Abstract
Transforming growth factor beta 1 (TGF-β1) is secreted as a latent complex, which consists of latency associated peptide (LAP) and the mature ligand. The release of the mature ligand from LAP usually occurs through conformational change of the latent complex and is therefore considered to be the first step in the activation of the TGF-β signaling pathway. So far, factors such as heat, pH changes, and proteolytic cleavage are reportedly involved in this activation process, but the precise molecular mechanism is still far from clear. Identification and characterization of the cell surface proteins that bind to LAP are important to our understanding of the latent TGF-β activation process. In this study, we have identified Heat Shock Protein 90 β (HSP90β) from the cell surface of the MG63 osteosarcoma cell line as a LAP binding protein. We have also found that MG63 cells secrete HSP90β into extracellular space which inhibits the activation of latent TGF-β1, and that there is a subsequent decrease in cell proliferation. TGF-β1-mediated stimulation of MG63 cells resulted in the increased cell surface expression of HSP90β. Thus, extracellular HSP90β is a negative regulator for the activation of latent TGF-β1 modulating TGF-β signaling in the extracellular domain.
Keywords: TGF-β1, Extracellular HSP90β, Osteosarcoma, Latent TGF-β activation, MG63 cell
Introduction
Transforming growth factor-beta 1 (TGF-β1) is a multifunctional cytokine that regulates cell proliferation, migration and differentiation, as well as synthesis of the extracellular matrix [1–5]. TGF-β1 is abundant in skeletal tissue, where it is stored in the bone matrix and promotes bone formation [6, 7]. Recent findings showed that there is an abnormally high expression of TGF-β in osteosarcoma, which is the most common bone cancer in young adults [8, 9].
TGF-β1 is transcribed as a proprotein consisting of three distinct parts: the signal peptide, the latency-associated peptide (LAP), and the mature peptide. The disulfide-bonded homodimer, in which the LAP and the mature peptide are non-covalently linked, is secreted either alone (as a small latent complex: SLC), or in combination with a latent TGF-β binding protein (LTBP) as a large latent complex (LLC) [10–12]. In order for TGF-β1 to bind to its receptor and activate the downstream signaling pathway, it is necessary that the mature ligand be released from the latent complex. Many mechanisms for the activation of latent TGF-β1 have been described, including binding to ανβ6 integrin or thrombospondin-1 (TSP-1), and proteolytic processing by plasmin or metalloproteinases (MMPs) [13, 14]. Interestingly, a recent report indicated that ανβ6 integrin, one of the major activators of the latent TGF-β1 complex, requires LTBP1 for its activity. This suggests that ανβ6 integrin prefers LLC but not SLC [15].
Most types of cells secrete TGF-β1 in the form of LLC; in contrast, bone cells secrete most of the TGF-β1 as SLC [7]. Thus, in bone cells such as osteosarcoma and osteoblasts, the activation mechanisms of latent TGF-β1 are considered to be regulated differently. Therefore, to identify novel regulators of latent TGF-β1 activation in bone cells, we have isolated extracellular proteins that bind to recombinant LAP from the osteosarcoma cell line, which is made up of MG63 cells. We found that heat shock protein 90 beta (HSP90β) bound to the TGF-β1 latent complex and inhibited the TGF-β1 signaling pathway. Furthermore, we observed that HSP90β was secreted into the extracellular space in MG63 cells, and this secretion was mediated by TGF-β1 stimulation. In the present study, we provide the first evidence that extracellular HSP90β secretion is induced by TGF-β signaling, and that extracellular HSP90β interacts with the TGF-β1 latent complex in MG63 cells and inhibits the TGF-β signaling pathway through a negative feedback loop.
Material and methods
Tissue Culture
MG63 cells (ATCC) and 293 EBNA cells (Invitrogen, Carlsbad, CA) were maintained in DMEM (Dulbecco’s modified Eagle’s medium; Invitrogen), supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 100 units/ml penicillin, and 100 mg/ml streptomycin (Invitrogen) at 37°C in a humidified, 5% CO2, 95% air atmosphere.
Isolation of LAP binding proteins
To prepare the recombinant LAP protein, LAP TGF-β1 cDNA was amplified and cloned into the pCEP4-Mul-PURD expression vector [16, 17]. The recombinant LAP expression vectors were then transfected into 293/EBNA cells and the N-terminal His-tagged recombinant LAP proteins were purified as described previously [16]. The cell surface proteins of MG63 cells were extracted using ProteoExtract® Native Membrane Protein Extraction Kit (EMD Chemicals Inc., Gibbstown, NJ). The extracted proteins were then mixed with or without His-tagged-LAP proteins, and protein complexes were precipitated by nickel-charged magnetic beads (Qiagen, Valencia, CA). The precipitants were washed and loaded onto SDS-PAGE gel under reducing conditions and the gels were stained with silver staining. The specific band in the precipitants mixed with recombinant LAP was identified by mass spectrometry performed by Alphalyse, Inc., Palo Alto, CA.
