Abstract
The DM domain proteins Doublesex- and MAB-3–related transcription factors (DMRTs) are widely conserved in metazoan sex determination and sexual differentiation. One of these proteins, DMRT1, plays diverse and essential roles in development of the vertebrate testis. In mammals DMRT1 is expressed and required in both germ cells and their supporting Sertoli cells. Despite its critical role in testicular development, little is known about how DMRT1 functions as a transcription factor or what genes it binds and regulates. We combined ChIP methods with conditional gene targeting and mRNA expression analysis and identified almost 1,400 promoter-proximal regions bound by DMRT1 in the juvenile mouse testis and determined how expression of the associated mRNAs is affected when Dmrt1 is selectively mutated in germ cells or Sertoli cells. These analyses revealed that DMRT1 is a bifunctional transcriptional regulator, activating some genes and repressing others. ChIP analysis using conditional mutant testes showed that DNA binding and transcriptional regulation of individual target genes can differ between germ cells and Sertoli cells. Genes bound by DMRT1 in vivo were enriched for a motif closely resembling the sequence DMRT1 prefers in vitro. Differential response of genes to loss of DMRT1 corresponded to differences in the enriched motif, suggesting that other transacting factors may modulate DMRT1 activity. DMRT1 bound its own promoter and those of six other Dmrt genes, indicating auto- and cross-regulation of these genes. Many of the DMRT1 target genes identified here are known to be important for a variety of functions in testicular development; the others are candidates for further investigation.
Keywords: chromatin immunoprecipitation, DM domain, development, transcription
Proteins related to Doublesex (DSX) of Drosophila control sex determination and sexual differentiation in a broad array of metazoan animals and comprise the only family of deeply conserved sexual regulators so far identified (1). These proteins share the DM domain, a highly intertwined zinc finger DNA-binding motif first identified in DSX and the related Caenorhabditis elegans sexual regulator MAB-3 (male abnormal 3) (2–4). The vertebrate DM domain gene Dmrt1 and its close orthologs act as primary sex-determining genes in vertebrate clades including fish, amphibians, and probably birds, each with an independently evolved chromosomal sex determination mechanism (5–7). Thus Dmrt1 homologs frequently are recruited or retained to determine sex as new sex determination mechanisms arise. The targets of DM domain gene regulation also have important evolutionary roles: In insects the evolution of cis-regulatory elements bound by DSX appears to have driven divergence of sex-specific characters and thus may help drive speciation (8, 9).
Functional studies in the mouse have demonstrated that Doublesex and MAB-3 related transcription factor 1 (DMRT1) is essential for testicular differentiation (10). In mice DMRT1 is expressed in premeiotic germ cells and in Sertoli cells (the somatic “nurse” cells for the male germ line). Cell type-specific gene targeting revealed requirements for Dmrt1 in both cell types and showed that DMRT1 controls many aspects of testicular development, including differentiation, proliferation, migration, and pluripotency of germ cells, and also proliferation and differentiation of Sertoli cells (11, 12).
Despite the critical importance of DM domain genes in metazoan development, relatively little is known of how they regulate transcription. DNA binding-site preferences have been determined in vitro for a number of DM domain proteins, and most bind very similar consensus elements, generally as homodimers or heterodimers with other DM domain proteins (13–15). MAB-3 acts as a transcriptional repressor, as does the male isoform of DSX, but the female isoform of DSX can activate transcription (9, 14, 16). A fusion of the C-terminal domain of Xenopus DMRT1 to the yeast GAL4 DNA-binding domain can activate reporter gene transcription in transfected cells, whereas in the fish Medaka Dmrt1a and its paralog Dmrt1bY/DMY can repress reporter transcription in transfected cells (17, 18). Finding the in vivo targets of DMRT1 and determining how their expression is regulated are critical to a better understanding of how DMRT1 directs testicular development and function.
Here we have investigated transcriptional regulation by DMRT1 in cultured mammalian cells and in vivo in the mouse testis. In transfected cells DMRT1 caused different transcriptional responses depending on promoter context and cell type; we therefore focused on DMRT1 regulation in vivo. Using ChIP and microarray analysis (“ChIP-chip”), we identified peaks of DMRT1 binding associated with promoter-proximal regions of 1,439 genes in the juvenile mouse testis and used motif searches to identify sequence elements tightly associated with DMRT1 binding. By combining conditional targeting of Dmrt1 in germ cells and Sertoli cells with ChIP and mRNA expression analysis, we examined cell type-specific DNA binding and regulation of target genes. The genes regulated by DMRT1 include many known testis regulators and candidate regulators, and we found that genes in several canonical pathways were enriched among DMRT1 targets. Given the conserved role of DM domain genes in sexual differentiation across a broad swath of metazoans and the almost complete lack of known downstream targets, finding the targets of DMRT1 and understanding its mode(s) of transcriptional regulation should have wide implications for understanding the control and evolution of sexual differentiation.
Results
DMRT1 Can Activate or Repress Reporter Transcription in Cultured Cells.
