Summary
Filamentous fungi produce a vast array of small molecules called secondary metabolites, which include toxins as well as antibiotics. Co-regulated gene clusters are the hallmark of fungal secondary metabolism, and there is a growing body of evidence that suggests regulation is at least, in part, epigenetic. Chromatin-level control is involved in several silencing phenomena observed in fungi including mating type switching, telomere position effect, silencing of ribosomal DNA, regulation of genes involved in nutrient acquisition, and as presented here, secondary metabolite cluster expression. These phenomena are tied together by the underlying theme of chromosomal location, often near centromeres and telomeres, where facultative heterochromatin plays a role in transcription. Secondary metabolite gene clusters are often located sub-telomerically and recently it has been shown that proteins involved in chromatin remodeling, such as LaeA, ClrD, CclA and HepA mediate cluster regulation.
Introduction
For many years it has been known that chromosomal location and histone modification have profound effects on gene transcription in a variety of organisms from yeast to humans. Filamentous fungi produce many bioactive small molecules, or secondary metabolites, that range from beneficial antibiotics to harmful toxins. Genes responsible for production of these secondary metabolites are typically clustered and co-regulated [1]. Interestingly, the order and location of biosynthetic genes within a cluster is important for their regulation. Additionally, secondary metabolite gene clusters have a tendency to be located near the ends of chromosomes in areas termed sub-telomeric [2••,3•] – a region where chromatin modifiers impact transcription of these clustered genes. Here we review the importance of location, both specific locations of genes within a cluster, the chromosomal location of the entire cluster itself, and putative epigenetic forces on the genetic regulation of secondary metabolite gene clusters in fungi. We offer a view that secondary metabolite clusters are located in regions of facultative heterochromatin, which can be silenced and activated by both canonical and novel chromatin-mediated mechanisms.
Hallmarks of gene silencing in fungi
Eukaryotic organisms have evolved orchestrated mechanisms to regulate their large gene networks for proper development and appropriate environmental responses. In recent years, much interest has been focused on epigenetic and small RNA regulation of gene expression. Common to all eukaryotes, fungi possess several cellular devices important in gene silencing and activation. Early research in Saccharomyces cerevisiae identified the silent mating type loci (HML/HMR), which subsequently opened the door to an extensive body of work on positional effects in fungi as well as higher eukaryotes [4]. A key finding from the S. cerevisiae work was that exogenous genes were repressed when integrated at the silent mating type loci, thus indicating that the repression was due to positional effects [5]. The mating type switching phenomenon has also been reported in fission yeast, Schizosaccharomyces pombe, where repetitive border elements facilitate the silencing effect [6].
An additional silencing mechanism is termed telomere position effect (TPE). This phenomenon was first reported in yeast and occurs when sub-telomerically located genes are repressed [7]. In fungi, TPE has been demonstrated in S. cerevisiae [8], Sc. pombe [9], Candida glabrata [10], Neurospora crassa [11], and recently, Aspergillus nidulans [12•, Palmer et al., unpublished results]. The extent of TPE is variable at the 32 yeast telomeres [13], but generally extends 20 Kb indicating several hundred genes are regulated by TPE [14].
A commonality in the above instances of positional silencing of gene expression is the involvement of chromatin-level control, commonly termed the histone code, where residues on the histone tails are modified, which in turn results in alterations of chromatin structure [15]. Chromatin can exist in two states: euchromatin is transcriptionally active and characterized by low nucleosome density, while heterochromatin is transcriptionally silent and contains densely packed nucleosomes. Heterochromatin that can become activated under particular circumstances is sometimes referred to as facultative heterochromatin as illustrated by developmentally timed gene expression in Drosophila [16]. Histone tail residues that are hyperacetylated and methylated at lysine 4 of histone 3 (H3K4) are associated with gene transcription and euchromatin, while hypoacetylation and methylation lysine 9 of histone 3 (H3K9) are associated with gene silencing and heterochromatin [17]. These generalities are not rigid, however, as H3K4 methylation is also associated with silencing in yeast sub-telomeric and rDNA regions [18]. A few examples of chromatin-mediated control affecting aspects of development in fungi are listed in Table 1.
