Abstract
The endosomal sorting complexes required for transport (ESCRTs) catalyze one of the most unusual membrane remodelling events in cell biology. ESCRT-I and ESCRT-II direct membrane budding away from the cytosol by stabilizing bud necks without coating the bud and without being consumed in the buds. ESCRT-III cleaves the bud necks from their cytosolic face. ESCRT-III-mediated membrane neck cleavage is crucial for many processes, including the biogenesis of multivesicular bodies, viral budding, cytokinesis, and probably autophagy. Recent studies of ultrastructures induced by ESCRT-III overexpression in cells and the in vitro reconstitution of the budding and scission reactions have led to breakthroughs in understanding these remarkable membrane reactions.
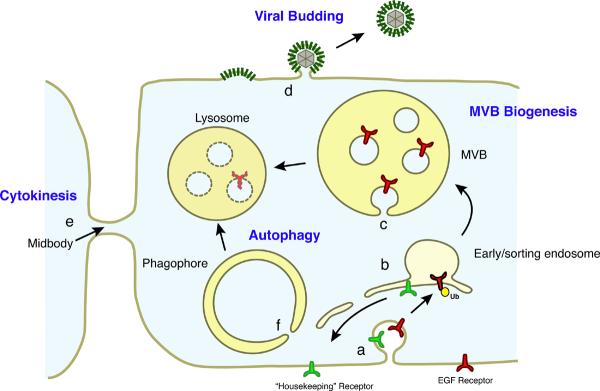
Understanding how membranes delimit and organize cell functions is one of the main challenges in cell biology. Membrane-bound proteins are actively sorted to different points in the cell through tightly choreographed processes, many of which centre on the early endosome (Fig. 1). Some proteins, including housekeeping receptors, are typically recycled back to their points of action in the plasma membrane or Golgi. Others, such as the signalling epidermal growth factor receptor (EGFR) and lysosomal hydrolases, are sorted to the lysosome1. This endolysosomal sorting is mediated by budding portions of the endosome's limiting membrane into the lumen of the endosome. These membrane buds are cleaved through a membrane scission reaction to form intralumenal vesicles (ILVs) within the endosome. Late endosomes filled with ILVs are referred to as multivesicular bodies (MVBs) or sometimes multivesicular endosomes2–4. Once formed, MVBs fuse with one another and the lysosome in a process directed by the HOPS and CORVET complexes5 and their contents are degraded.
Figure 1.
Biological roles of the ESCRTs. a. Plasma membrane proteins such as EGFR are ubiquitylated and endocytosed following their stimulation. The initial endocytosis of ubiquitylated receptors does not require the ESCRTs. b. The early endosome is a branch point from which housekeeping receptors and other cargoes not bound for the lysosome are recycled, again not requiring the ESCRTs. c. MVB biogenesis involves the formation of membrane buds, the sorting of ubiquitylated cargo into the buds and cleavage of the buds to form ILVs. MVB biogenesis requires all of the ESCRT complexes. d. HIV-1 budding needs ESCRT-I, ESCRT-III, VPS4 and, to a lesser extent, ALIX. Other ESCRT-dependent viruses have varying requirements for specific components of the ESCRT machinery, but all require ESCRT-III and VPS4. e. Cytokinesis in animal cells requires ESCRT-I, ESCRT-III, VPS4 and ALIX, which have been shown to localize to the midbody during cytokinesis in the appropriate time and place to carry out the final cleavage of the membrane neck, separating the two daughter cells. Cytokinesis in the Crenarchaea requires homologues of ESCRT-III and Vps4. f. Autophagy requires all of the ESCRT complexes, similar to MVB biogenesis, although it is not clear whether their roles in autophagy are direct or indirect. There is an intriguing analogy between the closure of the phagophore neck and the formation of ILVs, making it tempting to speculate that the ESCRTs are involved in closing this neck. However the presence of ESCRTs at the phagophore neck has never been visualized.
Because budding in the endolysosomal pathway is directed away from the cytosol, it differs fundamentally from well-studied conventional vesicle budding reactions that are directed into the cytosol. Many such reactions are directed by a complex but conceptually straightforward mechanism in which coat proteins impose curvature on the cytosolic face of the bud. The best-known of these coat proteins are clathrin and the COP complexes, which cooperate with scission factors to cleave membrane tubes from the outside and thereby release vesicles into the cytosol. Dynamin, the most-studied scission factor, achieves membrane scission in an intuitively clear (but still debated) reaction6. The formation of ILVs requires a fundamentally different machinery in which cytosolic factors promote the formation and cleavage of buds and the necks connecting them to the limiting membrane from the inside of the membrane neck.
Studies in the yeast Saccharomyces cerevisiae have been integral for identifying the factors involved in forming MVBs in the endosomal pathway. In yeast, the equivalent of the mammalian lysosome is the vacuole, the biogenesis of which is linked to that of the MVBs and is regulated by vacuolar protein sorting (VPS) genes. A subset of the VPS genes, known as class E genes, encode proteins that are directly involved in MVB biogenesis7. Indeed, deleting any of these genes leads to the formation of an abnormal multicisternal endosome lacking internal vesicles, referred to as a class E compartment. Many of these proteins are core subunits of four complexes that have been dubbed the endosomal sorting complexes required for transport (ESCRTs) and include ESCRT-0, ESCRT-I, ESCRT-II and ESCRT -III (Table 1)8–10. The rest of the class E VPS proteins include the Vps4 AAA+ ATPase, which provides the only direct energy input to the pathway by hydrolysing ATP, regulatory proteins that modulate ESCRT disassembly and the multifunctional ESCRT-associated protein Bro1 (known in metazoa as ALIX).
Table 1.