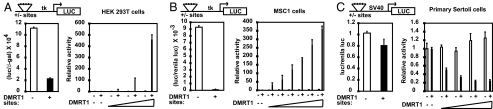
To investigate whether DMRT1 can activate or repress transcription, we used reporter plasmids containing four copies of an in vitro-defined DMRT1 consensus binding site (15) and performed transient transfections into immortalized and primary cell types. In HEK293 cells, addition of DMRT1 binding sites to an HSV thymidine kinase TATA box reporter reduced the basal activity by about 4-fold, indicating repression by an endogenous activity (Fig. 1A). Adding DMRT1 very strongly and specifically activated the reporter containing DMRT1 binding sites (by up to 400-fold), indicating that activation depends on the presence of both DMRT1 and its binding sites. In a mouse Sertoli cell line (MSC1), transcription of the reporter with binding sites was repressed more strongly by an endogenous activity. In these cells DMRT1 also caused elevated transcription of the reporter containing DMRT1 binding sites, but the amounts of DMRT1 tested did not elevate transcription levels beyond the background activity of the reporter lacking binding sites (Fig. 1B).
Fig. 1.
Transcriptional regulation by DMRT1 in transfected cells. (A) (Left) Luciferase expression in 293T cells transfected with 200 ng pLUCMCS luciferase reporter with or without DMRT1 binding sites. (Right) 293T cells transfected with each reporter alone (−) or with 0.02 ng, 0.1 ng, or 0.5 ng of DMRT1 expression vector. (B) MSC1 cells. Experiment as in A except amounts of DMRT1 expression vector in the right panel are 0 ng, 0.5 ng, 1 ng, 2.5 ng, 5 ng, 10 ng, or 50 ng. (C) Primary rat Sertoli cells. Experiment as in A except 100 ng of pGL3promoter reporter was used and amounts of DMRT1 expression vector are 2 ng, 5 ng, 10 ng, and 25 ng. In the right panels in A–C, reporter expression in the absence of exogenous DMRT1 is set to 1.
Because the reporter behaved differently in the two immortalized cell types, we turned to primary rat Sertoli cells. In these cells DMRT1 had no effect on transcription of the reporter with the HSV TATA box but repressed a reporter containing the SV40 minimal promoter and four DMRT1 binding sites (Fig. 1C). From these data we conclude that DMRT1 can activate or repress transcription and that its regulatory activity is highly sensitive to both cell type and promoter context. These results highlight the importance of analyzing DMRT1 activity in vivo.
Promoter-Proximal Regions Bound in Vivo by DMRT1 in the Juvenile Mouse Testis.
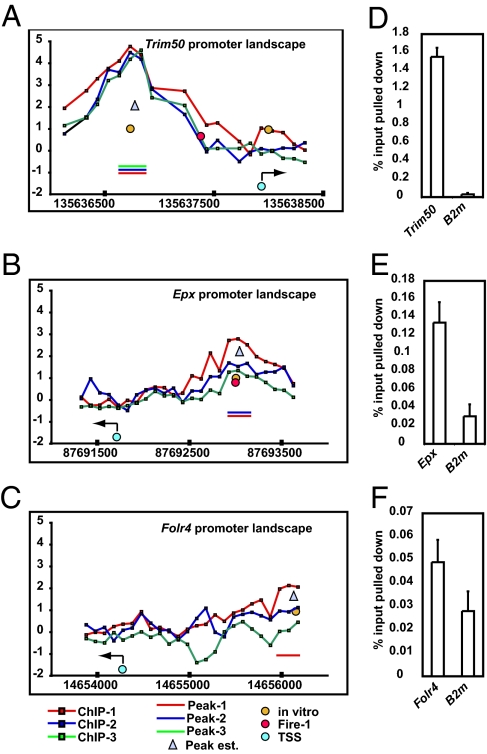
When this work was initiated, no direct targets of DMRT1 regulation had been reported in any species. We therefore used ChIP to identify genes bound by DMRT1 in the mouse testis at postnatal day 9 (P9), when DMRT1 is highly expressed in both Sertoli and germ cells. We first used ChIP display (19) to identify a small number of candidate target sites. Among these target sites was the androgen receptor (Ar), which is misregulated in Dmrt1 mutant testes (11). We confirmed the binding of DMRT1 to the Ar promoter by quantitative ChIP (qChIP) and used this interaction to optimize ChIP conditions. We then performed three independent ChIP-chip experiments, hybridizing DMRT1-associated chromatin fragments to a mouse promoter microarray containing 2.5 kb for each of 17,354 promoter-proximal regions in the mouse genome. To identify genes bound by DMRT1, we used a sliding window to detect DMRT1-specific enrichment present in promoter-proximal regions at three consecutive probes in all three replicates (n = 706), in two of the replicates (n = 292), or in just one experiment (n = 374). A representative example of each type is shown in Fig. 2 A–C. In total, this analysis identified 1,372 regions of significant DMRT1 association. After accounting for bidirectional promoters and multiple transcription start sites, this identification represents association with the proximal promoters of 1,439 unique genes (Materials and Methods and Dataset S1). We validated the ChIP-chip data by qChIP analysis of 42 promoter-proximal regions (detected in one, two, or three replicates) and were able to confirm all of them (Fig. 2 D–F and Fig. S1). Strikingly, among the genes whose promoters were bound by DMRT1 in multiple replicates were Dmrt3, Dmrt5 (Dmrta2), Dmrt7 (Dmrtc2), Dmrt8.1 (4921520P21Rik), and Dmrt8.2, as well as Dmrt1 itself. Dmrt4 (Dmrta1) was bound in one replicate; Dmrt6 (Dmrtb1) was not represented on the array. These data together with our previous finding that DMRT proteins bind very similar DNA motifs (15) strongly suggest cross-regulation and autoregulation in this gene family.