Solid phase binding assay
96-well, flat bottom microtiter plates (Inmunolon 2HB, Thermo Fisher Scientific, Waltham, MA) were coated with a 0.1 µg/well of recombinant HSP90β (SignalChem, Vancouver, BC) and incubated overnight at 4 °C. After washing, plates were blocked with 3% BSA and then recombinant LAP was added in triplicate in a serial dilution with PBS and incubated overnight at 4 °C. The next day, plates were washed and then incubated with anti-LAP antibodies (R&D, Minneapolis, MN AF246A)(1:3000) for 90 min at RT. Anti-goat antibodies conjugated with HRP (1:1000) in PBS were incubated for 1 h at RT. After washing, 3, 3, 5,5’-tetramethyl benzidine substrate solution (TMB, Pierce, Rockford, IL) was added to the wells and incubated for 30 min at RT. After the addition of 2 N H2SO4 to stop the colorimetric reaction, optical density was measured at 450 nm using a microplate reader. Control plates were prepared by coating them with 0.1 µg/well of BSA instead of HSP90β, and used for binding assays along with HSP90β-coated wells.
Co-immunoprecipitation assay
293/EBNA cells and His-tagged LAP-stably-transfected 293/EBNA cells were transfected with an HA-HSP90β expression vector. The medium was replaced 6 h later, and 48 h later the medium was collected and centrifuged at 1000 × g for 1 min. One half of the collected medium was mixed with Ni-NTA agarose beads and centrifuged at 1000 × g for 1 min, and then precipitants were washed by 0.1% Triton-PBS and then 10 mM imidazole PBS. Finally, precipitants were eluted by 500 mM imidazole and then reduced with DTT. The rest of the collected medium was mixed with anti-HA antibody bound to Protein G beads (Invitrogen), and precipitants were washed by 0.1% Triton-PBS and eluted by boiling with a reducing agent. Both eluted samples were loaded onto SDS-PAGE, electrophoresed, and transferred onto PVDF membranes (Invitrogen). The membranes were incubated with anti-HA to detect HSP90β, or anti-His to detect LAP, respectively. The immunoreactive proteins were identified using Supersignal chemiluminescent substrate (Pierce).
Biotinylation of cell surface proteins and lysis of cells
MG63 cells were washed twice with ice-cold PBS and incubated with 0.25 mg/ml cell-impermeable Sulfo-NHS-SS-biotin (Pierce) in PBS for 30 min on ice. Cells were then washed with quenching solution and washed twice with PBS before being scraped into an ice-cold lysis buffer containing a protease inhibitor cocktail (Complete-Mini; Roche, Basel, Switzerland). The cells were then sonicated and placed on a shaker for 2 h. Samples were centrifuged at 12,000 × g for 20 min and the pellets were discarded. The protein concentration of the resulting supernatant was determined using a BCA kit (Pierce).
Streptavidin pull-down
Streptavidin beads (50 µl; Invitrogen) were washed three times with PBS. Lysed biotinylated samples were added to the beads (50 µg total proteins), and volumes of up to 1 ml were made with lysis buffer and mixed on a shaker overnight. Beads were then centrifuged at 1000 × g for 1 min. They were washed three times with 1 % Triton-PBS. A loading buffer was added to the washed beads and then heated at 70 °C for 12 min, and beads were centrifuged at 1000 × g for 1 min. The supernatant was loaded onto SDS-PAGE.
Luciferase activity
MG63 cells were transfected with both the (CAGA)12-luciferase reporter plasmid [18] and the pRL-TK control vector utilizing LipofectAMINE 2000 with LTX (Invitrogen). After 24 h, they were pre-treated with recombinant HSP90β or anti-HSP90β antibody, and then treated with 200 ng/ml latent TGF-β1 (R&D). After 24 h, cells were lysed and assayed for luciferase activity using the Dual Luciferase Reporter Assay System (Promega Corporation, Madison, WI), and luciferase activities were normalized based on Renilla luciferase expression from the pRL-TK control vector.
Cell-proliferation assay
MG63 cells were plated at a density of 2.5 × 103 cells/well in a 96-well plate and cultured in 0.1% FBS medium. After 24 h, cells were incubated with 200 ng/ml latent TGF-β1 with various concentrations of recombinant HSP90β. Before and after 48 h, cells were lysed and the total active cell number was compared by CellTiter-GloTM Luminescent Cell Viability Assay (Promega), which is based on the quantitation of ATP levels present in the cells.
Real-Time PCR
RNA was extracted from MG63 cells using the TRIzol method. RNA samples were quantified and reverse-transcription reactions were performed using Superscript III reverse transcriptase (Invitrogen). All Real-Time PCR reactions were prepared with iQ SYBR Green supermix and carried out in a PTC-200 thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA).
