Abstract
Isothermal titration calorimetry (ITC) is a useful technique to study RNA-protein interactions, as it provides the only method by which the thermodynamic parameters of free energy, enthalpy, and entropy can be directly determined. This chapter presents a general procedure for studying RNA-protein interactions using ITC, and gives specific examples for monitoring the binding of Caenorhabditis elegans GLD-1 STAR domain to TGE RNA and the binding of Aquifex aeolicus S6:S18 ribosomal protein heterodimer to an S15-rRNA complex.
Keywords: Calorimetry, ribonucleoprotein complex, thermodynamics, RNA-protein interaction, binding
1. Introduction
1.1 Isothermal Titration Calorimetry (ITC)
There are many studies in which ITC has been used to monitor protein-ligand, protein-protein, and protein-nucleic acid interactions(1–3). ITC measures the heat (q) evolved or consumed as aliquots of one reagent are added to a second reagent in a calorimetric cell. The reaction heat as a function of concentration is analyzed to obtain the complete thermodynamic characterization (ΔH, ΔG, ΔS) of the binding reaction (4). A full titration experiment can usually be completed in 2 hours.
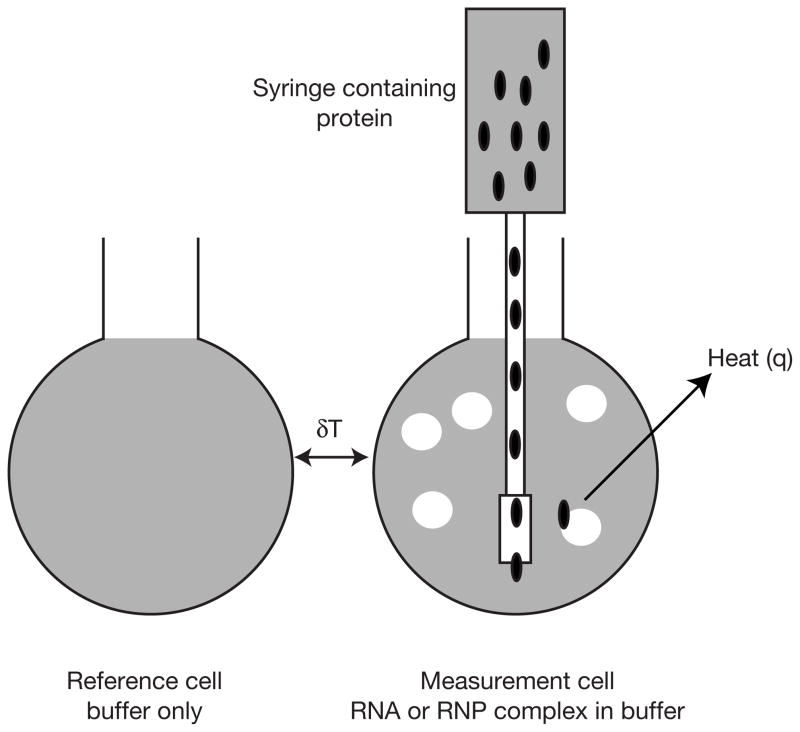
As shown in Figure 1, within an adiabatic chamber the calorimeter contains a measurement and a reference cell(5). The measurement cell is filled with a solution of a macromolecule in the buffer of choice. The reference cell is filled with either water or the same buffer in which the macromolecule is dissolved. The tip of a syringe containing the ligand (either a small molecule or another macromolecule) is inserted in the measurement cell. Aliquots of the ligand are added to the measurement cell. The instrument maintains a small constant temperature difference between the measurement and reference cells. In response to binding of ligand to macromolecule in the measurement cell, which produces or consumes heat, the feedback system compensates to maintain the constant temperature difference between the measurement and sample cells. The signal measured during the titration is the rate of heating as a function of time, with a pulse corresponding to the addition of aliquots of ligand to the measurement cell. Integration of these pulses as a function of time yields a plot of q as a function of injection number, or molar ratio of ligand to macromolecule.
Figure 1.
Schematic of the measurement and reference cells inside the adiabatic housing of the ITC. The measurement and reference cells have an active volume of about 1.4 ml. The instrument maintains a constant temperature difference (dT) between the measurement and reference cells. Aliquots of a ligand solution are added to the measurement cell using a syringe controlled by a motorized injection system. Binding of the ligand to the macromolecule releases or consumes heat.