Fig. 2.
Binding of DMRT1 to promoter-proximal sequences in the testis. (A–C) Representative ChIP-chip data from three independent ChIP-chip experiments in P9 testes (ChIP-1, -2, and -3) showing promoter-proximal regions with peaks called from all three experiments (A), from two experiments (B), or from one experiment (C). The y axis is log(2) enrichment of ChIP sample relative to input at each probe position. Positions of called peaks are indicated by horizontal bars; transcription start (TSS) is shown with the direction of transcription is indicated by an arrow. Fire-1: locations of motifs defined by FIRE that resemble the in vitro derived DMRT1 binding site; in vitro: Profit-defined, 75% or better, matches to the DMRT1 in vitro DNA-binding consensus; Peak est: average position of called peak centers. (D–F) DMRT1 qChIP confirmation of ChIP-chip peaks. Negative control was b2-microglobulin (B2m), a gene expressed in both Sertoli and germ cells at P9 but not bound by DMRT1.
DMRT1 DNA Binding in Vivo Is Mediated by a Specific Motif.
DMRT1 could be recruited to DNA by direct binding of a DNA recognition element or indirectly through interaction with another DNA-associated protein. To investigate whether target genes contain a consensus motif related to the previously determined DMRT1 in vitro consensus element, we used the Profit algorithm (http://bioweb.pasteur.fr/docs/EMBOSS/profit.html). We searched all promoter-proximal regions (Dataset S1), using a weighted matrix from the in vitro DNA binding-site selection (15). Seventy-five percent of promoter-proximal regions bound by DMRT1 in all three experiments contained a motif with at least 75% similarity to the in vitro-derived binding site. The DMRT1-bound promoter regions were unambiguously enriched for in vitro binding motifs relative to promoter regions not bound by DMRT1 (P = 2.06 × 10−80). We also examined promoters bound only in one or two of the experiments and found that these promoters also show enrichment of the in vitro element, although with lesser P values (Table S1).
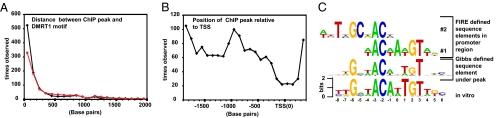
To test further the relevance of the in vitro motif, we asked whether this sequence is closely associated with peaks of DMRT1 binding. We measured the distance between the peak center and the nearest predicted in vitro motif for each of the 1,039 peaks of DMRT1 binding that contained a motif (Fig. 3A) and found that in vitro motifs were closely associated with peaks of DMRT1 binding. We also examined the location of each peak center relative to the transcription start site for each bound promoter. There was no positional bias of DMRT1 binding except against the transcription start site (Fig. 3B).
Fig. 3.
DMRT1 binding and enriched DNA motifs. (A) Distances from DMRT1 binding peak estimate to nearest Profit consensus element, binned in 100-bp increments (black). Distances from DMRT1 binding peak estimate to highest scoring FIRE motif (FIRE-1 shown in C; z-score: 215), binned in 100-bp increments (red). (B) Position of DMRT1 binding peak estimates relative to the transcription start, binned in 100-bp increments. (C) Alignment of the in vitro-defined DMRT1 site to the Gibbs-defined motif and the two most strongly enriched FIRE-defined motifs enriched within DMRT1-bound promoter-proximal regions.
We next took an unbiased approach, using the Gibbs Recursive Sampler algorithm (20) to find sequence motifs enriched within 250 bp of the center of DMRT1 binding peaks. This approach detected a motif almost identical to the in vitro-defined motif in 77% of DMRT1 binding peaks (Fig. 3C). The only enriched motif falling under the peak of DMRT1 binding is a close match to the DMRT1 in vitro binding consensus, suggesting that DMRT1 is recruited primarily to target genes by direct association with its previously defined DNA element. However, we cannot exclude the possibility that DMRT1 is recruited indirectly to some regulatory elements by association with other less common motifs that would not have been found by the Gibbs Recursive Sampler.
To find additional motifs associated with DMRT1 regulation, we next used the FIRE algorithm (21) to search the entire 2,500 bp of each bound promoter-proximal region. FIRE compares the bound promoter regions with the nonbound promoter regions. This comparison identified two motifs that overlap the DMRT1 in vitro consensus element (Fig. 3C). Computational time constraints limited FIRE searches to eight-nucleotide motifs, and this limitation probably is why FIRE detected two overlapping motifs. We calculated the distance from the most enriched motif (FIRE-1) to the peak center and found that it was associated closely with DMRT1 binding peaks (Fig. 3A). FIRE also detected a GC-rich element, suggesting that DMRT1 target genes are enriched for CpG islands, and indeed the bound genes were more likely to have CpG islands than would be expected by chance (P = 2.2 × 10−24; Materials and Methods).