Table 1.
Selected examples of chromatin-level control affecting aspects of fungal development
| Developmental aspect: | Organism: | Phenotypic Description: | Reference: |
|---|---|---|---|
| Nitrate and Proline utilization | Aspergillus nidulans |
|
[50,51]. |
|
| |||
| Adhesion | Candida glabrata |
|
[52] |
|
| |||
| Growth and Reproduction Defects | Neurospora crassa |
|
[53,54] |
| Aspergillus fumigatus |
|
[55] | |
Regulation of Secondary Metabolite Gene Clusters in Fungi
An unexpected finding upon inspection of several fungal genomes was the presence of vast numbers of secondary metabolite gene clusters [19]. Although most remain undefined, research on select gene clusters is quite robust and serves to illustrate several important points on the regulation of secondary metabolite gene clusters. The reader is directed to recent reviews detailing non-heterochromatic regulatory mechanisms employed to regulate these clusters [1,19–21]. Briefly, many clusters contain cluster specific transcription factors, often C6 zinc binuclear cluster proteins such as AflR for aflatoxin/sterigmatocystin biosynthesis in Aspergillus spp. [22] or Tri6 for trichothecene biosynthesis in Fusarium spp. [23] that function to activate biosynthetic genes in their respective cluster. Secondary metabolite clusters are also activated, and sometimes shut down, in response to a variety of environmental stimuli that include but are not limited to light, pH, carbon source, nitrogen source, ROS and temperature (Figure 1) [24]. Environmental stimuli are translated to the nucleus through signal transduction cascades, such as the mitogen activating protein kinase cascade (MAPK) and the cAMP mediated PkaA cascade [25–29] and have been linked to activation of specific broad domain regulator factors including CreA (carbon metabolism), AreA (nitrogen metabolism) and PacC (pH sensor) [1].
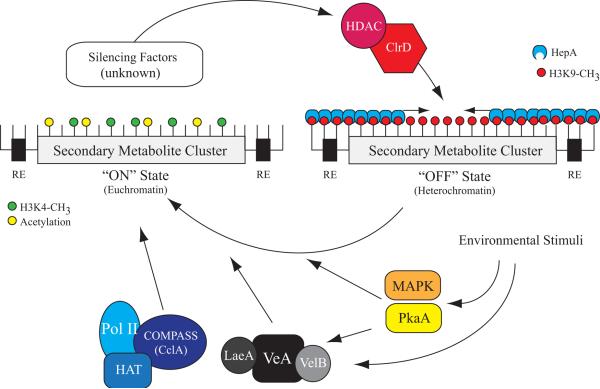
Figure 1. A proposed model for chromatin mediated control of secondary metabolite gene clusters.
Secondary metabolite gene clusters are often flanked by repetitive elements (RE) and located in sub-telomeric regions of the genome. The epigenetic marks of H3K4 methylation (H3K4-CH3) and general histone acetylation have been shown to be associated with active gene transcription [17]. Thus, histone acetyltransferases (HAT) and the H3K4 methylation protein complex (COMPASS) are involved in initiation of transcription through RNA polymerase II (Pol II) [18]. Environmental stimuli are translated by signal transduction cascades, including but not limited to MAPK and PkaA, to trigger production of secondary metabolites [19]. These signals work independently and dependently through the LaeA containing velvet complex [25,26]. On the other hand, in several eukaryotic systems heterochromatin protein 1 has been shown to bind H3K9 methylation (H3K9-CH3) and is associated with gene silencing. In Aspergillus nidulans, null mutants of the H3K9 methyltransferase (ClrD) and heterochromatin protein 1 (HepA) result in derepression of the ST gene cluster [40••]. Currently, the genetic components involved in initiation of heterochromatin at secondary metabolite gene clusters is unknown, RNAi mediated heterochromatin formation could function this way as well as DNA binding repressors.