1.2 RNA-Protein Interactions
There is a great range in the size and complexity of RNA-protein complexes. These span simplest case involving a single protein binding to an isolated fragment of RNA to the large ribonucleoprotein complexes which involve multiple proteins binding to one or more RNAs.
The great variability observed in RNA-protein interactions makes ITC an ideal choice to study these interactions. In contrast to most other methods of detecting binding of RNA and proteins, there is no inherent size limitations to the macromolecules under investigation. Fluorescence anisotropy (6,7) requires that one of the two interacting components be much smaller than the other and surface plasmon resonance (8) requires that one of the two components be immobilized. Gel mobility shift assays require that the migration of an RNP complex in the gel is sufficiently different than the free RNA, which is less likely when complexes between small proteins and large, highly structured RNAs are studied. Gel shift assays have the requirement that the RNP complex is kinetically stable during electrophoresis, which is often not the case for weak binding complexes. ITC does not require labeling or immobilization of either macromolecule, is performed under equilibrium conditions, and can be performed under a wide variety of buffer conditions.
1.3 Experimental design
The amount of material required is an important factor to consider when performing an ITC experiment. Unlike gel mobility shift assays or fluorescence anisotropy measurements, in which the labeled molecule is often present in trace quantities, ITC requires nanomole quantities of each reactant. To obtain a reliable value for the association constant of an interaction, the appropriate concentrations of reactants must be used. As described by Wiseman et al (5), the parameter c, which equals the association constant (Ka) times the total concentration of the reactant in the calorimeter cell (Mtot), should lie between 1 and 1000. A more reliable estimation of Ka can be obtained if the value of c is kept between 10 and 500. A sufficient concentration of titrant should be used so that its concentration at the completion of the titration will be 1.5–2 × n (the number of binding sites per molecule) times the concentration of reactant in the cell. For a titration in which a total of 250 μl of protein is added to a reaction cell containing approximately 1.5 ml of RNA, the protein should be 10–20n times the concentration of RNA.
1.4 Biological Systems
Two different systems are chosen to demonstrate both a simple protein-RNA interaction and a protein-RNP interaction.
Germline development in Caenorhabditis elegans. C. elegans GLD-1 belongs to the highly conserved STAR/GSG family of RNA-binding proteins which play a central role in metazoan development. STAR/GSG proteins are composed of a single KH domain flanked by two regions homologous to the murine quaking gene, Qua1 and Qua2. STAR domain proteins form functional homodimers in cells via the Qua1 domain, and the KH and Qua2 regions form an extended RNA interaction surface. GLD-1, a C. elegans germline developmental regulator, binds to a 28 nucleotide sequence (TGE) in the 3′-UTR of tra-2 mRNA and recruits a complex that silences translation. The protocol here describes monitoring the binding of the GLD-1 STAR domain homodimer to the 28 nucleotide TGE RNA(9).
Assembly of the platform domain of the 30S ribosomal subunit from the hyperthomophilic bacterium Aquifex aeolicus(10). There is ordered binding of five ribosomal proteins (S6,S8, S11, S15, and S18) to the central domain of 16S rRNA during assembly. S6 and S18 form a heterodimer with a Kd of 8.7 nM. The S6:S18 heterodimer binds to an S15-rRNA RNP complex. The protocol here describes monitoring the binding of a pre-formed S6:S18 heterodimer to an RNP complex containing ribosomal protein S15 bound to a fragment of the central domain of 16S rRNA.
2. Materials
10–20 nmoles of purified RNA (2 ml at 5–10 μM) for each titration
100 nmoles of each purified protein (1 ml at 100 μM) (see note 1)
Dialysis membrane of the appropriate MWCO (3500–30000)
-
Buffer solutions
For A. aeolicus ribosomal proteins and central domain rRNA: 20 mM potassium-HEPES, pH 7.6, 330 mM potassium chloride, 10 mM magnesium chloride
For C. elegans GLD-1 protein and TGE RNA: 20 mM Tris-HCl, pH 8.0, 25 mM NaCl, 1 mM DTT
Isothermal titration calorimeter
The protocol was written for the MCS-ITC model calorimeter made by Microcal, Inc. (Northampton, MA). This model uses a circulating water bath to allow cooling of the unit below ambient temperature. The newer VP-ITC calorimeter does not require the circulating water bath to provide cooling and experiments can be performed between 4° C and 80° C. These experiments can be performed on the VP-ITC using only slight modifications of the following procedures.