DMRT1 Target Genes Regulated at P9.
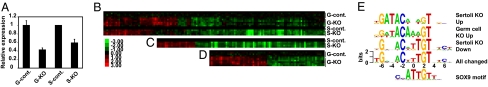
To narrow the analysis of DMRT1 targets to those most likely to mediate juvenile testis development, we asked which genes bound by DMRT1 at P9 display abnormal expression in Dmrt1 mutant testes at this stage. DMRT1 is expressed in Sertoli cells and germ cells and is required in both (10–12). To examine transcriptional regulation by DMRT1 in each cell type, we used conditional gene targeting to delete Dmrt1 separately in germ cells [Ngn3-cre, active in a fraction of neonatal spermatogonia (22)] or Sertoli cells [Dhh-cre, active in embryonic Sertoli cells (23)]. We then performed microarray-based mRNA expression profiling of 23,849 transcripts and identified mRNAs with expression changes of at least 1.5-fold (P < 0.05) between control and cre testes. Total Dmrt1 expression was reduced significantly in each cell type-specific mutant (Fig. 4A). Deletion of Dmrt1 in Sertoli cells affected 852 mRNAs (3.6%). and deletion in germ cells affected 759 mRNAs (3.2%) (Fig. S2 A and B). Among genes represented on both the expression and the promoter microarrays (18,141), we found that 146 of 1,434 DMRT1-bound genes (10%) were misexpressed when Dmrt1 was deleted in either cell type at P9 (Fig. 4B and Fig. S3). Of these 146 genes, 32 (22%) were misexpressed in both cell type-specific mutants. Conversely, of the transcripts misexpressed in either cell type when Dmrt1 is deleted, 146 of 1,432 (10%) had DMRT1 associated with their 5′ proximal region. Quantitative RT-PCR (qRT-PCR) for a representative set of DMRT1 target transcripts confirmed 19 of 20 expression changes (Fig. S4).
Fig. 4.
Misexpressed DMRT1 target genes at P9 and motifs enriched near DMRT1 peak estimates. (A) Expression of Dmrt1 in mutant testes germ knockouts (G-KO) and Sertoli cell knockouts (S-KO) compared with controls (G-cont and S-cont). (B) Compilation heat map of genes that are misexpressed in testes mutant for Dmrt1 in Sertoli cells, germ cells, or both. (C) Heat map of genes that are misexpressed in testes mutant for Dmrt1 in Sertoli cells. (D) Heat map of genes that are misexpressed in testes mutant for Dmrt1 in germ cells. Thresholds for inclusion: at least 1.5-fold difference between control and Cre testes and P < 0.05. High-resolution versions with gene names are shown in Fig. S3. (E) Motifs found to be enriched within 250 bp of the DMRT1 peak estimates by the Gibbs Recursive Sampler in the different classes of misregulated DMRT1 target genes. From top to bottom, these motifs were found in 54%, 53%, 52%, and 77% of promoter regions queried. No enriched motif was found for the germ cell knockout down class. Bottom motif is SOX9 binding consensus (31).
Functional Classes of DMRT1 Target Genes.
To investigate whether DMRT1 direct targets are enriched for specific functional classes, we used Ingenuity pathway analysis. This analysis revealed significant enrichment in several canonical signaling pathways, including androgen, focal adhesion kinase, IL-6, phosphatase and tensin homolog (PTEN), and insulin-like growth factor 1 (IGF-1) (Fig. S5 and Table S2). The association of these canonical pathways with DMRT1 DNA binding is consistent with previous functional analyses showing that androgen receptor expression is reduced in Dmrt1 mutant testes and that Dmrt1 mutant germ cells have abnormal pluripotency, a property that is also controlled by PTEN and IGF-1 (24, 25).
We also examined the DMRT1 targets manually to explore which cellular processes DMRT1 directly regulates to control testis development and function (Table 1). These targets included genes involved in many aspects of cell biology, cell signaling, and regulation of gene expression, including many known to be required for testicular development or function. Only a subset of these genes (written in bold in Table 1) was misregulated at P9, but many are likely to be misregulated at other stages. Indeed, we have found that changes in some of these genes (e.g., Sox2, Cdkn2d/p19INK4d) likely underlie Dmrt1 phenotypes in the embryonic testis (12).
Table 1.