Results
Isolation and characterization of LAP binding protein in MG63 cells
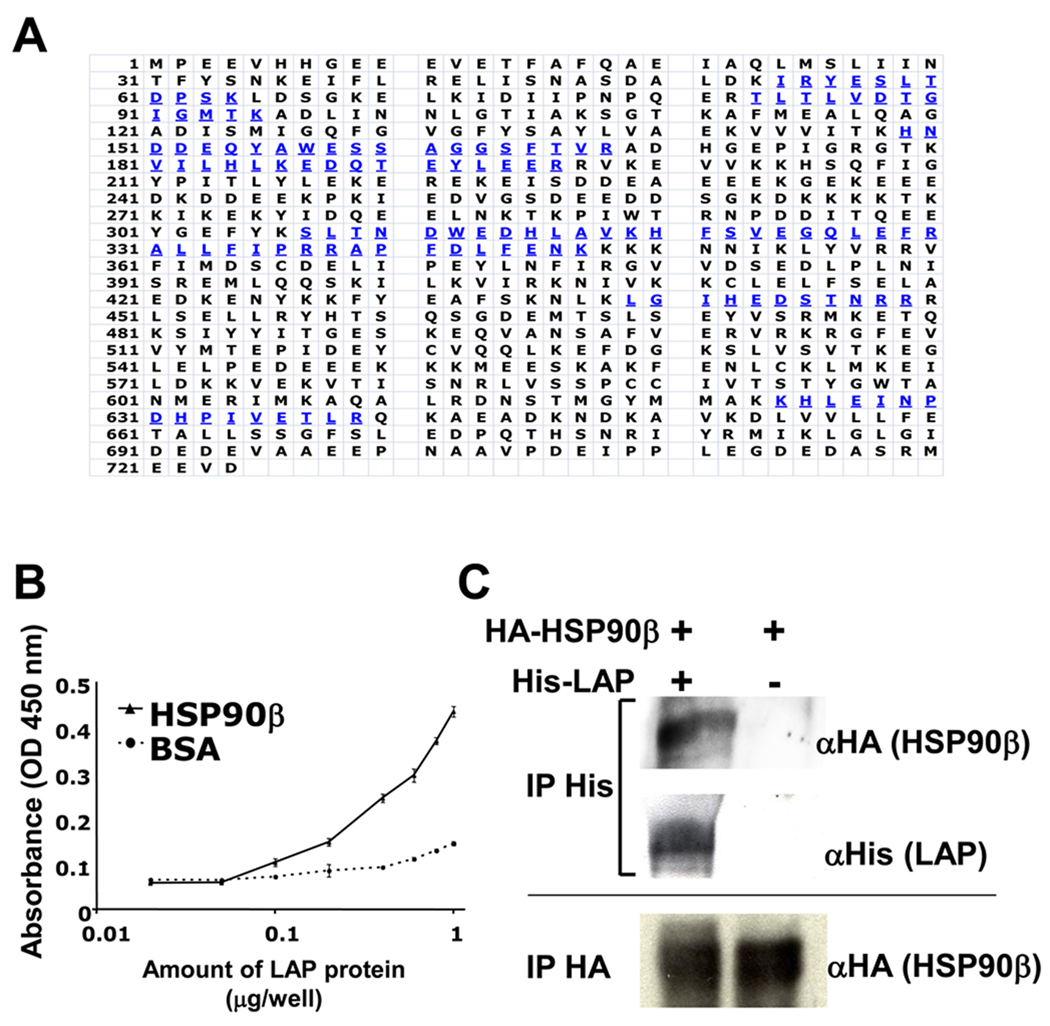
To identify the cell surface proteins that can bind to LAP, we incubated the customized recombinant LAP protein with the cell surface proteins of MG63 cells, and found one intense band of about 90 kDa (data not shown). We then analyzed this band with mass spectrometry and amino acid sequencing, which revealed its identity to be HSP90β (Fig. 1A). Next, we confirmed HSP90β and recombinant LAP binding by solid phase binding assay (B) and co-immunoprecipitation assay (C). As shown in Fig. 1B, the binding of LAP to HSP90β was dose-dependent, suggesting that HSP90β can bind to LAP directly. Furthermore, as shown in Fig. 1C, we performed a co-immunoprecipitation assay using the 293/EBNA cells, which were transfected with His-LAP and HA-HSP90β. We detected an HA-HSP90β-specific band in the His-LAP precipitants only when 293/EBNA cells were transfected with His-LAP and HA-HSP90β, but not transfected with HA-HSP90β alone.
Figure 1. Isolation of HSP90β as LAP binding protein from MG63 cells.
A, The single intense band around 90 kDa is collected and trypsinized, and then sequenced by mass spectrometry. Blue amino acids were the sequences identified by mass spectrometry. B, 96-well plates were coated with HSP90β or BSA (100 µg/well). Coated plates were then incubated with serial concentrations of LAP proteins. The amount of binding LAP protein was determined by the anti-LAP antibody. Each value represents the mean of triplicate determinations; bars, SD. C, 293/EBNA and 293/EBNA-His-LAP cells were transiently transfected with HSP90β-HA. Supernatant was immunoprecipitated (IP) with the anti-His antibody (1st and 2nd rows) or anti-HA antibody (3rd lane), and then immunoblotted with anti-HA antibody (1st and 3rd rows) or anti-His antibody (2nd row).