5% solution of Contrad 70 (Decon Labs, Inc., Bryn Mawr, PA).
3. Methods
3.1 Sample and equipment preparation
Below is a step-by-step protocol for running an ITC experiment to study the binding of a single protein to RNA. To study the binding of a protein to an RNP complex, the only modification required is the formation of the RNP complex prior to degassing the samples (Section 3.1.4, following step 1, see note 4)
3.1.1 RNA synthesis
Synthesis and purification of the RNA is beyond the scope of this paper. The RNA can be obtained either by chemical synthesis (for RNAs shorter than 50 nucleotides) or by transcription from a DNA template using T7 RNA polymerase (11) (for RNAs of any length).
For studies of ribosome assembly, the fragments of 16S rRNA were transcribed using T7 RNA polymerase from a plasmid DNA template.
TGE RNA for C. elegans GLD-1 STAR studies was produced by chemical synthesis (Dharmacon) and deprotected and lyophilized as per the manufacturer’s protocol.
RNA produced by in vitro transcription methods should be purified on a denaturing polyacrylamide gel. The RNA product is visualized by UV shadowing and the band excised and RNA eluted by electroelution (Schleicher & Schuell). Concentrate the RNA by precipitation with ethanol and dissolve in an appropriate volume of dialysis buffer to yield a 20 μM solution.
3.1.2 Protein purification
The protein to be used in the titration should be purified to near homogeneity. At a minimum, it must be free of contaminating proteases and nucleases. Since the value of the fit parameters depends highly on the accurate determination of the active protein concentration, a very pure sample is better for the ITC experiment. GLD-1, S6, S18, and S15 can be purified as previously described (9,10).
Prepare solutions of proteins that are at least 1.5X the final desired concentration to allow for dilution that may occur during dialysis.
3.1.3 Dialysis of proteins and RNA
Prepare 4 liters of the appropriate buffer (A or B) and chill to 4° C. Dialyze RNA and protein against this buffer overnight at 4° C (see note 2).
Equilibrate the calorimeter at the experimental temperature by first setting the circulating water bath 5 to 10°C below the desired temperature for the experiment. Set the thermostat to the desired temperature (30°C or 40°C) and allow to equilibrate at least 12 hours.
Following dialysis of the proteins and RNA, retain the buffer for washing of the injection syringe, reference and measurement cells of the ITC, and for control titrations and dilution of RNA or proteins (as necessary).
Determine the concentration of RNA and proteins and dilute as necessary using dialysis buffer (see note 3).
3.1.4 Loading samples in calorimeter
Anneal the RNA by heating to 95°C for 2 minutes and place on ice for 5 minutes (see note 4).
Degas 10 ml of dialysis buffer and the RNA sample for 7–10 minutes with a vacuum pump.
Wash the reaction and reference cells with a 5% Contrad 70 solution and rinse thoroughly with water. Fill the reference cell with buffer or water.
Rinse the measurement cell with buffer before adding the RNA sample slowly to avoid introducing any air bubbles to the sample.
Fill the 250 μl injection syringe with either GLD-1 or S6:S18 heterodimer solution (See note 5).
Insert the injection syringe into the reaction cell, taking care to avoid bending the needle.
3.2 Titration experiment
Set the run parameters for the experiment: temperature (30°C for GLD-1 STAR/TGE RNA or 40°C for S6:S18/S15-RNA complex), number of injections (26), reference power (10 μcal/s), initial delay (60 sec), time between injections (240 sec), stirring speed (400 rpm), concentration in the cell (5 μM for TGE RNA, 10 μM for S15-RNA complex) and the syringe (110 μM for GLD-1 STAR, 100μM for S6:S18), and the injection volume (first injection 2 μl, next 25 injections 10 μl) (see note 6).
Start the experiment. The MCS-ITC can be set up to automatically proceed through all steps of the experiment. The instrument will proceed through a thermal equilibration step which, depending on how closely the temperature of the sample placed in the reaction cell matches the experimental temperature, generally takes between 10 and 30 minutes. Following thermal equilibration, the injection mechanism engages the syringe and stirring commences. This is followed by a mechanical equilibration until a stable baseline is achieved, which generally takes an additional 10 to 15 minutes.