Functional classes of DMRT1-bound genes
| Functional class | DMRT1-bound genes |
| Cellular junction formation and function | Claudin 11, Vinculin, Gja3,Gje1,lgsf4a(Cadm1) |
| Redox regulation | Pdla5, P4ha1, Gsta3, Gsta4 |
| Regulation of steroid metabolism and function | Osbpl1a, Pkarsa, Dhcr7,Sex hormone binding globulin (Shbg) |
| Nuclear hormone receptors | Ar, Sf1 (Nr5a1), LXRalpha (Nr1h3), Fxrb (Nr1h5) |
| Nuclear hormone receptor coactivators | Ncoa2 and Ncoa4 |
| Retinoic acid metabolism and response | Adh4, Aldh1a1, Stra8 |
| Pluripotency regulators | Sox2 and Utf1 |
| Insulin related growth factor signaling | Igf1r, Igf2r, Igfbp3, Igfbp6 |
| FGF signaling | Fgf10, Fgf18,Fgfr2 |
| Cell cycle regulators | Cdk2, cdk7, Cdkn2c, Cdkn2d, Chek1, E2f1, E2f4, E2f8 |
| Matrix metalloproteases | Adamst19,Adamst20, Mmp16, Mmp24 |
| MAP kinases | Map2k3, Map2k6, Map3k7ip1, Mapk1, Mapk4, Mapkapk3 |
| Hox gene function | Meis1, Hoxb13, Hoxb4,Hoxb7, Hoxc4, Hoxc9, Hoxd1 |
| Dmrt family members | Dmrt1, Dmrt3,Dmrt4 (Dmrta1), Dmrt5 (Dmrta2), Dmrt7 (Dmrtc2),Dmrt8.1(4921520P21Rik), andDmrt8.2 |
| Others | Sohlh1, Bmpr1a, Vrk1, Gfra3, col4a3 |
Genes in bold are misregulated at P9.
DMRT1 Activates Transcription of Some Direct Target Genes and Represses Others.
We next examined the data to investigate whether DMRT1 regulates its direct targets by activation, repression, or both and whether regulation is similar in Sertoli cells and germ cells. Deleting Dmrt1 in either cell type caused some DMRT1-associated genes to increase in mRNA expression and others to decrease (Fig. 4B and Fig. S3A). This finding strongly suggests that in both cell types DMRT1 activates transcription of some genes and represses others. In Sertoli cells, more putative direct targets decreased than increased in expression upon removal of Dmrt1, suggesting that DMRT1 primarily activates transcription in this cell type (Fig. 4C and Fig. S3B). In germ cells we detected fewer putative target genes with altered expression (Fig. 4D and Fig. S3C), probably because Ngn3-cre is not active in some of the undifferentiated spermatogonia that express DMRT1 (22); nevertheless, loss of DMRT1 caused gain or loss of expression of significant numbers of genes in germ cells at this stage.
Correlation Between DMRT1 Transcriptional Activity and Associated DNA Motifs.
To investigate whether the sequence of the DMRT1 binding site may influence DMRT1 transcriptional activity and cell-type specificity, we used the Gibbs Recursive Sampler (Materials and Methods and Fig. 4E). Genes with decreased expression in the Sertoli cell mutant had an enriched motif very similar to that enriched in the complete set of binding peaks. Genes with increased expression in the Sertoli or germ cell mutants also had a close match to this motif, but with extra preferences at positions −4 and −5. The motif associated with genes whose expression increased in mutant Sertoli cells contained a potential binding site for GATA binding protein 1 (GATA1) (GATAC) which is bound well by GATA1 in vitro (26). This analysis suggests that differences in the binding site may underlie the difference in regulation by DMRT1, perhaps by altering its DNA-binding mode or association with coregulators.
Cell Type Specificity of Binding and Transcriptional Regulation by DMRT1.
We next asked whether DMRT1 regulates individual genes in the same direction in Sertoli cells and germ cells. Many misexpressed direct targets (31/146; 21%) showed either increased or decreased expression in both cell types upon loss of DMRT1. Although most mRNAs were significantly affected in only one cell type at the expression and significance thresholds chosen, the majority of genes showed the same direction of misexpression (Fig. 4B and Fig. S3A). Several genes showed opposite changes in expression when DMRT1 was deleted in the two cell types, but only one of these, Emb, met our significance thresholds in both cell types. This result indicates that DMRT1 not only can activate or repress transcription in both cell types, as shown above, but also may affect transcription of an individual gene differentially in the two cell types.
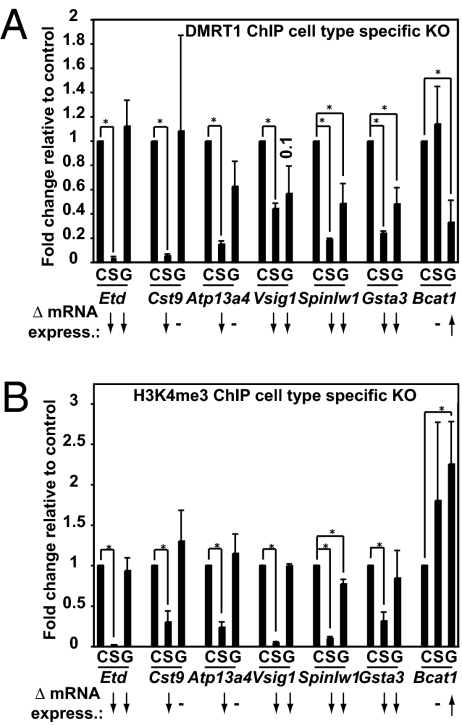
To investigate whether cell type-specific regulation by DMRT1 could result from cell type-specific DNA binding, we, used qChIP to analyze conditionally targeted testes at P9. As described above, 31 genes showing misexpression when DMRT1 was mutated were affected similarly in both cell types. For such genes (Vsig1, Spinlw1, Gsta3), removing DMRT1 from either cell type reduced binding, indicating that these promoters are bound by DMRT1 in both cell types (Fig. 5A). DMRT1 bound other genes only in Sertoli cells (Etd, Cst9, Atp13a4) or germ cells (Bcat1), based on specific reduction of DMRT1 binding in Dhh-cre– or Ngn3-cre–targeted testes, respectively. Because Ngn3-cre targets only a fraction of germ cells at P9, even when loss of DMRT1 binding in germ cells was highly specific, the loss was not complete. We conclude that cell type-specific binding of DMRT1 can explain some cases of differential regulation in the two cell types. In the case of Etd, DMRT1 binding appeared to be Sertoli cell specific, but expression changes were detected in both cell types (Fig. 5A). This finding indicates either cell nonautonomous effects or the presence of germ cell-specific DMRT1 binding sites outside the region examined. This issue can be resolved when a more efficient germ cell cre transgene is available and ChIP-seq is used to analyze a broader region of DNA.