ESCRT complex subunits and associated proteins
| Complex | yeast proteins | metazoan protein* | Motifs and Domains | Activity of ESCRT Complex | Biochemical Role(s) of Individual Protein Components | Biological Role(s) |
|---|---|---|---|---|---|---|
| ESCRT-0 | Vps27 | Hrs (Hgs) | VHS, FYVE, UIM (yeast), DUIM (metazoan), PTAP, GAT domain and coiled-coil core, clathrin-binding | Clustering of ubiquitinated cargo | Binds PI(3)P-containing membranes, ESCRT-I, Ub, clathrin | MVB biogenesis Autophagy |
| ESCRT-0 | Hse1 | STAM1, 2 | VHS, UIM, SH3, GAT domain and coiled-coil core | Clustering of ubiquitinated cargo | Binds Ub, deubiquitinating enzymes | MVB biogenesis Autophagy |
| ESCRT-I | Vps23 | VPS23 (TSG101) | UEV, proline-rich linker, stalk, headpiece | Membrane budding (with ESCRT-II) | Binds Ub and ESCRT-0 in MVB pathway Binds PTAP motifs in viral Gag proteins Adaptor for midbody protein CEP55 |
MVB biogenesis Autophagy Viral budding Cytokinesis |
| ESCRT-I | Vps28 | VPS28 | headpiece, Vps28 CTD | Membrane budding (with ESCRT-II) | ESCRT-II binding | MVB biogenesis Autophagy Viral budding Cytokinesis |
| ESCRT-I | Vps37 | VPS37A,B,C,D | basic helix, stalk, headpiece | Membrane budding (with ESCRT-II) | Membrane binding; mammalian VPS37 binds ESCRT-III protein IST1 | MVB biogenesis Autophagy Viral budding Cytokinesis |
| ESCRT-I | Mvb12 | MVB12A,B | Stalk, Ub-binding C-terminus | Membrane budding (with ESCRT-II) | Needed to stabilize ESCRT-I oligomer, contacts each of other subunits; C-terminus binds Ub | MVB biogenesis Autophagy Viral budding Cytokinesis |
| ESCRT-II | Vps22 | VPS22 (EAP30) | basic helix, two winged-helix domains | Membrane budding (with ESCRT-I) | Membrane binding | MVB biogenesis Autophagy |
| ESCRT-II | Vps25 | VPS25 (EAP20) | Two winged-helix domains | Membrane budding (with ESCRT-I) | Binds to ESCRT-III (Vps20) to recruit it to bud neck and initiate its activity | MVB biogenesis Autophagy |
| ESCRT-II | Vps36 | VPS36 (EAP45) | GLUE, NZF1 (yeast), NZF2 (yeast), two winged helix domains | Membrane budding (with ESCRT-I) | Binds phosphoinositide-containing membranes, Ub, and ESCRT-I binding | MVB biogenesis Autophagy |
| ESCRT-III | Vps20 | VPS20 (CHMP6) | Myristoylated; MIM2 | Membrane scission | Initiates ESCRT-III assembly in MVB biogenesis by binding Snf7, initiates membrane scission, MIM2 binds Vps4 for subsequent disassembly | MVB biogenesis Autophagy |
| ESCRT-III | SNF7 (Vps32) | SNF7A (CHMP4A), SNFB (CHMP4B), SNFC (CHMP4C) | MIM2 | Membrane scission | Main driver of membrane scission Recruitment of deubiquitinating enzyme (yeast) Binds Bro1 domain containing-proteins including Bro1/ALIX |
MVB biogenesis Autophagy Viral budding Cytokinesis |
| ESCRT-III | Vps24 | VPS24 (CHMP3) | MIM1 | Membrane scission | Completion of membrane scission Recruitment of AMSH deubiquitinating enzyme (mammals) |
MVB biogenesis Autophagy Viral budding Cytokinesis |
| ESCRT-III | Vps2 | VPS2A (CHMP2A), VPS2B (CHMP2B) | MIM1 | Membrane scission | Recruitment of Vps4 and initiation of ESCRT-III disassembly | MVB biogenesis Autophagy Viral budding Cytokinesis |
| ESCRT-III related | Did2 (Vps46) | DID2A (CHMP1A), DID2B (CHMP1B) | MIM1 | Regulating membrane scission and ESCRT-III disassembly | Recruitment of Vps4 Recruitment of microtubule severing enzyme spastin (mammalian DID2B) |
MVB biogenesis Viral budding Cytokinesis |
| ESCRT-III related | Vps60 | CHMP5 | None defined | Regulating membrane scission and ESCRT-III disassembly | Binds Vta1 with high affinity | MVB biogenesis Cytokinesis |
| ESCRT-III related | Ist1 | IST1 | MIM1, MIM2 | Regulating membrane scission and ESCRT-III disassembly | Binds Vps4, Vta1, Did2, ESCRT-I | MVB biogenesis Cytokinesis |
| ESCRT-III related | None | CHMP7 | MIM1, MIM2 | Regulating membrane scission and ESCRT-III disassembly (putative) | Tandem ESCRT-III domains, binds SNF7B and deubiquitinating enzyme UBPY | MVB biogenesis Viral budding |
| Vps4-Vta1 | Vps4 | VPS4A, VPS4B (SKD1) | MIT, AAA | ESCRT-III disassembly | AAA+ ATPase, acts as assembled oligomer on ESCRT-III | MVB biogenesis Autophagy Viral budding Cytokinesis |
| Vps4-Vta1 | Vta1 | VTA1 (LIP5) | MIT, VSL | ESCRT-III disassembly | Binds Vps4 to promote assembly of oligomer and ESCRT-III recycling; also binds MIM motifs | MVB biogenesis Autophagy Viral budding Cytokinesis |
| Other | Bro1 (Vps31) | ALIX (AIP1) | Bro1, V, PRD | Multiple modulatory and targeting functions | Recruitment of ESCRT-III (Snf7) for scission Recruitment of deubiquitinating enzymes Adaptor for midbody protein CEP55 Adaptor for viral YPXL motifs ALIX has many other interactions with apoptosis factors and cytoskeleton with currently unclear significance for ESCRT pathway |
MVB biogenesis Viral budding Cytokinesis |
Alternative names are provided in brackets.
In addition to MVB biogenesis, the ESCRTs are required for cytokinesis11–13, budding of HIV-1 and other enveloped viruses14,15 and macroautophagy16,17 (Fig. 1). All of these pathways involve the cleavage of membrane necks with the same unconventional topology as in MVB biogenesis (discussed below).
Until 2 years ago, the mechanisms of membrane remodelling by the ESCRTs were almost purely a matter of conjecture. Step by step analysis of ESCRT function using classical approaches in vivo was hampered by the fact that deletion of any of the required pathway components induces the formation of aberrant class E compartments, which are devoid of internal vesicles18–20. Furthermore, the structure of the class E compartment is the same, regardless of whether it is induced by deletion of genes with primary roles in cargo sorting, membrane budding, membrane scission or ESCRT recycling. However, recent in vitro studies have provided new insight into the function of each of the major ESCRT complexes and moved forward our thinking about the underlying mechanisms of membrane remodelling. Particularly important advances have come from electron microscopic analysis of membrane tubules formed by overexpressed or purified ESCRTIII subunits21–24, reconstitution of budding and scission in giant unilamellar vesicles (GUVs)25–27 and computational analyses28,29.
In this Review we describe the mechanisms of membrane deformation, budding and scission associated with the ESCRTs. The reader is directed elsewhere for excellent reviews of the cell biology30–32, biochemistry and structural biology33–36 of these proteins. We organize our discussion around the steps required to deform and then remodel the membrane to create lumenal vesicles within the MVB. Although vesicle formation in MVB biogenesis is at least in part initiated by ubiquitin moieties attached to cargo molecules, ubiquitin is not strictly required for the mechanics of budding into MVBs and has not been shown to be crucial for neck cleavage in any of the ESCRT-dependent pathways. The central role of ubiquitin in directing cargo into the ESCRT pathway has recently been reviewed elsewhere37 and so will not be considered in detail here.
Organization of the ESCRT complexes
ESCRT-I and ESCRT-II are directly involved in membrane budding, while ESCRT-III is responsible for membrane scission. The structural organization of these three complexes, which of all the ESCRTs and ESCRT-associated proteins are the most germane to the focus of the review, is described in detail below.