The first hint that locality of secondary metabolite genes plays a role in their regulation came from characterization of one of the biosynthetic enzymes of the aflatoxin (AF) cluster in A. parasiticus, where localization of the ver-1 gene outside of the AF cluster resulted in 500 fold lower expression than ver-1 located inside the cluster [30]. Similarly, the AF biosynthetic enzyme nor-1 was not expressed when located at two different positions outside of the AF cluster, which led to the conclusion that positional effects are important for expression of AF biosynthetic genes [31]. Insight into a mechanism controlling positional regulation of AF genes came with the discovery of LaeA, a global regulator of secondary metabolism in filamentous fungi [32–35, B. Tudzynksi et al., personal communication]. Recently, LaeA has been shown to be part of the velvet complex, consisting of LaeA-VeA-VelB, that functions to regulate development and secondary metabolism in response to light [36••]. LaeA regulation of gene clusters was found to be location dependent as placement of aflR outside of the sterigmatocystin (ST) cluster removes it from LaeA regulation, and conversely, placement of non-cluster gene in the ST cluster puts it under LaeA control [37].
Although the precise function of LaeA remains enigmatic, several studies link LaeA activity with chromatin modifications. Recent data illustrates that mutations in Aspergillus histone modifying genes activate silent or poorly expressed gene clusters and, significantly, can partially remediate loss of secondary metabolite production in ΔlaeA strains (Table 2). Three deletion mutants that produce increased levels of secondary metabolites target the H3K9 residue including HdaA, a histone deacetylase (HDAC) [38,39], HepA (heterochromatin protein 1) and ClrD (H3K9 methyltransferase) [40••]. The latter two mutations resulted in decreased H3K9 methylation inside ST cluster, which corresponded to increased ST production. In the same study, ChIP analysis showed that secondary metabolite deficient ΔlaeA strains contain increased H3K9 methylation in the ST cluster [40••]. Furthermore, HDAC inhibitors have been reported to increase secondary metabolite production in several fungi [38,41•]. Finally, again supporting a role for chromatin-level control, the order in which AF biosynthetic genes are transcribed mirrors increased histone H4 acetylation patterns in the AF cluster [42•]. While these results confirm that histone modifications are directly linked with secondary metabolite cluster activation, it remains unclear if LaeA directly or indirectly modifies chromatin structure. It has long been speculated that LaeA could directly change chromatin structure through methylation of histones [32,37], however, a substrate for methylation by LaeA remains to be identified.
Table 2.
Genes involved in chromatin-mediated control of secondary metabolism in Aspergillus nidulans
| Gene | Function | Secondary Metabolism Phenotype* | Reference |
|---|---|---|---|
| hepA | Heterochromatin protein 1 | ΔhepA results in increased production of ST | [40••] |
| clrD | H3K9 methyltransferase | ΔclrD results in increased production of ST, partial remediation of ST in ΔlaeA background | [40••] |
| hdaA | Histone deacetylase | ΔhdaA results in increased production of ST and PN. Partial remediation of ST/PN in ΔlaeA background | [38] |
| cclA | H3K4 methyltransferase (part of COMPASS complex) | ΔcclA resulted in production of secondary metabolites from cryptic clusters | [43••] |
| laeA | unknown | ΔlaeA results in loss of several secondary metabolites (ST, PN, TQ) and increased H3K9 methylation in the ST cluster | [37,40] |
ST = sterigmatocystin, PN = penicillin, TQ = terrequinone A
Chromosomal Location of Secondary Metabolite Gene Clusters
As mentioned earlier, methylation of H3K9 is associated with heterochromatin, while methylation of H3K4 is more commonly associated with euchromatin and transcription. However, the COMPASS complex, which methylates H3K4 in yeast, is also associated with homothallic mating type silencing, ribosomal DNA silencing, and sub-telomeric gene expression in this fungus [18]. Paralleling these observations, it was shown that a mutant defective in a component of the COMPASS complex activates silent secondary metabolite clusters in A. nidulans [43••]. These studies led to the discovery of the gene clusters responsible for producing emodin, F9775A/F9775B, and monodictyphenone in addition to shedding light on genome mining techniques leading to discovery of cryptic gene clusters [43••–45]. Moreover, these advances have led to “chemical epigenetic mining” where incorporation of exogenous acetylase/methylase inhibitors or activators have led to identification of novel fungal metabolites [41•,46,47••,48]. These data suggest that cryptic or silent secondary metabolite gene clusters are located in regions of facultative heterochromatin and can be turned on when chromatin structure is changed.