Fig. 5.
DMRT1 and H3K4me3 ChIP. (A) qChIP showing the effect on binding of DMRT1 to specific promoters upon deletion of Dmrt1 from either Sertoli cells (S) or germ cells (G) relative to control (C, undeleted Dmrt1flox/flox) testis. (B) qChIP showing the effect on H3K4me3 at specific promoters upon deletion of Dmrt1 from either Sertoli cells (S) or germ cells (G) relative to control (C, undeleted Dmrt1flox/flox) testis. Arrows indicate the observed significant expression changes. *P < 0.05.
We next asked whether DMRT1-dependent changes in gene expression correlate with chromatin modification, focusing on H3K4me3 as an example of a modification commonly associated with transcriptional activation (Fig. 5B). We examined four genes (Etd, Cst9, Atp13a4, and Vsig1) that were bound primarily in Sertoli cells and strongly down-regulated by loss of DMRT1 in Sertoli cells. In each case, deletion of DMRT1 in Sertoli but not germ cells caused a dramatic loss of H3K4me3 association in the region 100–200 bp 3′ to the transcriptional start site. We also examined two genes (Spinlw1 and Gsta3) whose expression was reduced by DMRT1 loss in both cell types. For these genes, H3K4me3 association was reduced severely in mutant Sertoli cells but was reduced only modestly in mutant germ cells, probably because of the presence of wild-type undifferentiated spermatogonia in the Ngn3-cre deletion mutant. We examined one gene (Bcat) whose expression was up-regulated specifically in germ cells and found that H3K4me3 association increased specifically upon deletion of Dmrt1 in germ cells. Thus DMRT1-dependent changes in gene expression correlate closely with changes in an activating chromatin mark.
Discussion
We have analyzed the transcriptional function of DMRT1 in cultured cells and in vivo and used ChIP to identify promoter-proximal regions bound by DMRT1 in vivo. These results permit a number of general conclusions about how DMRT1 regulates testicular gene expression and highlight questions for future investigation. At P9 DMRT1 bound to promoter-proximal regions of 1,439 genes, out of 17,354 promoter-proximal regions queried. The data presented here already have provided an important resource for investigation of testicular development and function (12), and we provide public access to these data in the form of an interactive database at http://www.dmrt1.umn.edu.
The number of promoter-proximal regions bound by DMRT1 in the neonatal testis might seem large. Some binding sites may not be functionally relevant, but phenotypic analysis has shown that DMRT1 controls many separate aspects of testicular development and function (10–12). The large number of genes bound by DMRT1 and the diversity of functional classes to which they belong suggest that many of these processes are controlled directly by DMRT1. As detailed above, DMRT1 directly regulates genes required for Sertoli cell differentiation, cell-cycle control, tight-junction dynamics, germ cell differentiation, and pluripotency. Other direct and indirect targets doubtless mediate DMRT1 function not only in the juvenile testis but also in the embryonic and adult testis.
Most of the Dmrt genes were bound by DMRT1, and several (Dmrt4/Dmrta1, Dmrt6/Dmrtb1, Dmrt8.1/4921520P21Rik, Dmrt8.2) were misregulated in mutant testes, supporting the idea that these genes may cross-regulate extensively. This regulation may be conserved in vertebrates, because expression of Dmrt1bY appears to be negatively autoregulated and cross-regulated by its autosomal paralog Dmrt1a in Medaka (18).
Reporter analysis in cultured cells indicated that DMRT1 can activate or repress transcription depending on cell type and promoter structure. This variability underscores the importance of analyzing transcriptional regulation in the intact organ. We found that DMRT1 both activates and represses transcription in vivo, based on loss or gain of expression of direct targets in Dmrt1 mutant testes. Most genes had the same response to loss of DMRT1 in germ cells and Sertoli cells, but a small number were regulated differentially in the two cell types, and some genes exhibited DMRT1 binding only in one cell type. These results strongly imply that other transacting factors modulate the binding and activity of DMRT1 to achieve gene-specific and cell type-specific function. These factors may be other DNA-binding transcriptional regulators, modifiers of DMRT1 (e.g., kinases), chromatin-modifying enzymes, or a combination of these factors. An important future goal will be to find these factors.