ESCRT-I
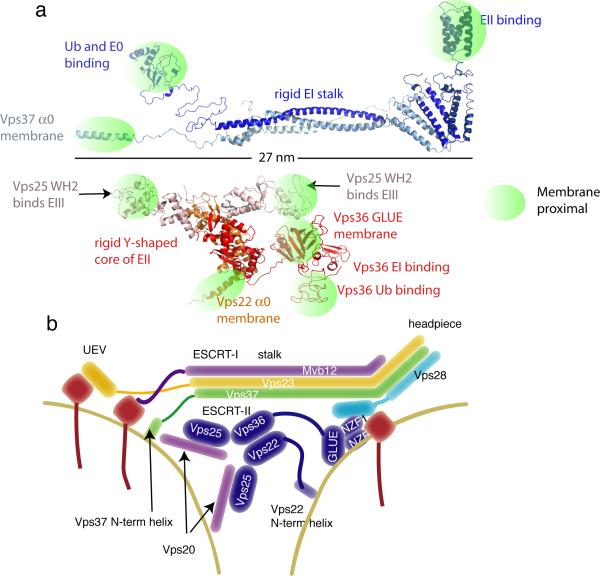
ESCRT-I is a constitutively assembled heterotetramer consisting of one copy each of the subunits Vps23, Vps28, Vps37 and Mvb12 (Fig. 2a,b)38–41. The complex is centred around a 13 nm-long stalk and a 5-nm long headpiece41. Projecting from the stalk corresponding to the N-termini of Vps23 and Vps37 is the region that contains the UEV domain of Vps23, which binds to ESCRT-042–44, the PTAP motifs of HIV-1 Gag and certain other viral proteins45, and to ubiquitylated cargo10,46,47. A short carboxy-terminal ubiquitin-binding domain in Mvb12 is also found at this end of the stalk48. Also at the same end, the basic amino-terminal helix of Vps37 is involved in electrostatic interactions with acidic membrane lipids41, and the linker between the UEV domain and the stalk contains a GPPXXY motif that targets ESCRT-I to midbodies during cytokinesis12,13,49. At the opposite end, a C-terminal helical domain of Vps28 projects to bind to the ESCRT-II complex50–52.
Figure 2.
ESCRT-I and ESCRT-II. a. Composite structures of ESCRT-I and II modelled on the basis of crystal structures of the core complexes and flexibly attached domains. Regions with known functions that bring them close to the membrane are highlighted to show that these regions are concentrated at either end of the ESCRT-I stalk and at the three tips of the Y-shaped ESCRT-II scaffold. Above the scale bar at left, Ub and ESCRT-0 bind to the UEV domain of ESCRT-I, and the N-terminal basic helix of ESCRT-I contributes to membrane binding. Above the scale bar at right, the C-terminal domain of Vps28 binds to ESCRT-II, at least in yeast. Below the scale bar at left and right, the WH2 domains of both molecules of Vps25 bind to the ESCRT-III subunit Vps20. At bottom center, the basic helix 0 of Vps22 and the GLUE domain of Vps36 contribute to membrane binding, and in yeast, the NZF inserts into the GLUE domain bind to ubiquitin and ESCRT-I. b. Schematic version of the structures.
ESCRT-II
ESCRT-II is also a constitutively assembled heterotetramer, each copy of which contains one molecule of Vps22 and Vps36 and two molecules of Vps25. The complex has a rigid Y-shaped core, and the overall structure has a maximum dimension of ~15 nm53–55 (Fig. 2a,b). Vps22 and Vps36 have an intimate and extensive interface, and the two copies of Vps25 associate with the Vps22–Vps36 subcore through smaller interfaces. Both copies of Vps25 are essential for ESCRT-II function53,56. A short PPXY motif at the N-terminus of the first winged helix (WH) motif of Vps25 binds to structurally analogous pockets in the C-terminal WH motifs of Vps22 and Vps36. The C-terminal WH domains of Vps25 bind to, recruit and activate the most upstream ESCRT-III subunit, Vps2026,54,57. The N-terminus of Vps22 contains a basic helix that interacts non-specifically with acidic lipids and is important for membrane recruitment of ESCRT-II55. The N-terminus of yeast Vps36 is complex. Yeast Vps36 contains a variant PH domain known as a “GLUE domain”, into the sequence of which two Npl4-type zinc finger domains (NZF) have been inserted52. In yeast Vps36, the GLUE domain binds to 3-phosphoinositides52, the NZF1 binds to ESCRT-I51, and NZF2 binds to ubiquitylated cargo58. Human VPS36 has a simpler architecture, in which the GLUE domain has no insertions and binds directly to both 3-phosphoinositides and ubiquitin59–61.
ESCRT-III
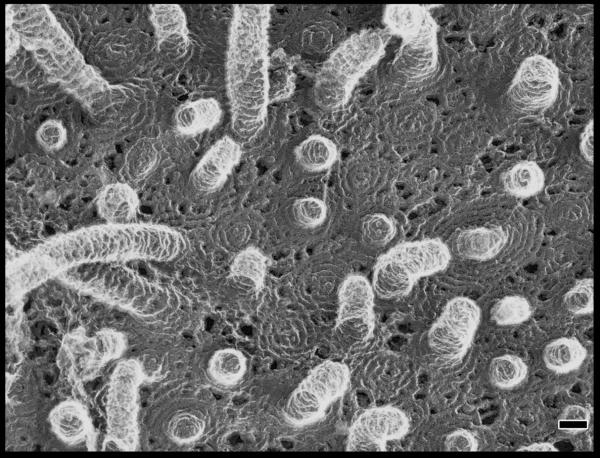
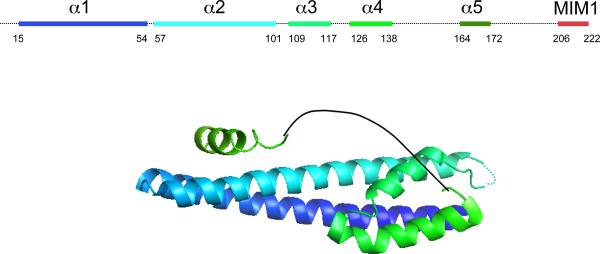
Unlike the other ESCRT complexes that are stable heteropolymers, ESCRT-III is a dynamic polymer of ESCRT-III proteins that does not have a clearly defined or unique composition. In yeast ESCRT-III consists of four core subunits, Vps20, Snf7, Vps24 and Vps29, and three peripheral or regulatory subunits, Did2, Vps60 and Ist1 (Table 1). This list of ESCRT-III subunits expands to include twelve proteins in humans. The human proteins include three isoforms of SNF7 and two each of DID2 and VPS2. In yeast, all seven proteins seem to work together in MVB biogenesis, and the various isoforms of the human proteins may function similarly and form alternative complexes. Studies of ESCRT-III complex in yeast suggest a model in which Vps20 initiates polymer assembly, Snf7 extends the polymer and is its major constituent, and Vps24 and Vps2 act to cap the polymer, thereby limiting its size62. The three peripheral subunits seem to assemble later than the four core subunits and may play a part in regulating complex disassembly. A representative ESCRT-III polymer created by overexpressing human SNF7 together with an inactive mutant of VPS4B in cultured cells is shown in Fig. 3a. All ESCRT-III proteins share a common overall architecture; representative crystal structures show that they are a 7 nm-long α-helical hairpin buttressed by several short helices (Fig. 3b)24,63,64. ESCRT-III proteins also have the common behaviour of cycling between a closed monomeric state in the cytosol and an open polymerized state on the endosomal membrane (see below)65,66 (Fig. 3c).
Figure 3.
