete worm Capitella sp. I (JGI gene model estExt_fgenesh1_pm.C_80002, BLAST E = 1e-20), the sea anemone Nematostella vectensis (XP_001640906, BLAST E = 5e-17), the basal eumetazoan Trichoplax adhaerens (XP_0002114382, BLAST E = 2e-19), and the protist Trichomonas vaginalis (XP_001580162, BLAST E = 2e-11). In the oomycete Phytophthora sojae, FAP232 is predicted to be part of a larger protein (JGI ID 142169, BLAST E = 1e-19). Trypanosoma brucei Tbl1.01.4880 has weaker homology to FAP232 (BLAST E = 9e-06) but the two proteins identified each other as best matches in reciprocal BLAST searches, indicating that the trypanosome protein also is likely to be an orthologue of FAP232. Interestingly, homologues of FAP232 appear to be absent from the genomes of centric diatoms, apicomplexan parasites, the moss Physcomitrella, C. elegans, D. melanogaster, and Ciona intestinalis, all of which have cilia or flagella. No sequences similar to FAP232 were found in non-ciliated organisms (Saccharomyces cerevisiae and Arabidopsis thaliana).
Fig. 6.
Alignment of C. reinhardtii FAP232/IFT25 with homologues from D. rerio, M. musculus, and H. sapiens. The alignment was generated by ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
DISCUSSION
Eukaryotic flagella and cilia are complex structures consisting of more than six hundred different proteins. A current challenge to investigators of these organelles is to determine where each protein is located within the flagellum, and to identify its function there. Because only about 100 of these proteins have been characterized to this extent, there is a need for methods to rapidly and efficiently determine the flagellar locations of the uncharacterized proteins and obtain enough preliminary information to prioritize them for further study. The C. reinhardtii flagellar proteome provided some guidance, as many proteins could be assigned to the membrane, matrix, or axonemal compartments. Approaches for learning more about the locations and functions of proteins on a large scale include transposon-based random epitope-tagging, and expression of proteins fused to a tag that can then be used to localize the proteins by fluorescence microscopy [for review see O'Rourke et al., 2005]. For example, Andersen et al. [2003] and Kilburn et al. [2007] tagged candidate centrosomal and basal body components with GFP and determined their subcellular locations by immunofluorescence microscopy. Similarly, Van Damme et al. [2004] tagged selected Arabidopsis thaliana proteins potentially involved in cytokinesis with GFP, and then observed their subcellular locations.
We have taken a similar approach with putative C. reinhardtii flagellar proteins, but used an HA tag rather than a GFP tag. The HA tag was chosen for several reasons. First, in our hands, an HA-tagged protein is much more likely to be expressed in C. reinhardtii than is a GFP-tagged protein, possibly because of the smaller size of the HA tag. Second, the smaller tag may be advantageous when the epitope-tagged protein is expressed in a wild-type background and has to compete with the endogenous protein for binding at the correct site, as is the case when a mutant for the target gene has not yet been identified. Third, although functional expression of proteins tagged with GFP has been accomplished for several soluble flagellar proteins, this has not been reported yet for axonemal components, where steric constraints may be high and interfere with the assembly of a protein fused to a large tag. Finally, a monoclonal antibody to HA is available that has a very high specificity for the HA tag when used in C. reinhardtii.
The HA-tagged versions of all ten proteins described here were expressed following transformation into wild-type C. reinhardtii, and all were localized to the flagella by immunofluorescence microscopy. The epitope-tagged proteins generally showed a spotted distribution along the flagella. This pattern of distribution was not observed for tubulin (see Fig. 4B) and is unlikely to represent the native distribution for other axonemal proteins because the axoneme has uniform ultrastructure with a 96-nm periodicity over most of its length. Inasmuch as all of the tagged proteins initially were expressed in the presence of the endogenous wild-type protein, it is likely that the punctate distribution of the axonemal HA-tagged proteins is due to a mixture of tagged and wild-type proteins along the axoneme. Because there are only a limited number of binding sites for the proteins along the axoneme, competition between the endogenous and tagged proteins for these binding sites results in a speckled distribution of the tagged protein. In support of this hypothesis, strong and continuous labeling along the axoneme was observed when FAP134-HA was expressed in a mutant that does not express wild-type FAP134 [Fig. 2B]. Similarly, the incorporation of PACRG-HA into the axoneme increased and became less punctate when the expression of both the endogenous and the HA-tagged PACRG were decreased by introduction of an RNAi inverted-repeat construct (Fig. 2C, D; see Supplementary Fig. 1). In contrast to the result in Fig. 1, where the PACRG-HA was localized most strongly to the distal part of the flagellum, the tagged protein was incorporated preferentially into the proximal and/or middle regions of the flagellum in the RNAi cells. The likely explanation for this is that the endogenous protein, when present in normal amounts, outcompetes the tagged protein for binding sites in the proximal part of the flagellum.