Are other DM domain proteins bifunctional transcriptional regulators? Although DM domain proteins occur in a wide range of metazoans, target genes have been identified only for DSX and MAB-3. DSX is alternatively spliced into sex-specific isoforms. There is evidence that these isoforms have opposite activities, with DSX-F activating and DSX-M repressing transcription, whereas MAB-3 appears exclusively to repress transcription (9, 14, 16). However, only a handful of DSX and MAB-3 targets have been identified, so it is unclear whether any of these proteins has additional activity. Thus, although there is precedence for activation or repression by DM domain proteins, so far only DMRT1 has been shown to be bifunctional.
Analysis of DMRT1-associated regulatory regions identified at least one close match to the in vitro-derived DNA-binding motif in 70% of these regions. In addition Gibbs motif analyses identified at least one motif closely related to the in vitro-defined DMRT1 consensus element under 77% of peaks of DMRT1 association. This result suggests that DMRT1 binds DNA mainly by direct interaction, primarily via the previously identified binding motif. DMRT1 may be recruited to the other sites of association by indirect binding or via direct binding to DNA motifs that do not closely match the in vitro consensus. The ability of DMRT1 to heterodimerize on DNA with other DM domain proteins (15) raises the possibility that that it may bind some target genes together with other DMRT proteins. Likewise, the ability of different DMRT proteins to bind the DMRT1 recognition motif (15) suggests that some of the target genes identified here may be regulated in other tissues by other DMRT proteins.
DNA-binding motifs associated with DMRT1-bound target genes varied between different classes of DMRT1-regulated genes. These differences could play a role in differential activity of DMRT1: A single-nucleotide difference in a κB binding site can determine which coactivators associate with NF-κB dimers (27). These differences also could affect DNA binding of other transcription factors. Indeed, the motifs with a central A/T pair contain a consensus SRY box containing gene 9 (SOX9) binding site (Fig. 4E). This sequence may allow some target genes to be regulated by either DMRT1 or SOX9. This idea also potentially could explain how Dmrt1 and Sox9 appear to have swapped roles in the regulatory hierarchy of the vertebrate testis, with Sox9 acting upstream in mammals and Dmrt1 upstream in other vertebrates (28). It will be important to test whether SOX9 binds to any of these sites in the testis. DMRT1 and GATA1 are coexpressed in Sertoli cells, and the co-occurrence of DMRT1 and GATA1 sites in more than half of genes directly repressed by DMRT1 in Sertoli cells may indicate a functional relationship between these two transcriptional regulators.
The promoter-proximal sequences probed in these experiments represent less than 3% of the genome, and thus DMRT1 probably binds many regions that were not queried. It will be important to perform genome-wide ChIP analysis to determine whether many additional genes are bound by DMRT1 in vivo and whether DMRT1 binding is biased to particular regions of target genes.
Prior to this work no DMRT1 direct target genes were known, and the mode of DMRT1 transcriptional regulation was undetermined. Among the many targets we identified are a number of known regulators of testicular differentiation as well as many candidate regulators. It will be important to study further how the targets of DMRT1 in Sertoli cells and germ cells contribute to testicular function and how the constellation of target genes changes during testicular development. In addition, some of the genes we have identified are likely candidates for modulation of spermatogenesis and for contraceptive design. Finally, it remains to be determined whether homologs of the DMRT1 target genes, like homologs of DMRT1 itself, regulate sexual development in other metazoan phyla.
Materials and Methods
Cell Transfection.
293T cells were transfected by CaPO4 precipitation. MSC1 and primary Sertoli cells were transfected using Lipofectamine reagent (Life Technologies).
Mouse Breeding.
Mixed-background Dmrt1flox/flox male mice were bred to mixed-background Dmrt1flox/flox or Dmrt1−/− females containing a Dhh-cre (23) or Ngn3-cre (22) transgene. Male cre-positive offspring were compared with cre-negative littermates. Mouse protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee.
ChIP and ChIP-Chip.
An antibody (10) was purified using Affigel 15 resin (Bio-Rad) coupled to DMRT-GST fusion protein. A detailed ChIP protocol is provided in SI Materials and Methods. The genomic products of three biological DMRT1 ChIP replicates were amplified by ligation-mediated PCR (29) modified to use DNATerminator (Lucigen Corp.) for DNA end polishing. Amplified material was labeled by NimbleGen and hybridized to Mouse ChIP 385K RefSeq Promoter Arrays (05545021001; NimbleGen) together with matched amplified input DNA. For validation, qPCR was performed on unamplified ChIP-recovered DNA using gene-specific primers directly flanking the ChIP peak (SI Materials and Methods ). Negative controls were one or more of the following genes: B2M, Btg2, Otx2.
Gene Expression Analysis.
RNA was isolated from P9 testes using TRIzol reagent (Life Technologies), further purified with the RNeasy MinElute cleanup kit (74204; Qiagen), and RNA quality was verified with a 2100 Bioanalyzer (Agilent Technologies). RNA was reverse transcribed and hybridized to an MM8_60mer_expr array (NimbleGen A4543-00-01). Expression array data analysis methods are provided in SI Materials and Methods. For qRT-PCR one microgram of total RNA isolated from P9 testes using TRIzol reagent (Life Technologies) was reverse-transcribed using M-MLV reverse transcriptase (Life Technologies) and amplified using FastStart SYBR green (Roche). All samples were normalized to Hprt expression.