Upon completion of the experiment, the titrated RNA-protein complex should be removed from the reaction cell and the cell should be cleaned thoroughly.
Perform an additional experiment in which protein at the same concentration used in step 1 is titrated into a measurement cell containing buffer. The heat evolved by dilution of the concentrated protein solution will be subtracted from the data for titration of protein into RNA (see Note 7).
3.3 Data analysis
-
The Origin software provided by Microcal (Northampton, MA) can be used for data analysis. For both examples presented here, model describing a single set of identical sites is adequate to describe the data (5,12). Assuming a 1:1 interaction in which a macromolecule (M) binds to a ligand (X), the following binding equilibrium exists:
(1) (2) It follows that:(3) and(4) At the start of the titration, the macromolecule concentration is Mt in a volume Vo. As aliquots of ligand are added during the titration the total volume will be Vo + ΔV. The concentration of macromolecule is reduced during the titration, so that:(5) and similarly,(6) The total heat content, Q, of the solution contained in Vo is:(7) in which θ is the fraction of sites on M occupied by X, n is the number of sites, and Vo is the active cell volume.(8) In which Ka is the association constant, and Mt and Xt are the bulk concentration of macromolecule (in Vo) and ligand, respectively.
The data is plotted as the differential heat evolved for each injection of an aliquot of ligand X to the sample in the measurement cell versus the molar ratio of Xt/Mt. The differential heat is described by:(9) The heat released from the ith injection is:(10) In fitting data with the single set of identical sites model, initial guesses are made for the values of Ka, ΔH and n. ΔQ(i) is calculated for each injection and compared to the measured values. The values of Ka, ΔH and n are varied until the best fit of the data is obtained using standard Marquardt methods (13) (See note 8). The free energy change for the interaction is calculated using the relationship:(11) The entropy change (ΔS) is obtained using the standard thermodynamic expression:(12) -
The value of n obtained using this model should be close to the stiochiometry of the interactions. For interactions with a 1:1 stiochiometry, n should typically lie between 0.9 and 1.1 (or 1.8–2.2 for 2:1 stiochiometry). Large deviations from these values indicate one of two possibilities:
For n<1, either the concentration of RNA in the cell has been overestimated, the concentration of protein has been underestimated, or not all of the RNA is in an active state.
For n>1, either the concentration of protein in the syringe has been underestimated, the concentration of RNA has been underestimated, or not all the protein is active.
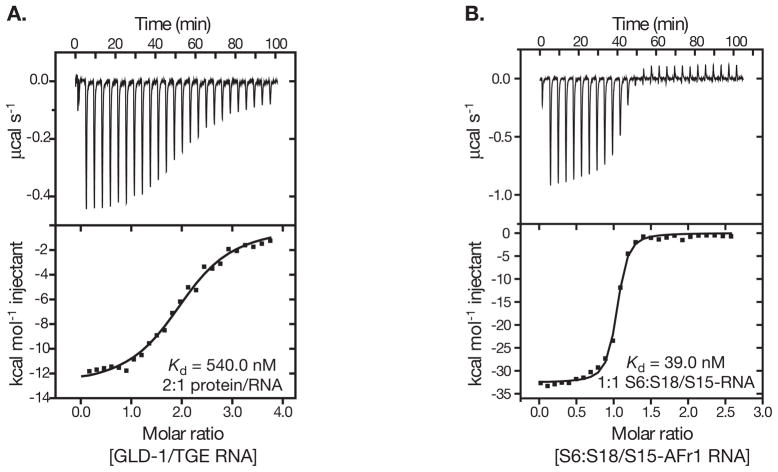
Figure 2.
(A) Titration of TGE RNA (4.9 μM) with GLD-1 STAR (95 μM) at 30 °C. The data was fit to a single binding site model yielding a ΔHobs of −11.7 kcal mol−1, K of 1.81 × 106 and n of 2.3. (B)Titration of S15-rRNA (10 μM) complex with S6:S18 heterodimer (100 μM) at 40°C. The data was fit to a single binding site model yielding a ΔHobs of − 32.6 kcal mol−1, K of 2.56 × 107 and n of 1.0.
Acknowledgments
M.I.R. was supported by Research Scholar Grant #PF-01-087-01-GMC from the American Cancer Society. S.P.R. is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-1723). This research was supported by grants from The Skaggs Institute for Chemical Biology and the National Institutes of Health (NIH, GM53320 and GM53757).