HSP90β is expressed on the cell surface and extracellular space of MG63 cells
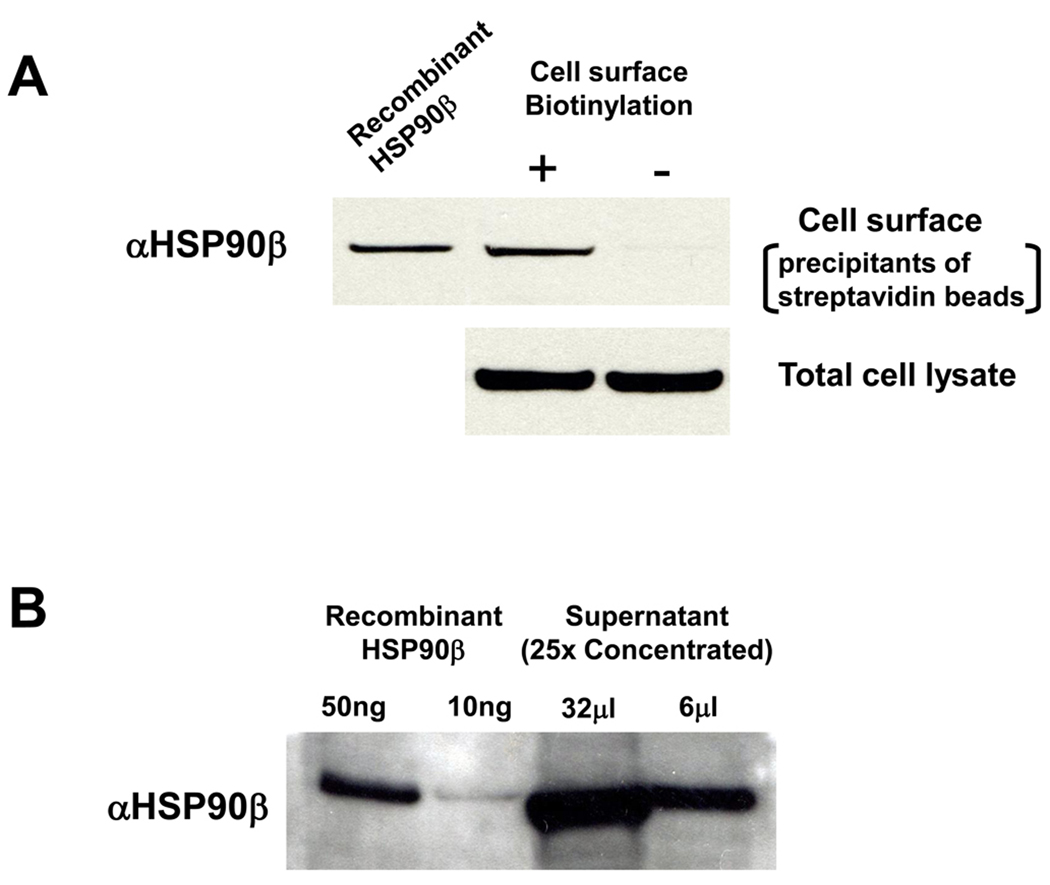
HSP proteins are known to be present inside the cells. In order to investigate whether HSP90β was present on the surface of MG63 cells, we examined the cell surface expression of HSP90β using the biotinylation method, as described above. HSP90β was detected in the precipitants of streptavidin beads only when cell surface proteins were biotinylated (Fig. 2A). We loaded the total cell lysate to show the amounts of proteins used for biotin-streptavidin precipitate. We could not detect HSP90α on the MG63 cell surface when utilizing the same method (data not shown). Finally, immunoblotting of the conditioned medium of MG63 cells with HSP90β antibodies showed that HSP90β was present in the medium (Fig. 2B).
Figure 2. Secreted HSP90β expression in MG63 cells.
A, Surface proteins were biotinylated, and collected by streptavidin beads (upper panel). Total cell lysate (lower panel), which represents the extracellular and intracellular HSP90β. Surface proteins and total lysate were immunoblotted with anti-HSP90β. Recombinant HSP90β (100 ng/ml) was also loaded as positive control B, Serum-free conditioned medium (25 × concentrated) from MG63 cell culture and recombinant HSP90β were immunoblotted with anti-HSP90β antibodies.
HSP90β inhibition of latent TGF-β1-induced transcription and cell proliferation
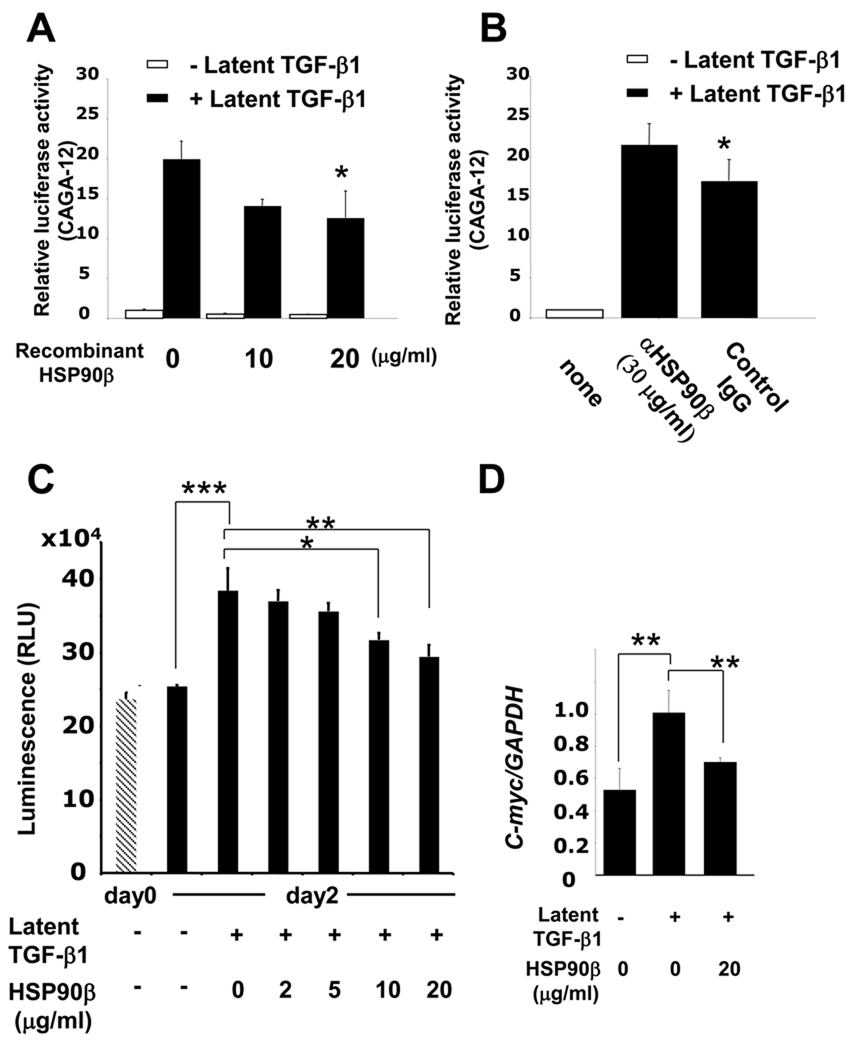
To explore the functional significance of the interaction between HSP90β and LAP, we investigated the effects of HSP90β on latent TGF-β1-mediated transcriptional responses. For this purpose, we used the TGFβ-smad3-responsive reporter CAGA12-Lux assay [18], and found that the MG63 cells treated with recombinant HSP90β (20 µg/ml) displayed a twofold decrease in latent TGF-β1-induced transcription (Fig. 3A). We also investigated the role of endogenous extracellular HSP90β in regulating transcriptional activation by TGF-β1. As shown in Fig. 3B, addition of the HSP90β specific antibody to a conditioned medium resulted in a 1.4-fold increase in TGF-β1-induced transcription, confirming the inhibitory effects of HSP90β. To confirm the inhibitory role of extracellular HSP90β in latent TGF-β1-induced transcription, we analyzed its effect on the latent TGF-β1-induced proliferation of MG63 cells. The cell numbers of MG63 cells were counted 48 h after the addition of latent TGF-β1 with or without HSP90β. As shown in Fig. 3C, cell numbers were significantly increased by latent TGF-β1 treatment at day 2 (1.5-fold). This effect was inhibited by the co-addition of HSP90β in a dose-dependent manner, with a significant inhibition at 10 and 20 µg/ml of HSP90β. Next, we examined mRNA expression of C-myc, a common pro-proliferative factor, in MG63 cells. As shown in Fig. 3D, latent TGF-β1 induced the C-myc expression level approximately twofold. However, the co-addition of HSP90β (20 µg/ml) significantly inhibited C-myc expression.
Figure 3. HSP90β binding to LAP affects latent TGFβ mediated signaling and cell proliferation.