ESCRT-III. a. Assembled ESCRT-III polymer is visualized in this electron micrograph showing the filaments underlying buds and tubules emerging from the top of a cell overexpressing human SNF7 and an inactive mutant of VPS4B. Image is reproduced, with permission, from21. The SNF7-containing filaments are seen because of post-fixation extraction with detergent. b. Helical organization of individual ESCRT-III proteins based on the observed structure of human VPS2433,58. Helices α1– α4 form the core of the protein responsible for membrane binding and polymer assembly and are regulated by autoinhibitory sequences in α5 and the C-terminal MIM1 (not resolved in the CHMP3 structure). All ESCRT-III proteins contain the α1– α4 core and the α5 autoinhibitory sequence. Vps2, Did2, and Vps24 contain a C-terminal MIM1 motif, whereas Vps20 and Snf7 contain a C-terminal MIM2 motif. Ist1 has both MIM1 and MIM2 motifs at its C-terminus. Both types of MIM bind to the MIT domain of Vps4, but they do so at separate sites and do not compete with one another. c. Cycle of ESCRT-III assembly and VPS4-mediated disassembly. ESCRT-III proteins are in a `closed' and monomeric conformation in the cytosol, with intramolecular autoinhibitory interactions preventing membrane binding and polymer assembly. In their `open' conformation, these autoinhibitory interactions are released, and the subunits bind to each other and to the membrane as they assemble into functional ESCRT-III polymers.
-This could be redrawn with colors that fit better w/ rest of figs
Assembly of the ESCRT-III polymer is energetically favourable but also highly regulated. The closed cytosolic conformation of each ESCRT-III protein is maintained by intramolecular autoinhibitory interactions in which sequences in the C-terminal α5 and α6 helices hide residues along the hairpin surface, which are responsible for membrane binding and polymer assembly. Once the ESCRT-III proteins bind the membrane, interactions between the subunits require hydrophobic residues near the tip of the helical hairpin24,63,64. Analysis of variability in crystal contacts in various ESCRT-III structures and a low resolution cryo EM reconstruction have led to some reasonable inferences about which contacts are important in polymer assembly, but these need to be confirmed by higher resolution EM analysis and additional functional corroboration.
Membrane budding
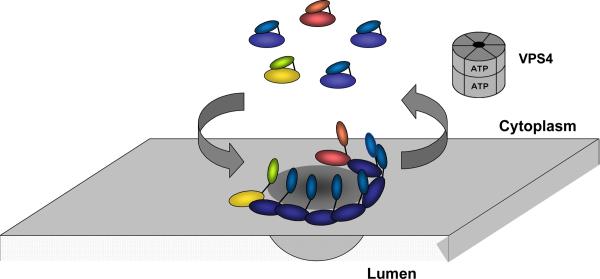
The first step in creating a lumenal vesicle is to deform the membrane into a bud. In this Review, we use the term bud to describe a nascent vesicle (or virion) that it still attached to the limiting membrane — in other words, prior to scission — and the term ILV to refer to a detached vesicle subsequent to membrane scission. In vitro analyses using purified proteins and GUVs show that large buds can be formed in vitro by recruiting either ESCRT-I and ESCRT-II together27, or the Vps20 and Snf7 subunits of ESCRT-III25,57, to an appropriate membrane surface. ESCRT-I and ESCRT-II colocalize almost exclusively to the necks of such buds and seem to act by stabilizing the bud neck (Fig. 4a, b). ESCRT-I and ESCRT-II are extremely potent inducers of bud formation and are effective even at concentrations of 15 nM, below their estimated concentrations of ~75 nM in yeast cells. The ESCRT-III subunits Vps20 and Snf7 can also induce bud formation, but only at superphysiological concentrations (600 nM for Snf7). When ESCRT-I and ESCRT-II induce bud formation in vitro, ESCRT-III subunits are recruited only to the bud necks and are not substantially present in the lumen, consistent with physiology. However, Vps20- and Snf7-induced buds contain ESCRTs in the lumen of the bud, in contrast to the physiological situation in which ESCRTs are recycled, and therefore not concentrated in buds. The mechanism by which ESCRT-III subunits proceed to carry out scission at the necks of ESCRT-I- and ESCRT-II-induced buds is discussed in detail below.
Figure 4.
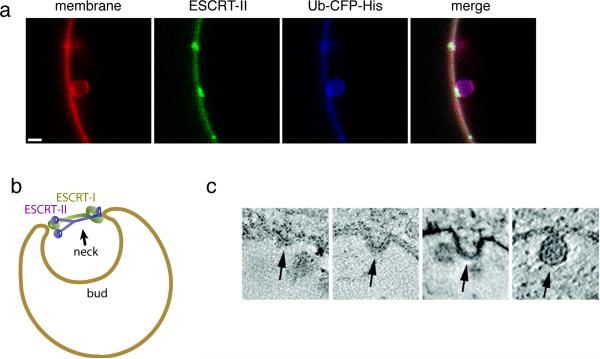
Membrane budding by ESCRTs a. Buds induced by ESCRT-I and II. Image reproduced, with permission, from27. The first panel at left shows the membrane labelled with rhodamine-PE. The second and fourth panels show that ESCRT-II (Alexa-488) is localized exclusively to the neck of the bud. The third panel shows that ubiquitin (CFP) is localized throughout the bud, but with some additional concentration at the neck coinciding with ESCRT-II. The same has been found for ESCRT-I, Vps20 and Snf7 (not shown). The scale bar is 2 microns. b. Model for bud stabilization by ESCRT-I and II. In this model, the various membrane-proximal domains of ESCRT-I and –II as shown in Fig. 2 bind to one side of the rim of the bud neck. The rigid stalk of ESCRT-I and the rigid Y-shaped core of ESCRT-II stabilize the opening of the bud neck. c. Putative pathway of bud formation in vivo, derived from a series of images of fixed, negatively stained cells, and arranged into a putative time series, reproduced by permission from67.
Recent in vitro studies support the idea that ESCRT-I and ESCRT-II play the central part in membrane budding. Physiological concentrations of ESCRT-I and ESCRT-II, but not ESCRT-III, drive bud formation in reconstituted reactions in GUVs. In these in vitro studies buds formed by ESCRT-I and ESCRT-II on the GUVs, but not those formed by ESCRT-III, exclude ESCRTs from the lumen on the bud, consistent with physiology. Finally, bud necks formed by ESCRT-I and ESCRT-II have the ability to recruit ESCRT-III, which in this setting then cleaves the membrane necks with the appropriate topology (that is, from the surface contiguous with the inside of the membrane neck). Importantly, however, all of these complexes share a propensity to localize at and stabilize membrane necks. Structures of neck-like assemblies of ESCRTIII have been visualized by EM21,22 (Fig. 3a). However, the structure of the ESCRT-I and ESCRT-II membrane neck assembly is currently unknown; elucidating these structures will be crucial to further understand the pathway and mechanism by which ESCRTs promote membrane budding.
Bud formation by ESCRT-I and ESCRT-II involves the docking of the interacting motifs of the two complexes51 such that in an ESCRT-I-ESCRT-II supercomplex, the membrane binding sites on the two complexes are rigidly separated by at least 18 nm. The dimensions of the physiological bud neck are not well characterized, as these structures are short-lived during the normal budding process in cells and are rarely detected by EM67. Although it seems that buds begin with wide-necked membrane invaginations (Fig. 4c), there is currently limited experimental information on the pathway of budding. In any case, the bud neck diameter can reasonably be expected to be equal to or smaller than the diameter of the final ILV, ~25 nm in yeast and ~50 nm in human cells. This suggests a potential mechanism for stabilizing the formation of a bud neck of these dimensions (Fig. 4b), in that the length of the complexes is similar to the likely initial dimensions of the neck. Clearly the kinetic mechanism and energy landscape of the budding trajectory will require further analysis, and much remains to learned about the mechanism underlying bud formation.