In the human pathogen A. fumigatus, null mutants of LaeA display reduced pathogenicity in murine models of invasive aspergillosis [35,49]. An interesting feature of the LaeA regulon was revealed by microarray analysis in A. fumigatus, which suggested there was a tendency for LaeA regulated secondary metabolite clusters to be located in sub-telomeric regions [3•]. This observation was recently substantiated by expression profiling in A. fumigatus, which revealed sub-telomeric regions, including toxin genes, were highly up regulated when exposed to the murine lung compared to normal laboratory growth [2••]. There is striking overlap between secondary metabolite clusters regulated by LaeA and the sub-telomeric regions differentially regulated upon exposure to the murine model [2••]. Taken together, these data imply that sub-telomeric location of secondary metabolite clusters may be important for their genetic regulation and biological function.
A conserved feature of sub-telomeric DNA sequences, including secondary metabolite gene clusters, is the presence of repetitive elements (RE) composed of active transposable elements or transposon relics. Because active transposons have the potential to be disruptive in the genome, organisms employ complex regulatory mechanisms to limit their expression, such as RNAi mediated heterochromatin formation [6]. A possible role for transposon regulation of a sub-telomeric gene clusters was recently reported for the penicillin (PN) gene cluster [12•]. The PN cluster consists of only three genes and is located ~ 30 Kb from the telomere of chromosome VI. Disruption of large areas of repetitive DNA sequences resulted in mutants producing significantly less PN. Characterization of one area, a 3.7 Kb repeat termed PbIa (penicillin boundary element Ia) containing two transposons/transposon relics, showed its removal decreased PN production, whereas control strains harboring marker gene insertions to either side of PbIa had no effect on PN production. Subsequent trans-complementation experiments were unable to restore PN production. In contrast, deletion of the HDAC HdaA in the ΔPbIa background was able to restore production of PN, suggesting that a transposon mechanism of secondary metabolite cluster expression could involve localized chromatin modifications [12•].
Conclusions
This review highlights work suggestive of epigenetic regulation of secondary metabolite gene clusters in filamentous fungi. Recently there has been an increase in the number of examples of gene cluster regulation mediated by chromatin remodeling enzymes, including chemical epigenetic approaches. These studies reveal the importance of positional effects, both location effects within a cluster and chromosomal location effects on cluster regulation. Future studies are warranted to tease out the molecular mechanisms of epigenetic regulation. Interesting questions remain to be answered: Which happens first - chromatin remodeling leading to transcription factor activation or transcription factor binding leading to chromatin remodeling? Does RNAi have a role in chromatin-mediated regulation of secondary metabolism? What role do repetitive elements that flank gene clusters have in regulation? Does LaeA directly or indirectly modify chromatin structure?
Acknowledgements
This work was in part funded by the National Institute of Health (1R01 AI065728-01) to NPK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Yu JH, Keller NP. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol. 2005;43:437–458. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]
- 2••.McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008;4:e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study analyzes the transcriptome of the pathogenic mold Aspergillus fumigatus during invasion of the murine lung. Up regulated transcripts inside the lung were biased towards genes located in the sub-telomere, of which several were secondary metabolite gene clusters. This paper describes a strong overlap between LaeA regulated secondary metabolite gene clusters and transcripts during colonization of an animal host.
- 3•.Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]; Microarray analysis in Aspergillus fumigatus indicated that LaeA regulates ~ 50% of secondary metabolite gene clusters and highlighted a strong tendency for LaeA regulated clusters to be located in sub-telomeric regions.