ChIP on Chip Data Analysis.
A region was considered positive for DMRT1 binding if it exceeded a defined signal intensity threshold (SI Materials and Methods). This analysis identified 1,372 regions of significant DMRT1 association. Peak centers were defined and linked with neighboring transcription start sites and their associated genes (Dataset S1). Because of bidirectional promoters and alternative transcription start sites, these peaks of DMRT1 binding correspond to 1,477 promoter regions and 1,439 unique genes (SI Materials and Methods).
CpG Island Analysis.
We found that 10,749 of 18,936 total promoters (56.8%) have CpG islands within 2 kb of the transcriptional start site. Of the 1,477 transcription start sites with peaks of DMRT1 binding, 1,035 had CpG islands within 2 kb (70.1%). The P value for likelihood of this distribution occurring randomly is 2.2 × 10−24 (Fisher's exact test).
Interactive Web Site.
ChIP data and mRNA expression data are contained in an interactive Web site based on a previously described data-driven Web application platform (30), with the addition of the capability to access and search the promoters bound by DMRT1 as well as to view DMRT1-binding data in the context of individual promoter landscapes (SI Materials and Methods). The resulting Web site containing all data is accessible at http://www.dmrt1.umn.edu/.
Supplementary Material
Acknowledgments
We thank members of the V.J.B. and D.Z. laboratories for helpful discussions, Dr. Howard Towle for critical reading of the manuscript, Dr. Shosei Yoshida (National Institute for Basic Biology, Okazaki, Japan) for Ngn3-cre mice, Dr. Dies Meijer (Erasmus University) for providing Dhh-cre mice, Dr. Michihiko Ito (Kitasato University) for providing pLUCMCS with DMRT1 binding sites, Dr. Baruch Frenkel for providing ChIP display methods before publication, Dr. Ken-ishi Morohashi for advice on gonadal ChIP methods, and the Minnesota Supercomputing Institute for computational resources. This work was supported by the University of Minnesota Masonic Cancer Center, Minnesota Medical Foundation, the Leukemia and Lymphoma Society (A.M.), and National Institutes of Health Grants GM59152 (to D.Z.), HD055763 (to L.L.H.), and HD041056 (to L.L.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: An interactive database of ChIP-chip and mRNA expression data is at www.dmrt1.umn.edu. Expression array data are in the GEO database (GSE22510) and ChIP-chip data are at http://genome.ucsc.edu/goldenPath/customTracks/custTracks.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006243107/-/DCSupplemental.
References
- 1.Zarkower D. Establishing sexual dimorphism: Conservation amidst diversity? Nat Rev Genet. 2001;2:175–185. doi: 10.1038/35056032. [DOI] [PubMed] [Google Scholar]
- 2.Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 1993;12:527–535. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raymond CS, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 4.Zhu L, et al. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimoto S, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda M, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 8.Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7:e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol. 2007;307:314–327. doi: 10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krentz AD, et al. The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc Natl Acad Sci USA. 2009;106:22323–22328. doi: 10.1073/pnas.0905431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erdman SE, Chen HJ, Burtis KC. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics. 1996;144:1639–1652. doi: 10.1093/genetics/144.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- 15.Murphy MW, Zarkower D, Bardwell VJ. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. doi: 10.1186/1471-2199-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimoto S, et al. Expression and promoter analysis of Xenopus DMRT1 and functional characterization of the transactivation property of its protein. Dev Growth Differ. 2006;48:597–603. doi: 10.1111/j.1440-169X.2006.00894.x. [DOI] [PubMed] [Google Scholar]
- 18.Herpin A, et al. Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet. 2010;6:e1000844. doi: 10.1371/journal.pgen.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barski A, Pregizer S, Frenkel B. Identification of transcription factor target genes by ChIP display. Methods Mol Biol. 2008;455:177–190. doi: 10.1007/978-1-59745-104-8_14. [DOI] [PubMed] [Google Scholar]
- 20.Thompson W, Rouchka EC, Lawrence CE. Gibbs Recursive Sampler: Finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida S, et al. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol. 2004;269:447–458. doi: 10.1016/j.ydbio.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 23.Lindeboom F, et al. A tissue-specific knockout reveals that Gata1 is not essential for Sertoli cell function in the mouse. Nucleic Acids Res. 2003;31:5405–5412. doi: 10.1093/nar/gkg723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura T, et al. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development. 2003;130:1691–1700. doi: 10.1242/dev.00392. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, et al. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110:4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Morrish BC, Sinclair AH. Vertebrate sex determination: Many means to an end. Reproduction. 2002;124:447–457. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- 29.Oberley MJ, Tsao J, Yau P, Farnham PJ. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 2004;376:315–334. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- 30.Sarver AL, Phalak R, Thayanithy V, Subramanian S. S-MED: Sarcoma microRNA expression database. Lab Invest. 2010;90:753–761. doi: 10.1038/labinvest.2010.53. [DOI] [PubMed] [Google Scholar]
- 31.Mertin S, McDowall SG, Harley VR. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.