Membrane scission
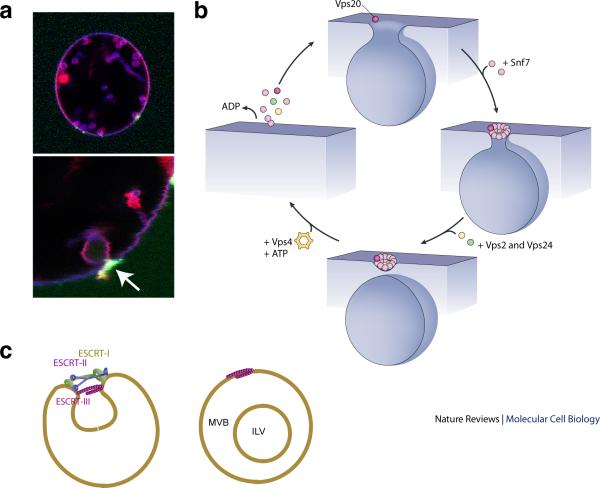
Once a bud is created, its separation from the parent membrane requires scission. The ESCRT-III complex catalyzes the scission of membrane necks25. The scission reaction occurring in yeast MVB biogenesis62 and reconstituted in GUVs25 involves the assembly of Vps20, Snf7 and Vps24, in that order and in an loosely defined ratio, at the membrane neck27 (Fig. 5a). Of all the ESCRT-III subunits, only Snf7, the most abundant ESCRT-III protein in yeast, is absolutely required for severing. Vps20 is present at lower concentrations than Snf7 and couples ESCRT-II to ESCRT-III26,54,56,57. The coupling of ESCRT-II to Vps20 is required for MVB biogenesis, but it does not seem to be required for ESCRT-II-independent pathways such as HIV-1 budding and cytokinesis (see below)13,68,69. Vps24 also operates at low concentrations, and its omission sharply slows but does not completely block scission25.
Figure 5.
Membrane scission by ESCRT-III. a. Imaging of Snf7 in the act of membrane scission at a bud neck in vitro, reproduced by permission from27. The merged image at top combines red (membrane, rhodamine), blue (ubiquitylated cargo, CFP) and green (Snf7, Alexa 488) channels. The lower panel shows a close-up of the membrane bud, with the arrow highlighting the localization of Snf7 to the bud neck. ESCRT-I, -II, and Vps20 are present but not labeled in this experiment. b. Cartoon for scission, inspired by spiral SNF7 structures21. At top, a molecule of Vps20 initiates ESCRT-III assembly. In yeast cells, at least two copies of Vps20 are required to initiate the scission-competent assembly of ESCRT-III53,56. At 3 o'clock, multiple Snf7 molecules57 polymerize downstream of Vps20. At bottom, one or a few molecules of Vps24 and Vps2 close the spiral, with only Vps24 minimally required for scission. At 9 o'clock, Vps4 breaks up the ESCRT-III polymer at the expense of ATP hydrolysis. c. Integration of budding and scission. After assembling in the bud neck as depicted in Fig. 4, the working model postulates that the ESCRT-III spiral assembles on the bud-proximal side of ESCRT-II to carry out scission.
Energy for the scission reaction is provided by the strong intersubunit and subunit–membrane contacts formed as ESCRT-III assembles on the membrane. An energetically plausible model for the structure responsible for scission29 (Fig. 5b) has been suggested that is inspired by the dome-shaped tip of helical lipid-coated protein tubules formed by VPS24-VPS2 in vitro29 and is consistent with the tubules seen emerging from cells overexpressing human SNF7 (Fig. 3a). Importantly, this model has as a starting point the presence of a membrane deformation or bud, which could be created by the combined action of ESCRT-I and ESCRT-II as discussed above. In this working model for scission29, the basic face of an ESCRT-III dome makes strong, favourable electrostatic contacts with the acidic membrane. The interaction energy per unit area between the membrane and the ESCRT-III dome becomes a crucial parameter in determining whether the energy barrier to scission and accompanying release of the vesicle can be overcome. Provided they are strong enough, the protein–membrane interactions draw the membrane to enfold the top of the dome. As the membrane envelops the top of the dome, the neck narrows to ~ 3 nm, and a scission intermediate is spontaneously formed. Although human VPS2 and VPS24 can form structures in vitro that would seem to have the potential to catalyze scission22,29, it has yet to be shown that they have membrane scission activity in the absence of SNF7. Indeed, in vitro and in vivo studies with yeast proteins suggest that Snf7 is absolutely required for severing of vesicles from the limiting membrane9,25,62.
It remains to be understood how each of the ESCRT-III proteins, which share similar structures and can form similar polymers in vitro either alone or in pairs, have what seem to be different roles in the scission reaction. Some differences can be attributed to the unique binding partners of the proteins, but more fundamental variations seem likely. A simple hypothesis, which has yet to be tested, would be that the scission-competent proteins (that is, Snf7 working in conjunction with Vps20 and Vps24) form stronger interactions with the membrane than do other ESCRT-III subunits.
Finishing up: ESCRT-III disassembly
A defining feature of the ESCRT pathway is the requirement for the AAA+ ATPaseVps4 to disassemble ESCRT-III polymers (Fig. 3c). This ATP hydrolysis reaction is the only energy input into the system and provides the sole thermodynamic driving force for ESCRT-mediated membrane budding and scission. The role of Vps4 seems to be conserved in all ESCRT-dependent processes, including viral budding, cytokinesis and autophagy.
Disassembly22,57,70 and recycling25 of ESCRT-III by Vps4 have been reconstituted in vitro, and early studies showed that without Vps4 activity the entire ESCRT machinery accumulates on membranes and that cargo processing through the late endosomal pathway is blocked71,72. The simplest model for Vps4 function is that it acts by disassembling ESCRT-III after membrane scission is complete. Deletion of Vps4 increases the levels of ESCRT-I and ESCRT-II that associate with the membrane, probably because ESCRT-II binds tightly to the ESCRT-III subunit Vps20, and ESCRT-I in turn binds to ESCRT-II. However, in contrast to ESCRT-III, the dissociation of ESCRT-I and ESCRT-II from the membrane does not strictly depend on Vps420.
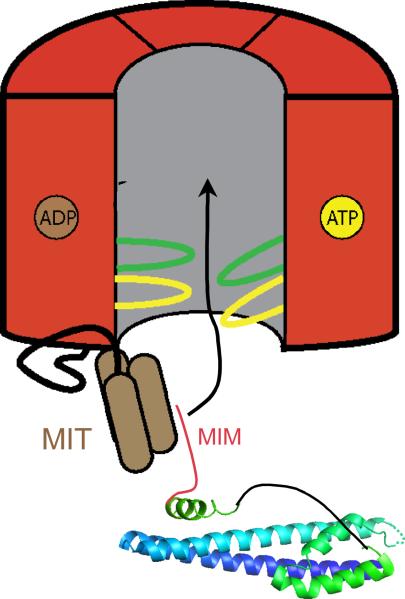
Structural and biochemical studies suggest that Vps4 functions similarly to other unfolding AAA+ ATPases as a ring-shaped dodecameric cylinder (Fig. 6a)73–78. Conserved amino acid residues in the central pore of Vps4 are required for function73. Current models posit that ESCRT-III subunits are pulled into and potentially through a central pore to release them from a membrane-bound ESCRT-III polymer into the cytosol as monomeric subunits (Fig. 6b)73–75. Initial binding of Vps4 to its ESCRT-III substrates requires the enzyme's N-terminal MIT domain, which interacts specifically with short α-helical MIT-interacting motifs (MIMs) at or near the C-termini of individual ESCRT-III proteins. Structural studies show that there are at least two types of MIM motifs (MIM1 and MIM2), which interact in different ways with the Vps4 MIT domain79–81. The MIM1 in the ESCRT-III protein Vps2 has the highest affinity for Vps4, and, correspondingly, Vps2 is essential for ESCRT-III disassembly in vitro25 and in vivo62, even though it is not required for membrane scission.