- 4.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 5.Schnell R, Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:1–8. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doheny JG, Mottus R, Grigliatti TA. Telomeric position effect--a third silencing mechanism in eukaryotes. PLoS One. 2008;3:e3864. doi: 10.1371/journal.pone.0003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tham W-H, Zakian VA. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene. 2002;21:512–521. doi: 10.1038/sj.onc.1205078. [DOI] [PubMed] [Google Scholar]
- 9.Nimmo ER, Cranston G, Allshire RC. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. Embo J. 1994;13:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castaño I, Pan S-J, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith KM, Kothe GO, Matsen CB, Khlafallah TK, Adhvaryu KK, Hemphill M, Freitag M, Motamedi MR, Selker EU. The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenetics Chromatin. 2008;1:5. doi: 10.1186/1756-8935-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Shaaban M, Palmer J, El-Naggar WA, El-Sokkary MA, Habib E-SE, Keller NP. Involvement of transposon-like elements inpenicillin gene cluster regulation. Fungal Genet Biol. 2010 doi: 10.1016/j.fgb.2010.02.006. doi:10.1016/j.fgb.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Secondary metabolite gene clusters are often flanked by repetitive DNA sequences. This recent study identifies a role for repetitive elements surrounding the penicillin gene cluster. Specifically, removal of repetitive DNA on either side of the cluster resulted in a reduction in penicillin production.
- 13.Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 15.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 16.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 18.Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Natural Product Reports. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 20.Fox EM, Howlett BJ. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol. 2008;11:481–487. doi: 10.1016/j.mib.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Georgianna DR, Payne GA. Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet Biol. 2009;46:113–125. doi: 10.1016/j.fgb.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes M, Keller NP, Adams TH. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol Microbiol. 1998;28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 23.Proctor RH, Hohn TM, McCormick SP, Desjardins AE. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl Environ Microbiol. 1995;61:1923–1930. doi: 10.1128/aem.61.5.1923-1930.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atoui A, Bao D, Kaur N, Grayburn WS, Calvo AM. Aspergillus nidulans natural product biosynthesis is regulated by mpkB, a putative pheromone response mitogen-activated protein kinase. Appl Environ Microb. 2008;74:3596–3600. doi: 10.1128/AEM.02842-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayram O, Sari F, Braus GH, Irniger S. The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol Microbiol. 2009;71:1278–1295. doi: 10.1111/j.1365-2958.2009.06606.x. [DOI] [PubMed] [Google Scholar]
- 27.Roze LV, Beaudry RM, Keller NP, Linz JE. Regulation of aflatoxin synthesis by FadA/cAMP/protein kinase A signaling in Aspergillus parasiticus. Mycopathologia. 2004;158:219–232. doi: 10.1023/b:myco.0000041841.71648.6e. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu K, Hicks JK, Huang TP, Keller NP. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics. 2003;165:1095–1104. doi: 10.1093/genetics/165.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tag A, Hicks JK, Garifullina G, Ake C, Phillips TD, Beremand M, Keller NP. G-protein signalling mediates differential production of toxic secondary metabolites. Mol Microbiol. 2000;38:658–665. doi: 10.1046/j.1365-2958.2000.02166.x. [DOI] [PubMed] [Google Scholar]
- 30.Liang SH, Wu TS, Lee R, Chu FS, Linz JE. Analysis of mechanisms regulating expression of the ver-1 gene, involved in aflatoxin biosynthesis. Appl Environ Microbiol. 1997;63:1058–1065. doi: 10.1128/aem.63.3.1058-1065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiou C-H, Miller M, Wilson DL, Trail F, Linz JE. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl Environ Microbiol. 2002;68:306–315. doi: 10.1128/AEM.68.1.306-315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kale SP, Milde L, Trapp MK, Frisvad JC, Keller NP, Bok JW. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet Biol. 2008;45:1422–1429. doi: 10.1016/j.fgb.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosalková K, García-Estrada C, Ullán RV, Godio RP, Feltrer R, Teijeira F, Mauriz E, Martín JF. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie. 2009;91:214–225. doi: 10.1016/j.biochi.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Sugui JA, Pardo J, Chang YC, Müllbacher A, Zarember KA, Galvez EM, Brinster L, Zerfas P, Gallin JI, Simon MM, et al. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot Cell. 2007;6:1552–1561. doi: 10.1128/EC.00140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]; Identification of the velvet complex, consisting of LaeA-VeA-VelB, in Aspergillus nidulans, which functions to control development and secondary metabolism in response to light. The complex has since been reported to be conserved in other fungi and represents a regulatory system that appears to be unique to filamentous fungi.