PACRG-HA Is Assembled on a Subset of Outer Doublets
The distribution of the PACRG-HA protein was further examined in disintegrated isolated axonemes. As expected from the work of Ikeda et al. [2007], the protein was present exclusively on the outer doublet microtubules and not on the central pair, which was visualized following trypsin plus ATP-induced extrusion from the axoneme. Surprisingly, in splayed axonemes, only a subset of doublets was labeled, suggesting that PACRG-HA is asymmetrically distributed around the axoneme. This distribution may be related to previously described differences between the outer doublets. Hoops and Witman [1983] reported structural asymmetries in the axoneme, specifically beak-like projections in the lumens of the B-tubules of the number 1, 5, and 6 doublets, and a bridge between the number 1 and 2 doublets. Rupp et al. [1996] reported a biased loss of outer dynein arms on a subset of outer doublets in the sup-pf-2 mutant. It will be of interest to determine if PACRG provides the biochemical basis for any of these differences between individual outer doublets.
FAP73-HA Is Enriched at a Single Spot Between the Two Basal Bodies
In addition to its punctuate distribution along the flagella, FAP73-HA was localized to a single spot near the basal bodies. Double staining with anti-IFT46 showed that FAP73-HA is concentrated between the two flagella-bearing basal bodies of C. reinhardtii. The region between the basal bodies is spanned by two proximal fibers and the massive distal connecting fiber, which contains the calcium-modulated protein centrin [Huang et al., 1988]. FAP73 is predicted to have a high probability of forming an α-helical coiled-coil, a structural motif common to components of the flagellar basal apparatus. In future experiments, it will be interesting to determine the localization of FAP73 at an ultrastructural level.
FAP232 Is a Novel IFT Particle Protein and Is Renamed IFT25
FAP232 and FAP173 were found exclusively and primarily, respectively, in the detergent-soluble fraction of the C. reinhardtii flagellar proteome. This fraction is enriched in components of IFT, the cellular machinery that assembles and maintains flagella. IFT particles consist of at least 18 different proteins, and studies in various organisms have identified additional proteins that are required for, control, or are moved by IFT. We therefore were interested in whether FAP232 and FAP173 are involved in IFT. Both proteins exhibited a labeling pattern similar to that of known IFT proteins – i.e, a punctate distribution along the flagella with brighter foci at the basal bodies – and in both cases the flagellar labeling was lost upon treatment of the cells with nonionic detergents, as also is the case for IFT proteins. However, FAP173-HA did not co-localize with the complex B protein IFT46. Moreover, it did not accumulate at the tips of regenerating flagella, as IFT components do. These results suggest that it is not an IFT component and instead represents a different, currently unknown, compartment or process in the flagellum. In contrast, FAP232-HA exhibited a 100% co-localization with IFT46 in steady-state flagella. It also accumulated at the tips of growing flagella. The HA tag also allowed us to follow FAP232 in biochemical experiments: FAP232-HA co-sedimented with IFT46 in sucrose density gradients, and antibodies to the HA tag co-immunoprecipitated IFT46. Conversely, antibodies to IFT46 co-immunoprecipitated FAP232-HA. We conclude that FAP232 is a component of IFT complex B, and rename it IFT25, consistent with the nomenclature for other C. reinhardtii IFT-particle proteins.
IFT25 Exhibits an Unusual Pattern of Evolutionary Conservation
Similar to many axonemal proteins, most of the known components of IFT are highly conserved and widely distributed among organisms having cilia or flagella. Putative orthologues of IFT25 are present in many flagellated protists and metazoans, including both protostomes and deuterostomes, but appear to be absent from the genomes of diatoms, the moss Physcomitrella, apicomplexan protists, ciliates, D. melanogaster, C. elegans, and C. intestinalis. This distribution suggests that IFT25 was part of the ancestral IFT machinery but was repeatedly lost during evolution in unrelated groups. One possibility is that the role of IFT25 has been taken over by a different protein in some species. Another possibility is that IFT25 has a more specialized role than general flagellar assembly and that its function is required in some species but not in others. Interestingly, one feature common to those organisms lacking IFT25 is that, as far as we know, their cilia and flagella are not disassembled as part of the cell cycle. This also is of interest given the apparent connection between IFT25 and IFT27 (see below).