Figure 6.
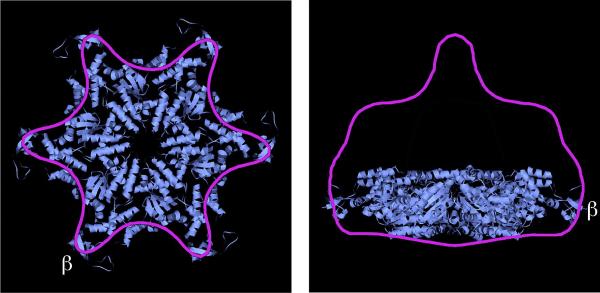
VPS4 structure and function. a. Bottom and side views of a proposed VPS4 hexamer placed into the cryo-EM reconstruction of full-length Vps4p, indicating assembly of Vps4p into a dodecamer. Reproduced with permission from66. b. Working model of VPS4 function, showing delivery of ESCRT-III substrate into the enzyme's central pore following interaction between the enzyme's MIT domains and MIM motifs present at the C-terminus of an ESCRT-III protein. Cycles of ATP hydrolysis lead to movements in loops lining the pore and are proposed to physically induce conformational changes in ESCRT-III proteins that release the proteins from the assembled polymer.
-Please redraw with color of cylinder matching color on
Fig. 6a (replace red with purple outline and blue fill)
The high concentration of MIM motifs present when ESCRT-III proteins assemble into polymers seems to be important not only for engaging individual Vps4 subunits but also for promoting their assembly into a functional AAA+ dodecamer. The assembly of Vps4 into a dodecamer seems to be a crucial regulatory step because the enzyme is an inactive monomer or dimer in the cytosol and seems to assemble only transiently into its oligomeric and active state on substrates73,75,82. Therefore, Vps4 is in a sense a substrate-activated enzyme. Direct visualization of a Vps4 oligomer during ESCRT-III disassembly has not been feasible to date, leaving some questions about whether Vps4 functions as the asymmetric dodecamer seen when ATP hydrolysis is blocked74,77 or in some other form. At least two and potentially as many as four additional proteins bind to Vps4 and have roles in regulating its oligomeric state and activity. The best-studied of these is Vta1 (Table 1), which is a dimer that binds to a unique β-domain within the Vps4 AAA module to promote Vps4 oligomerization and contributes MIT domains to a Vps4–Vta1 complex83. Additional proteins that regulate Vps4 include Did2, Ist1 and Vps60 (Table 1). Did2 is thought to be a positive regulator19 and Ist1 a negative regulator84,85 of Vps4 function, although the coordination of these proposed roles when everything is present together in vivo remains to be clarified. A recent study in yeast shows that these Vps4 regulatory proteins also modulate the size and number of lumenal vesicles20 by mechanisms that are still unclear.
An important outstanding question is how Vps4 interacts with and disassembles the abundant Snf7 from ESCRT-III polymers given that the two proteins (through the MIM2 motif in Snf7) have a low-affinity, yet functionally significant, interaction81,86. Mechanistically, it will be important to determine how much of a conformational change Vps4 induces in ESCRT-III proteins to disassemble ESCRT-III polymers. Will the reaction follow the paradigm established for protease-associated AAA+ ATPases in which molecules are unfolded by using repeated cycles of ATP hydrolysis to pull native proteins into and through the enzyme's central pore87? Or will Vps4 induce a more restricted conformational change that perturbs enough of the subunit to release it and thereby disassemble the ESCRT-III polymer? Will Vps4 function processively on ESCRT-III filaments or instead remodel targeted regions to more dynamically control ESCRT-III structure and function? Answers to these and many other questions promise to shed new light on this fascinating area of cell biology.
ESCRT machinery in other membrane remodelling events
The recent advances described above have led to the development of a working model of scission in MVB biogenesis, and have also furthered our understanding of the role of the ESCRT machinery in the other processes (Fig. 1). A seemingly general model is that ESCRT-III assembles at a membrane neck and cleaves it.
Cytokinesis
In cytokinesis, the narrow membrane neck between dividing cells must be severed as the final step in their separation. ESCRT proteins are thought to have a direct role in this process, as they have the ability to sever membrane necks and they localize at to the site of neck severing at the midbody. This reaction is conserved even in a subset of the Archaea, the genomes of which encode ESCRT-III and Vps4 orthologues but not ESCRT-0, ESCRT -I and ESCRT-II88,89. Studies have shown that cytokinesis requires ESCRT-III, Vps4, and to varying extents, ESCRT-I and ALIX, but not ESCRT-0 and probably not ESCRT-II. Specifically, the actomyosin contractile ring and microtubule dynamics create and extend the membrane neck to which the centrosomal protein of 55 kDa (CEP55) and then ESCRT-I and ALIX are recruited in turn; ESCRT-I and ALIX then recruit ESCRT-III12,13,49,90. However, other models pre-dating the discovery of the role of ESCRTs remain to be fully reconciled with this concept91.
Cytokinetic membrane neck cleavage by the ESCRTs has at least one unique requirement. The cytokinetic membrane neck surrounds the microtubule-based central spindle, so membrane cleavage is obstructed until the spindle is cleaved. Completion of cytokinesis requires the recruitment of the microtubule-severing enzyme spastin, which is thought to cleave the microtubules of the central spindle. Spastin is recruited by the human ESCRT-III subunit DID2B (Table 1)92,93. This illustrates how specialized ancillary functions of ESCRT-III can be coordinated with the principal function of membrane scission.
Viral budding
Once membrane-enveloped viruses have completed replication they exit the infected cell through budding. With a few exceptions, the most notable being the influenza virus, most membrane-enveloped viruses depend on the ESCRT machinery for budding94. By far the most intensively studied of all of the viral budding pathways is that of HIV-1, so we focus on this virus.
For HIV-1 budding, ESCRT proteins are recruited to nascent virions in a process required for their detachment from the plasma membrane, again consistent with a direct role for the ESCRT machinery in membrane severing. Similarly to cytokinesis, studies have shown that HIV-1 budding requires ESCRT-III, Vps4, ESCRT-I and the ESCRT-associated protein ALIX, but not ESCRT-0 and ESCRT-II68. The lack of ESCRT-0 and –II dependence makes sense in that the cargo clustering role of ESCRT-0 is not relevant in this context. The ESCRT-I recruitment function of ESCRT-0 and the bud formation function of ESCRT-II are both supplanted by HIV-1 Gag. The one aspect of the ESCRT-II-independence of HIV-1 budding that is puzzling is that it is not clear what supplants the ESCRT-III recruitment and activation function of ESCRT-II in this setting.