MG63 cells were transfected with (CAGA)12-luciferase reporter plasmid and the pRL-TK control vector. A, cells were treated with recombinant latent TGF-β1 and different concentrations of HSP90β for 24 h before lysis. B, cells were pre-treated with anti-HSP90β antibody or IgG control and then incubated with latent TGF-β1 for 24 h before lysis. C, MG63 cells were treated with 200 ng/ml latent TGF-β1 with various concentrations of recombinant HSP90β. Before and after 48 h of treatment, cell numbers were counted. D, MG63 cells were starved for 24 h and treated with 200 ng/ml latent TGF-β1 with various concentrations of recombinant HSP90β. Total RNA was extracted from the cells at 24 h after treatment and analyzed by quantitative real-time PCR. Each value represents the mean of triplicate determinations; bars, each value represents the mean of triplicate determinations; bars, mean + SD. n=3, *P < 0.05, **P < 0.01, ***P < 0.001.
TGF-β1 increases the cell surface localization of HSP90β in MG63 cells
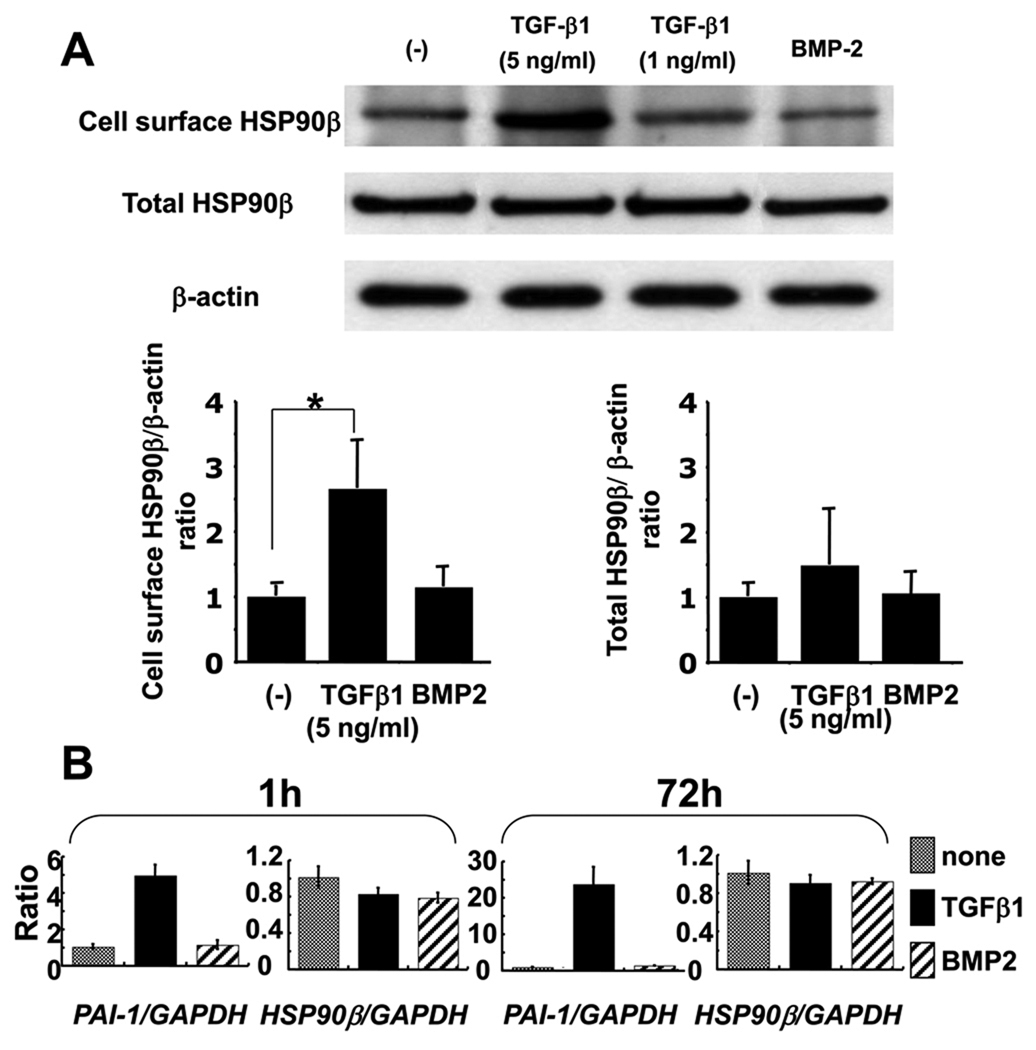
To analyze the effects of TGF-β1 on HSP90β expression in MG63 cells, we treated these cells with mature TGF-β1 and found that the cell surface expression level of HSP90β increased 2.6-fold. However, the total HSP90β expression level was not altered by the addition of TGF-β1. BMP2, another effective growth factor in MG63 cells, did not affect the cell surface or the total HSP90β expression level (Fig. 4A). We then performed real-time PCR analysis to investigate the TGF-β1 dependent regulation of HSP90β expression levels. We first examined PAI-1 expression, which is commonly known to be induced by TGF-β1. As expected, the addition of TGF-β1 (5 ng/ml), PAI-1 expression caused a drastic increase at both 1 h and 72 h after stimulation. However, the HSP90β level remained unchanged by this addition of TGF-β1 (5 ng/ml) at both stages (Fig. 4B).
Figure 4. TGF-β1 regulates cell surface expression of HSP90β.
A, MG63 cells were starved and then incubated with either TGF-β1 (5 or 1 ng/ml) or BMP2 (50 ng/ml). 3 days after incubation, cell surface proteins were biotinylated and cell surface proteins and total proteins were collected and immunoblotted, as described in Figure 2. The amount of the proteins used for cell surface precipitant and total HSP90β were quantified by the amount of β-actin. Each value represents the mean of triplicate determinations; bars, mean + SD. *P < 0.05 B, MG63 cells were starved and then incubated with TGF-β1 (5 ng/ml) or BMP2 (50 ng/ml). Total RNAs were extracted from the cells at 1 h and 72 h after treatment. PAI-1 and HSP90β expression levels were analyzed by quantitative real-time PCR. Each value represents the mean of triplicate determinations; bars, SD.
Discussion