The Human Homologue of IFT25 Is HSPB11
The human homologue of IFT25, which shares 37% identity (58% similarity) with the N-terminal 130 residues of C. reinhardtii IFT25, was originally identified as placental protein 25 (PP25) [Bohn and Winckler 1991]. Recently, the protein was shown to have sequence similarity to and properties of small heat shock proteins and was renamed small heat shock protein beta-11 (HSPB11, also known as C1orf41 protein, HSPCO34, and HSP16.2) (Bellyei et al. 2007]. X-ray crystallography and NMR spectroscopy have established the structure of HSPB11, which consists largely of short beta-strands (http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=30728) [Ramelot et al. 2008]. A similar structure has been observed for the galactose-binding domain of sialidase from Micromonospora viridifaciens (PDB ID 1EUT), other cell surface-attached carbohydrate-binding domains, and the human anaphase-promoting complex subunit 10 (PDB ID 1JHJ). Using western blotting and immunocytochemistry, HSPB11 has been localized in the cytoplasm, the nucleus, and the mitochondria of cultured cells [Bellyei et al., 2007]. However, it is not clear if the cells used in these studies were ciliated. The subcellular distribution of the FLAG-tagged mouse HSPB11/IFT25 homologue was examined in IMCD3 cells in the accompanying paper by Follit et al. [2009], who observed that it was localized to the cilium and centrosome. Follit et al. also observed that an anti-FLAG antibody co-immunoprecipitated FLAG-IFT25 and complex B proteins IFT20, IFT27, and IFT88 but not the complex A protein IFT140. Thus, mouse HSPB11/IFT25, like C. reinhardtii IFT25, appears to be an IFT complex-B component.
Several other chaperones and heat shock proteins have been identified in the flagella and cilia of C. reinhardtii and other organisms [Bloch and Johnson, 1995; Stephens and Lemieux, 1999, and see references cited therein]. For example, HSP70A, which appears to be relatively abundant in the flagellum [Pazour et al., 2005], has a punctate distribution along the flagellum that partially overlaps with that of the IFT motor kinesin-2 [Shapiro et al., 2005]. Radial spoke proteins RSP12 and RSP16 are predicted to function as molecular chaperones [Yang et al., 2006]; the latter is transported separately into the flagellum and then apparently joins with the radial spoke precursor complex during the latter's assembly onto the axoneme [Yang et al., 2005]. Some Bardet Biedl Syndrome proteins, which are associated with IFT, show sequence homology to chaperonins [Blacque and Leroux, 2006]. It is possible that IFT25 has a chaperone function during the loading and unloading of flagellar precursors from IFT particles, during the remodeling of IFT particles from anterograde to retrograde forms, or in the transport of cargo during IFT.
IFT25 and IFT27 Have Similar Phylogenetic Distributions and May Interact Physically and Functionally
Systematic mapping of human binary protein–protein interactions of 8,100 open reading frames using a stringent, high-throughput yeast two-hybrid system [Rual et al., 2005] identified only one interaction involving an established IFT complex B protein, namely IFT27 [Qin et al., 2007] as a putative binding partner of the human homologue of IFT25. As noted above, Follit et al. [2009] observed that the mouse homologue of IFT27 was co-immunoprecipitated with IFT25, although it was not demonstrated that this interaction is direct. Interestingly, the phylogenetic distribution of IFT27 is similar to that of IFT25. Like IFT25, IFT27 is present in many ciliated species including C. reinhardtii and various vertebrates but not in C. elegans and D. melanogaster. Taken together, these findings suggest that IFT25 and IFT27 may cooperate in some function related to assembly, maintenance, or disassembly of cilia and flagella.
CONCLUSIONS
We demonstrate the utility of a novel cassette for expressing selected C. reinhardtii proteins with an HA tag following insertion of the gene of interest into the vector and transformation of the vector into C. reinhardtii cells. In an analysis of PACRG and 9 uncharacterized proteins from the flagellar proteome, the HA-tagged versions of all 10 proteins were found to localize to the flagella. Further analysis of the HA-tagged proteins revealed that PACRG-HA was localized to a subset of outer doublet microtubules, FAP73 is concentrated in a single foci between the two basal bodies, and FAP232 is an IFT complex-B protein, which we rename IFT25. Homologues of the latter are present in a subset of ciliated organisms, suggesting that they participate in a specialized IFT function, possibly flagellar disassembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Douglas Cole (University of Idaho) and Hongmin Qin (Texas A & M, College Station) for generous sharing of antibodies. This work was supported by National Institutes of Health grant GM30626 and by the Robert W. Booth Fund at the Greater Worcester Community Foundation.