In the case of HIV-1 budding, the viral Gag protein can assemble and drive bud formation, but not scission, independently of the ESCRTs95,96.The ESCRTs, which are required for virion release, assemble to sever the membrane neck connecting the viral bud to the plasma membrane24,97. When ALIX recruits SNF7 for membrane scission it does so directly and can thereby activate ESCRT-III. It is still not clear whether ESCRT-I similarly recruits and activates ESCRT-III or if the link between these complexes is indirect. In a twist to the simple picture of ESCRT-independent budding and ESCRT-dependent scission, tomographic analysis shows that the viral capsid within the nascent uncleaved bud forms a more closed structure when ESCRT subunits are missing97. This suggests that ESCRT-mediated cleavage of viral buds occurs well before closure of the Gag lattice and may compete with complete Gag assembly. The mechanism of membrane scission in HIV-1 budding is probably similar to what occurs during cleavage of ILVs in MVB biogenesis. As above, the main difference is that VPS20 is dispensable, thus this step in the initiation of ESCRT-III assembly seems to be bypassed. It is easy to understand how SNF7 could be activated by ALIX without VPS20, as SNF7 binds directly to the Bro1 domain of ALIX98–101. It is less clear how SNF7 is activated downstream of ESCRT-I, and this is perhaps the most important gap in our understanding of this pathway.
Autophagy
Macroautophagy (autophagy for short) is now known to be responsible not only for bulk consumption of cytosol during starvation, but also for a range of processes of intense interest in medicine, including degradation of damaged mitochondria, breakdown of lipid droplets, degradation of potentially pathogenic inclusions of aggregated proteins, and regulation of cell growth and inflammation. All of these processes involve the formation of a unique double-membrane phagophore, which eventually forms an autophagosome102. The autophagosome is topologically equivalent to an MVB and contains a single large ILV filling the entire structure. Thus, the closure of the phagophore is topologically equivalent to ILV scission into an MVB. The entire ESCRT machinery is required for autophagy103, and it is tantalizing to speculate that phagophore closure operates by the same mechanism as ILV scission. However, because there is only one short-lived neck per phagophore, it is exceptionally difficult to analyze this event experimentally. There is currently no direct evidence for ESCRT recruitment to this neck. Other possible models for the role of ESCRTs in autophagy were recently discussed103. The fact that autophagy, similarly to MVB biogenesis, seems to require all of core ESCRT complexes, suggests a deeper relationship between the two pathways. It seems possible that the ESCRTs might have multiple roles in autophagy; perhaps a direct one in phagophore closure and an indirect one in maintaining a pool of MVBs to fuse with maturing autophagosomes.
Conclusions
Understanding of membrane deformation and scission by the ESCRTs has made stunning advances over the past 2 years. We now know that the ESCRTs have an intrinsic budding and scission activity that is focused on the neck of the nascent bud. These mechanisms differ markedly from familiar mechanisms in vesicular trafficking that depend on outer coats and in viral budding, in which Gag proteins form inner coats. The biophysical underpinnings of these new mechanisms are just beginning to be worked out by computational and other methods. Many questions, such as how the ESCRT-I and ESCRT-II complexes assembles in three dimensions to drive membrane budding, remain open. The coupling between the ESCRT-I- ESCRT-II and ESCRT-III (Fig. 5c) depends on the interaction of ESCRT-II with the ESCRT-III protein Vps20, but the nature of the membrane-bound assembly of all of the ESCRTs remains to be fully explored. The determinants for ILV size in vivo are also uncertain. Membrane mechanical properties are undoubtedly a factor in controlling ILV size, but kinetic factors such as the rate of cargo addition and vesicle scission are likely to matter as well. More sophisticated reconstitution studies may help to address the relative roles of kinetic and equilibrium factors in regulating ILV size.
The new mechanistic information also provides the foundation for understanding the role of ESCRTs in detaching viral buds and cleaving cytokinetic membrane necks. Whether the ESCRTs have a similar direct neck closure reaction in autophagy, as opposed to an indirect role of some sort, will be an important question to resolve. The fundamental mechanisms of the ESCRTs have progressed from an almost complete mystery just 3 years ago to a reasonably complete outline today. Elucidating these mechanisms will help to uncover the details of coupling and regulation of ESCRT-mediated budding in normal physiology and disease.
Glossary
- ESCRT
Endosomal Sorting Complex Required for Transport, one of four of the complexes 0-III responsible for membrane budding and scission in the MVB pathway, and sometimes loosely used to describe the associated pathway and proteins
- Ubiquitin
a conserved 76 amino acid protein that is covalently attached through its C-terminus to Lys residues of many proteins, including proteins trafficked to MVBs by ESCRTs
- MVB
multivesicular body, a late endosome containing intralumenal vesicles (ILVs)
- ILV
intralumenal vesicle, internal vesicles within MVBs
- Midbody
a dense protein structure located at the center of the narrow membrane neck connecting two dividing cells
- Autophagy
used here to refer to macroautophagy, the process of envelopment of cytosol and other material by the phagophore, which subsequently fuses with the lysosome
- Lysosome
in metazoa, the cellular organelle primarily responsible for degradation of unneeded or harmful materials and their recycling into biosynthetic precursors
- Vacuole
the yeast and plant counterpart of the lysosome
- GUV
giant unilamellar vesicle, synthetic vesicles of 5–50 microns in size, useful for visualizing membrane deformation by fluorescence microscopy
- Gag protein
the protein product of the HIV-1 gag gene, consisting of four functional segments, MA, CA, NC, and p6. The p6 segments contains the motifs PTAP and YPXL, which bind to ESCRT-I and ALIX, respectively
- AAA+ ATPases
ATPases associated with diverse cellular activities. Each enzyme contains one or two AAA motifs of 230 to 250 amino acids, including the Walker homology sequences of P-loop ATPases and regions of similarity unique to AAA proteins
Biographies
James Hurley has been a group leader in the Laboratory of Molecular Biology, NIDDK, NIH, Bethesda, USA, for 17 years. He carried out his doctoral work with Bob Stroud at the University of California, San Francisco, USA, and then did postdoctoral studies with Brian Matthews at the University of Oregon, Eugene, USA. His laboratory studies structural and molecular mechanisms in membrane biology, with a focus on subcellular sorting and transport of proteins and lipids.
Phyllis Hanson has been on the faculty in the Department of Cell Biology and Physiology at Washington University in St. Louis for 12 years. She did her doctoral work with Howard Schulman at Stanford University and then did postdoctoral studies with Reinhard Jahn at Yale University. Her laboratory studies the cell biology of membrane trafficking, with a special interest in processes regulated by chaperone-like AAA+ ATPases.
References
- 1.Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 3.Russell MRG, Nickerson DP, Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 2006;18:422–428. doi: 10.1016/j.ceb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Devel. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Pucadyil TJ, Schmid SL. Conserved Functions of Membrane Active GTPases in Coated Vesicle Formation. Science. 2009;325:1217–1220. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological Classification of the Yeast Vacuolar Protein Sorting Mutants - Evidence for a Prevacuolar Compartment in Class-E Vps Mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 9.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 10.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 11.Spitzer C, et al. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- 12.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 13.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita E, Sundquist WI. Retrovirus budding. Annu. Rev. Cell Devel. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 15.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in 'ESCRTing' HIV-1. Nat. Rev. Microbiol. 2007;5:912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 16.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 18.Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol. Biol. Cell. 2010;21:1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref. 21 used electron microscopy of ESCRT-III proteins overexpressed in cultured mammalian cells to show that Snf7 assembles into circular filaments that can be induced to create or stabilize negative curvature in buds and tubules that deform the membrane away from the cytoplasm.