Our studies demonstrate that HSP90β is identified as a LAP binding protein and extracellular HSP90β inhibits latent TGF-β1 activation. Our studies also show that TGF-β1 stimulation of MG63 cells results in an increase in the extracellular HSP90β level. This suggests that HSP90β modulates TGF-β1 signaling through a negative feedback loop in the extracellular compartment. HSP90β is one of the most abundant cellular chaperones required for the ATP-dependent refolding of denatured or “unfolded” proteins, and for the conformational maturation of a variety of key proteins involved in the growth response [19–21]. Interestingly, recent studies showed that HSP90 proteins could also be identified in the cell surface and supernatant of certain cells, and that the secreted HSP90 proteins carry out important extracellular functions, including the enhancement of cell migration, stimulation of immunological cytokine production, and activation of antigen presenting cells [22–26]. However, the kinds of HSPs, as well as their amounts, vary between cell lines. For instance, the fibrosarcoma cell line HT-1080 secretes HSP90α but not HSP90β [22], and the macrophages dominantly express HSP70s [27]. Importantly, there are no reports on HSP secretion by osteosarcoma cell lines. Thus, we report the presence of extracellular HSP90β expression in osteosarcoma-derived MG63 cells for the first time (Fig. 2).
We identified HSP90β as a LAP binding protein by protein-protein interaction assay (Fig. 1), and its binding was confirmed by solid phase binding assay and co-immunoprecipitation assay. We also identified that addition of recombinant HSP90β to culture medium had an inhibitory effect on the TGF-β1 dependent luciferase transcriptional activity and cell proliferation (Fig. 3). In this assay, we must consider the possibility that the exogenous recombinant HSP90β might be endocytosised and somehow influence intracellular TGF-β related molecules, resulting in inhibition of TGF-β1 signaling. However, unlike extracellular HSP90β, intracellular HSP90s are known to activate TGF signaling mainly through chaperoning TGF-β receptors [21]. As per the earlier report, TGF-β induced cell proliferation and C-myc expression in MG63 cells [28]. It is now clear that HSP90β can bind to LAP, resulting in inhibition of latent TGF-β1 activation and cell proliferation in MG63 cells. These facts strongly suggest that extracellular HSP90β directly binds to LAP and inhibits its cleavage, however we cannot exclude the possibility that extracellular HSP90β might inhibit the activity of LAP activators such as TSP-1, MMPs, and integrin, resulting in inhibition of LAP activation. Thus, the mechanisms of how HSP90β inhibits the release of the mature ligand from the latent TGF-β1 complex are still unclear. TSP-1 can bind to the VLAL site of LAP and release a mature ligand by conformational changes in the latent complex [29]. Moreover, knock-in mice with mutations in the RGD site of TGF-β1-LAP recapitulate the phenotypes of TGF-β1 null mice [30]. These observations indicate that LAP has several important sites for activation by other molecules. We believe that extracellular HSP90β might sequester these sites to prevent activators from binding to LAP, which would result in the inhibition of latent TGF-β1 activation.