- 37.Bok JW, Noordermeer D, Kale SP, Keller NP. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol Microbiol. 2006;61:1636–1645. doi: 10.1111/j.1365-2958.2006.05330.x. [DOI] [PubMed] [Google Scholar]
- 38.Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee I, Oh J, Keats Shwab E, Dagenais T, Andes D, Keller N. HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet Biol. 2009;46:782–790. doi: 10.1016/j.fgb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07051.x. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; H3K9 methylation and subsequent recruitment of heterochromatin protein 1 is associated with heterochromatin. Study of null mutants of the H3K9 methyltransferase (ClrD) and HP1 (HepA) orthologs in A. nidulans revealed that regulation of ST was altered. Subsequent ChIP analysis of the ST cluster shows that there is increased H3K9 methylation in ΔlaeA strains, indicating that chromatin-mediated regulation of the gene cluster. Additionally, ClrD and HepA were able to partially remediate the ST deficiency of ΔlaeA strains.
- 41•.Cichewicz RH. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Natural Product Reports. 2010;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]; An excellent review on the use of epigenetics in discovering novel metabolites from fungi.
- 42•.Roze LV, Arthur AE, Hong SY, Chanda A, Linz JE. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol Microbiol. 2007;66:713–726. doi: 10.1111/j.1365-2958.2007.05952.x. [DOI] [PubMed] [Google Scholar]; This study indicated that acetylation of histone H4 mirrored the transcriptional patterns of the aflatoxin gene cluster. These data suggest that aflatoxin is at least partially regulated an epigenetic mechanism.
- 43••.Bok JW, Chiang Y-M, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of cryptic secondary metabolites in A. nidulans produced by mutant of the COMPASS complex. COMPASS functions to methylate H3K4 and this study indicates that secondary metabolite gene clusters reside in areas of heterochromatin and by altering chromatin structure, different secondary metabolite clusters are activated.
- 44.Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Elizabeth Oakley C, Woo Bok J, Keller N, Oakley BR, Wang CCC. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2010;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang Y-M, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CCC, Oakley BR. Genetic characterization of the monodictyphenone gene cluster in Aspergillus nidulans. Appl Environ Microbiol. 2010 doi: 10.1128/AEM.02187-09. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Wägele H, Cichewicz RH. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J Ind Microbiol Biotechnol. 2009;36:1199–1213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- 47••.Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH. A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Org Biomol Chem. 2009;7:435–438. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]; This study was the first to utilize chemical epigenetics to identify a new secondary metabolite gene cluster, thereby shedding light on a relatively simple approach to identify secondary metabolites from fungi where genome sequences are not available.
- 48.Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- 49.Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Keller NP. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell. 2005;4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berger H, Basheer A, Böck S, Reyes-Dominguez Y, Dalik T, Altmann F, Strauss J. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol Microbiol. 2008;69:1385–1398. doi: 10.1111/j.1365-2958.2008.06359.x. [DOI] [PubMed] [Google Scholar]
- 51.Reyes-Dominguez Y, Narendja F, Berger H, Gallmetzer A, Fernandez-Martin R, Garcia I, Scazzocchio C, Strauss J. Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryot Cell. 2008;7:656–663. doi: 10.1128/EC.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol. 2005;8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 53.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 54.Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU. HP1 is essential for DNA methylation in Neurospora. Mol Cell. 2004;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- 55.Palmer JM, Perrin RM, Dagenais TR, Keller NP. H3K9 methylation regulates growth and development in Aspergillus fumigatus. Eukaryot Cell. 2008;7:2052–2060. doi: 10.1128/EC.00224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]