REFERENCES
- Adams M, Smith UM, Logan CV, Johnson CA. Recent advances in the molecular pathology, cell biology and genetics of ciliopathies. J Med Genet. 2008;45:257–267. doi: 10.1136/jmg.2007.054999. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Beales PL, Bland E, Tobin JL, Bacchelli C, Tuysuz B, Hill J, Rix S, Pearson CG, Kai M, Hartley J, et al. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet. 2007;39:727–729. doi: 10.1038/ng2038. [DOI] [PubMed] [Google Scholar]
- Bellyei S, Szigeti A, Pozsgai E, Boronkai A, Gomori E, Hocsak E, Farkas R, Sumegi B, Gallyas F., Jr Preventing apoptotic cell death by a novel small heat shock protein. Eur J Cell Biol. 2007;86:161–171. doi: 10.1016/j.ejcb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Blacque OE, Leroux MR. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch MA, Johnson KA. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J Cell Sci. 1995;108:3541–3545. doi: 10.1242/jcs.108.11.3541. [DOI] [PubMed] [Google Scholar]
- Bohn H, Winckler W. Isolation and characterization of five new soluble placental tissue proteins (PP22, PP23, PP24, PP25, PP26) Arch Gynecol Obstet. 1991;248:111–115. doi: 10.1007/BF02390087. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Xu F, Keady B, Pazour GJ. Characterization of Mouse IFT Complex B. Cell Motil Cytoskeleton. 2009;66 doi: 10.1002/cm.20346. in press [doi: 10.1002/cm.20346] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimer S, Teltenkotter A, Plessmann U, Weber K, Lechtreck KF. Purification and characterization of basal apparatuses from a flagellate green alga. Cell Motil Cytoskeleton. 1997;37:72–85. doi: 10.1002/(SICI)1097-0169(1997)37:1<72::AID-CM7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Mengersen A, Lee VD. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ikeda T, Morikawa K, Kamiya R. Axonemal localization of Chlamydomonas PACRG, a homologue of the human Parkin-coregulated gene product. Cell Motil Cytoskeleton. 2007;64:814–821. doi: 10.1002/cm.20225. [DOI] [PubMed] [Google Scholar]
- Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci. 2003;116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Witman GB. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J Cell Biol. 2007;176:473–482. doi: 10.1083/jcb.200611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke NA, Meyer T, Chandy G. Protein localization studies in the age of 'Omics'. Curr Opin Chem Biol. 2005;9:82–87. doi: 10.1016/j.cbpa.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA. Improving NMR protein structure quality by Rosetta refinement: A molecular replacement study. Proteins. 2008 doi: 10.1002/prot.22229. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Rupp G, O'Toole E, Gardner LC, Mitchell BF, Porter ME. The sup-pf-2 mutations of Chlamydomonas alter the activity of the outer dynein arms by modification of the gamma-dynein heavy chain. J Cell Biol. 1996;135:1853–1865. doi: 10.1083/jcb.135.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardi. Ann N Y Acad Sci. 1953;56:831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blocker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Shapiro J, Ingram J, Johnson KA. Characterization of a molecular chaperone present in the eukaryotic flagellum. Eukaryot Cell. 2005;4:1591–1594. doi: 10.1128/EC.4.9.1591-1594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, LaVoie M, Tam LW, Tousey S, Sanders M, Wu W, Borodovsky M, Lefebvre PA. The Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J Cell Biol. 2001;153:63–74. doi: 10.1083/jcb.153.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Rosenbaum JL. Multiple alpha- and beta-tubulin genes in Chlamydomonas and regulation of tubulin mRNA levels after deflagellation. Cell. 1981;24:81–88. doi: 10.1016/0092-8674(81)90503-1. [DOI] [PubMed] [Google Scholar]
- Stephens RE, Lemieux NA. Molecular chaperones in cilia and flagella: implications for protein turnover. Cell Motil Cytoskeleton. 1999;44:274–283. doi: 10.1002/(SICI)1097-0169(199912)44:4<274::AID-CM5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Van Damme D, Bouget FY, Van Poucke K, Inze D, Geelen D. Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J. 2004;40:386–398. doi: 10.1111/j.1365-313X.2004.02222.x. [DOI] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Yanagisawa HA, Yagi T, Kamiya R. Novel 44-kilodalton subunit of axonemal dynein conserved from Chlamydomonas to mammals. Eukaryot Cell. 2008;7:154–161. doi: 10.1128/EC.00341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Compton MM, Yang P. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol Biol Cell. 2005;16:637–648. doi: 10.1091/mbc.E04-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, Rosenbaum JL, Witman GB. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.