The expression level of the regulators of TGF-β signaling, such as Smad7 and asporin, is induced by TGF-β1 itself and works as a negative feedback loop [31, 32]. Thus, we considered the possibility that TGF-β1 may somehow affect the expression level or pattern of HSP90β. Interestingly, as shown in Fig. 4, TGF-β1 stimulation increased only the cell surface expression of HSP90β,. In order to elucidate the mechanisms of extracellular HSP90β up-regulation, we first analyzed the change in total HSP90β expression and concluded that TGF-β1 did not affect the total amount of HSP90β protein or mRNA levels. HSP90s are believed to not secrete through the typical secretion pathway due to a lack of signal peptides. Previous studies suggest that HSP90s were secreted into extracellular space through the exosome pathway [33]. We speculate that TGF-β1 stimulation activates HSP90β secretion through the exosome pathway without any change in the expression level of total HSP90β. Actually, in MG63 cells, the mRNA expression level of the tumor suppressor activated pathway-6 (TSAP6) was up-regulated through TGF-β1 stimulation (data not shown). TSAP6 is the transmembrane protein and is believed to be the essential factor in HSPs’ exosome secretion [34].
In summary, we identified HSP90β as an inhibitor of latent TGF-β1 activation. Since TGF-β1 is a potent growth factor, it is natural to speculate that TGF-β1 signaling is modulated by activators and inhibitors at every step. To the best of our knowledge, thus far there are no reports of proteins that inhibit the release of the mature ligand from a latent TGF-β1 complex. Extracellular HSP90β is the first molecule to be identified as an inhibitor of latent TGF-β1 activation. Further investigation of how extracellular HSP90β inhibits latent TGF-β1 activation, as well as which domains of HSP90β are critical for its inhibitory effect, may provide better insight into the precise roles of extracellular HSP90β.
Acknowledgements
We would like to thank Drs. Kathy Flanders, Lalage Wakefield and Yoshihiko Yamada for their critical reading of this manuscript, and Shelagh Powers for expert editorial assistance. This work was supported by the Intramural Division of the National Institute of Dental and Craniofacial Research, National Institutes of Health.
Abbreviations
- TGF-β1
Transforming growth factor-β 1
- LAP
Latency-associated peptide
- HSP90β
Heat shock protein 90β
References
- 1.Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acd Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moses HL, Serra R. Regulation of differentiation by TGF-beta. Curr Opin Genet Dev. 1996;6:581–586. doi: 10.1016/s0959-437x(96)80087-6. [DOI] [PubMed] [Google Scholar]
- 3.Massagué J, Blain SW, Lom RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;13:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 4.Verrecchia F, Mauviel A. Transforming growth factor-β signaling through the SMAD pathway: role in extracellular matrix gene expression and regulation. J Invest Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
- 5.Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 6.Seyedin SM, Thomas TC, Thompson AY, Rosen DM, Piez KA. Purification and characterization of two cartilage-inducing factors from bovine demineralized bone. Proc Natl Acd Sci USA. 1985;82:2267–2271. doi: 10.1073/pnas.82.8.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssens K, ten Dijke P, Janssens S, Van HW. Transforming growth factor-beta1 to the bone. Endocr Rev. 2005;26:743–774. doi: 10.1210/er.2004-0001. [DOI] [PubMed] [Google Scholar]
- 8.Franchi A, Arganini L, Baroni G, Calzolari A, Capanna R, Campanacci D, Caldora P, Masi L, Brand ML, Zampi G. Expression of transforming growth factor beta isoforms in osteosarcoma variants: association of TGF beta 1 with high-grade osteosarcomas. J Pathol. 1998;185:284–289. doi: 10.1002/(SICI)1096-9896(199807)185:3<284::AID-PATH94>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 9.Kloen P, Gebhardt MC, Perez-Atayde A, Rosenberg AE, Springfield DS, Gold LI, Mankin HJ. Expression of transforming growth factor-beta (TGF-beta) isoforms in osteosarcomas: TGF-beta3 is related to disease progression. Cancer. 1997;80:2230–2239. [PubMed] [Google Scholar]
- 10.Gentry LE, Lioubin MN, Purchio AF, Marquardt H. Molecular events in the processing of recombinant type 1 pre-pro-transforming growth factor beta to the mature polypeptide. Mol Cell Biol. 1988;8:4162–4168. doi: 10.1128/mcb.8.10.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazono K, Hellman U, Wernstedt C, Heldin CH. Latent high molecular weight complex of transforming growth factor beta 1. Purification from human platelets and structural characterization. J Bio Chem. 1988;263:6407–6415. [PubMed] [Google Scholar]
- 12.Wakefield LM, Smith DM, Flanders KC, Sporn MB. Latent transforming growth factor-beta from human platelets. A high molecular weight complex containing precursor sequences. J Bio Chem. 1988;263:7646–7654. [PubMed] [Google Scholar]