- 22.Lata S, et al. Helical Structures of ESCRT-III are Disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref. 22 reports the reconstitution of a tubular ESCRT-III structure from human VPS24 and VPS2, and showed that VPS4 could disassemble it from the inner surface of the tube.
- 23.Pires R, et al. A Crescent-Shaped ALIX Dimer Targets ESCRT-III CHMP4 Filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajorek M, et al. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009;16:754–U95. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref. 25 showed that the three earliest-assembling subunits of ESCRT-III, Vps20, Snf7, and Vps24, have an intrinsic membrane scission activity. The fourth subunit Vps2 is not required for scission, but is required for coupling to Vps4. This study also showed that the AAA ATPase Vps4 is required for recycling ESCRT-III but not required for scission.
- 26.Im YJ, Wollert T, Boura E, Hurley JH. Structure and Function of the ESCRT-II-III Interface in Multivesicular Body Biogenesis. Dev. Cell. 2009;17:234–243. doi: 10.1016/j.devcel.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by the ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref. 27 revealed that ESCRT-I and –II work together to induced membrane budding via assembly at the neck of the bud. This assembly recruits ESCRT-III selectively to the membrane neck, thus ensuring that none of the ESCRTs are consumed within the ILV.
- 28.Lenz M, Crow DJG, Joanny JF. Membrane Buckling Induced by Curved Filaments. Phys. Rev. Lett. 2009;103 doi: 10.1103/PhysRevLett.103.038101. [DOI] [PubMed] [Google Scholar]
- 29.Fabrikant G, et al. Computational model of membrane fission catalyzed by ESCRT-III. PLOS Comp. Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]; The computational analysis of the energetics of membrane scission in ref. 29 showed that the experimentally determined membrane affinity of ESCRT-III, when incorporated into an assembly of realistic dimensions, is sufficient to provide the energy required for membrane scission.
- 30.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 31.Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Curr. Opin. Cell Biol. 2009;21:568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem. Soc. Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 33.Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends. Biochem. Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 36.Hurley JH. ESCRT Complexes and the Biogenesis of Multivesicular Bodies. Curr. Opin. Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuffers S, Brech A, Stenmark H. ESCRT proteins in physiology and disease. Exp. Cell Res. 2009;315:1619–1626. doi: 10.1016/j.yexcr.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Chu T, Sun J, Saksena S, Emr SD. New component of ESCRT-I regulates endosomal sorting complex assembly. J. Cell Biol. 2006;175:815–823. doi: 10.1083/jcb.200608053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oestreich AJ, Davies BA, Payne JA, Katzmann DJ. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol. Biol. Cell. 2006;18:646–657. doi: 10.1091/mbc.E06-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtiss M, Jones C, Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol. Biol. Cell. 2006;18:636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostelansky MS, et al. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pornillos O, et al. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162:425–34. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bieniasz PD. The Cell Biology of HIV-1 Virion Genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teo H, Veprintsev DB, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J. Biol. Chem. 2004;279:28689–28696. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- 47.Sundquist WI, et al. Ubiquitin recognition by the human TSG101 protein. Mol. Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 48.Shields SB, et al. ESCRT ubiquitin binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kostelansky MS, et al. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–126. doi: 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gill DJ, et al. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J. 2007;26:600–612. doi: 10.1038/sj.emboj.7601501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teo HL, et al. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 53.Hierro A, et al. Structure of the ESCRT-II endosomal trafficking complex. Nature. 2004;431:221–225. doi: 10.1038/nature02914. [DOI] [PubMed] [Google Scholar]
- 54.Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: Crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev. Cell. 2008;14:902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional Reconstitution of ESCRT-III Assembly and Disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref. 57 provided an elegant demonstration using fluorescence spectroscopy that Vps20 and Snf7 undergo a series of conformational changes as ESCRT-III polymerizes on membranes.
- 58.Alam SL, et al. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slagsvold T, et al. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J. Biol. Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- 60.Alam SL, et al. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat. Struct. Mol. Biol. 2006;13:1029–1030. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- 61.Hirano S, et al. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat. Struct. Mol. Biol. 2006;13:1031–1032. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- 62.Teis D, Saksena S, Emr SD. Ordered Assembly of the ESCRT-III Complex on Endosomes Is Required to Sequester Cargo during MVB Formation. Dev. Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]; Ref. 62 defined the order in which ESCRT-III proteins are recruited to the endosome in yeast to be Vps20, Snf7, Vps24, Vps2 and quantified their relative abundance. Snf7 is the most abundant ESCRT-III protein in yeast and is shown by FRET to interact with itself.
- 63.Muziol T, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Xiao JY, et al. Structural Basis of Ist1 Function and Ist1-Did2 Interaction in the Multivesicular Body Pathway and Cytokinesis. Mol. Biol. Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zamborlini A, et al. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shim S, Kimpler LA, Hanson PI. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-terminal Domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 67.Murk JLAN, et al. Endosomal compartmentalization in three dimensions: Implications for membrane fusion. Proc. Natl. Acad. Sci. USA. 2003;100:13332–13337. doi: 10.1073/pnas.2232379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langelier C, et al. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 2006;80:9465–9480. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bajorek M, et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell. 2009;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghazi-Tabatabai S, et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scott A, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu ZH, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2 : 1 complex with Vta1p. J. Mol. Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. Journal Of Molecular Biology 377, 364-377 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref. 74 together with related studies provides the main structural underpinnings for the Vps4 disassembly mechanism. This study shows that the two hexameric rings of Vps4 are asymmetric, while the related ref. 73 showed that central pore residues are important for function and refs. 79–81 showed how Vps4 binds its ESCRT-III substrates.
- 75.Gonciarz MD, et al. Biochemical and Structural Studies of Yeast Vps4 Oligomerization. J. Mol. Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hartmann C, et al. Vacuolar protein sorting: Two different functional states of the AAA-ATPase Vps4p. J. Mol. Biol. 2008;377:352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 77.Landsberg MJ, Vajjhala PR, Rothnagel R, Munn AL, Hankamer B. Three-Dimensional Structure of AAA ATPase Vps4: Advancing Structural Insights into the Mechanisms of Endosomal Sorting and Enveloped Virus Budding. Structure. 2009;17:427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 78.Inoue M, et al. Nucleotide-Dependent Conformational Changes and Assembly of the AAA ATPase SKD1/VPS4B. Traffic. 2008;9:2180–2189. doi: 10.1111/j.1600-0854.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- 79.Obita T, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 80.Stuchell-Brereton M, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 81.Kieffer C, et al. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 82.Azmi IF, et al. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 83.Xiao J, et al. Structural basis of Vta1 function in the multi-vesicular body sorting pathway. Dev. Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shestakova A, et al. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide Translocation by the AAA plus ClpXP Protease Machine. Chem. Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Refs. 88 and 89 show that in the hyperthermophilic Crenarchaea, ancient homologs of ESCRT-III and Vps4 are required for completion of cell division. The Crenarchaea lack homologs of ESCRT-0, -I, and –II, so these studies strongly suggested that ESCRT-III and Vps4 might comprises the minimal membrane scission machinery.
- 90.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 2009;19:606–616. doi: 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 92.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ivanchenko S, et al. Dynamics of HIV-1 assembly and release. PloS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carlson LA, et al. Three-Dimensional Analysis of Budding Sites and Released Virus Suggests a Revised Model for HIV-1 Morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim J, et al. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fisher RD, et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 100.Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McCullough J, et al. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? J. Cell Sci. 2009;122:2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]