Footnotes
Only 500 μl of this sample will be used for the experiment and control titrations, but additional protein is needed so that the injection syringe can be filled without introducing bubbles. The needle contains two holes, one at the tip and another on the side. The hole on the side must be covered by protein sample during filling, necessitating the additional volume.
Depending on the buffer in which the proteins are stored, it may be necessary to prepare an additional 4 liters of dialysis buffer and change the buffer approximately 8 hours into the dialysis. This is particularly important if the protein is stored in a salt concentration that is much higher than in the measurement buffer or if a large volume of protein (>10 ml) is being dialyzed.
The concentration of the proteins used in this example was determined using the calculated extinction coefficient at 280 nm(14). Other methods may be used to determine concentrations of proteins, but inaccurate concentration determination will effect all parameters in the subsequent determination of ΔH, ΔG and n.
The RNA samples should be annealed prior to degassing, as the heating process will liberate more dissolved air. If an Aquifex aeolicus S15-RNA complex is to be titrated, form the complex before degassing by incubating the annealed RNA with an equimolar amount of protein for 5 minutes at 65 °C. The Aquifex aeolicus protein is stable under these conditions.
It is possible, though not recommended, to perform the titration with protein in the measurement cell and RNA in the syringe. The initial injections will create a situation in which protein is in great excess over RNA, which can produce complexes that are prone to aggregation/precipitation.
The injection parameters will vary based on the specific interaction being measured. The values given have yielded good results with both systems described here. The first injection is kept small because there is the possibility of some mixing of protein solution on the outside or in the tip of the needle during the equilibration process, resulting in an erroneous data point. The heat evolved during the first injection will not be included in the data fitting, but the amount of protein in this injection will be used to calculate the total ligand concentration in the reaction cell.
A control titration such as that described is not always necessary. If there is only a single binding site for the protein on the RNA, and it is fully saturated by the end of the titration, the heat observed following saturation can be used as the dilution reference.
For particularly high affinity interactions (Ka ≥ 1 × 109), the value of Ka may need to be held constant as it will not be well defined unless the value of c can be kept between 10 and 500. If a competitive inhibitor binds to the same site as a high-affinity binding ligand, one can determine the Ka of the high-affinity ligand using displacement isothermal titration calorimetry (15).
References
- 1.Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 2.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11:560–6. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 3.Weber PC, Salemme FR. Applications of calorimetric methods to drug discovery and the study of protein interactions. Curr Opin Struct Biol. 2003;13:115–21. doi: 10.1016/s0959-440x(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 4.Tame JRH, O'Brien R, Ladbury JE. Isothermal Titration Calorimetry Of Biomolecules. In: Ladbury JE, Chowdhry BZ, editors. Biocalorimetry: Applications of Calorimetry in the Biological Sciences. John Wiley & Sons Ltd; Chichester, NY: 1998. pp. 27–38. [Google Scholar]
- 5.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 6.LeTilly V, Royer CA. Fluorescence anisotropy assays implicate protein–protein interactions in regulating trp repressor DNA binding. Biochemistry. 1993;32:7753–8. doi: 10.1021/bi00081a021. [DOI] [PubMed] [Google Scholar]
- 7.Wilson GM, Sutphen K, Chuang K, Brewer G. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J Biol Chem. 2001;276:8695–704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 8.Katsamba PS, Park S, Laird-Offringa IA. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods. 2002;26:95–104. doi: 10.1016/S1046-2023(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 9.Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol. 2004;11:20–8. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- 10.Recht MI, Williamson JR. Central domain assembly: thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J Mol Biol. 2001;313:35–48. doi: 10.1006/jmbi.2001.5018. [DOI] [PubMed] [Google Scholar]
- 11.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 12.Indyk L, Fisher HF. Theoretical aspects of isothermal titration calorimetry. Methods Enzymol. 1998;295:350–64. doi: 10.1016/s0076-6879(98)95048-0. [DOI] [PubMed] [Google Scholar]
- 13.Marquardt D. An algorithm for least-squares estimation of nonlinear paramaters. Journal of the Society for Industrial and Applied Mathematics. 1963;11:431–441. [Google Scholar]
- 14.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 15.Sigurskjold BW. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal Biochem. 2000;277:260–6. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]