- 13.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 14.Blakytny R, Ludlow A, Martin GE, Ireland G, Lundm LR, Ferguson MW, Brunner G. Latent TGF-β1 activation by platelets. J Cell Physiol. 2004;199:67–76. doi: 10.1002/jcp.10454. [DOI] [PubMed] [Google Scholar]
- 15.Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165:723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hozumi K, Suzuki N, Nielsen PK, Nomizu M, Yamada Y. Laminin alpha1 chain LG4 module promotes cell attachment through syndecans and cell spreading through integrin alpha2beta1. J Biol Chem. 2006;281:32929–32940. doi: 10.1074/jbc.M605708200. [DOI] [PubMed] [Google Scholar]
- 17.de Vega S, Iwamoto T, Nakamura T, Hozumi K, McKnight DA, Fisher LW, Fukumoto S, Yamada Y. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. J Biol Chem. 2007;282:30878–30888. doi: 10.1074/jbc.M705847200. [DOI] [PubMed] [Google Scholar]
- 18.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearl LH, Prodromou C. Structure, function, and mechanism of the Hsp90 molecular chaperone. Adv Protein Chem. 2001;59:157–186. doi: 10.1016/s0065-3233(01)59005-1. [DOI] [PubMed] [Google Scholar]
- 20.Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrighton KH, Lin X, Feng XH. Critical role of chaperone HSP90 in TGF-β signaling. Proc Natl Acd Sci USA. 2008;105:9244–9249. doi: 10.1073/pnas.0800163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eustace BK, Sakurai T, Stewart JK, Yimlamai D, Unger C, Zehetmeier C, Lain B, Torella C, Henning SW, Beste G, Scroggins BT, Neckers L, Ilag LL, Jay DG. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6:507–514. doi: 10.1038/ncb1131. [DOI] [PubMed] [Google Scholar]
- 23.Sidera K, Samiotaki M, Yfanti E, Panayotou G, Pastsavoudi E. Involvement of cell surface HSP90 in cell migration reveals a novel role in the developing nervous system. J Bio Chem. 2004;279:45379–45388. doi: 10.1074/jbc.M405486200. [DOI] [PubMed] [Google Scholar]
- 24.Becker B, Multhoff G, Farkas B, Wild PJ, Landthaler M, Stolz W, Vogt T. Induction of Hsp90 protein expression in malignant melanomas and melanoma metastases. Exp Dermatol. 2004;13:27–32. doi: 10.1111/j.0906-6705.2004.00114.x. [DOI] [PubMed] [Google Scholar]
- 25.Calderwood SK, Mambula SS, Gray PJ, Theriault JR. Extracellular heat shock proteins in cell signaling. FEBS Lett. 2007;581:3689–3694. doi: 10.1016/j.febslet.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 26.Cheng CF, Fan J, Fedesco M, Guan S, Li Y, Bandyopadhyay B, Bright AM, Yerushalmi D, Liang M, Chen M, Han YP, Woodley DT, Li W. Transforming growth factor alpha (TGFalpha)-stimulated secretion of HSP90alpha: using the receptor LRP-1/CD91 to promote human skin cell migration against a TGF beta-rich environment during wound healing. Mol Cell Biol. 2008;28:3344–3358. doi: 10.1128/MCB.01287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker T, Hartl FU, Wieland F. CD40, an extracellular receptor for binding and uptake of Hsp70-peptide complexes. J Cell Biol. 2002;158:1277–1285. doi: 10.1083/jcb.200208083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuyama S, Iwadate M, Kondo M, Saitoh M, Hanyu A, Shimizu K, Aburatani H, Mishima HK, Imamura T, Miyazono K, Miyazawa K. SB-431542 and Gleevec inhibit transforming growth factor-beta-induced proliferation of human osteosarcoma cells. Cancer Res. 2003;63:7791–7798. [PubMed] [Google Scholar]
- 29.Young GD, Murphy-Ullrich JE. The tryptophan-rich motifs of the thrombospondin type 1 repeats bind VLAL motifs in the latent transforming growth factor-beta complex. J Biol Chem. 2004;279:47633–47642. doi: 10.1074/jbc.M404918200. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Mu Z, Dabovic B, Jurukovski V, Yu D, Sung J, Xiong X, Mungerm JS. Absence of integrin-mediated TGFbeta1 activation in vivo recapitulates the phenotype of TGFbeta1-null mice. J Cell Biol. 2007;176:787–793. doi: 10.1083/jcb.200611044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakao A, Afrakhte M, Morén A, Nakayama T, Christian JL, Heuchei R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGF-β-inducible antagonist of TGF-β signaling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima M, Kizawa H, Saitoh M, Kou I, Miyazono K, Ikegawa S. Mechanisms for asporin function and regulation in articular cartilage. J Biol Chem. 2007;282:32185–32192. doi: 10.1074/jbc.M700522200. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, Harris SL, Levine AJ. The Regulation of Exosome Secretion: a Novel Function of the p53 Protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 34.Amzallag N, Passer BJ, Allanic D, Segura E, Théry C, Goud B, Amson R, Telerman A. TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. J Bio Chem. 2004;279:46104–46112. doi: 10.1074/jbc.M404850200. [DOI] [PubMed] [Google Scholar]