Abstract
Class II major histocompatibility complex (MHC-II) proteins govern stimulation of adaptive immunity by presenting antigenic peptides to CD4+ T lymphocytes. Many allelic variants of MHC-II exist with implications in peptide presentation and immunity; thus, high-throughput experimental tools for rapid and quantitative analysis of peptide binding to MHC-II are needed. Here, we present an expression system wherein peptide and MHC-II are codisplayed on the surface of yeast in an intracellular association-dependent manner and assayed by flow cytometry. Accordingly, the relative binding of different peptides and/or MHC-II variants can be assayed by genetically manipulating either partner, enabling the application of directed evolution approaches for high-throughput characterization or engineering. We demonstrate the application of this tool to map the side-chain preference for peptides binding to HLA-DR1 and to evolve novel HLA-DR1 mutants with altered peptide-binding specificity.
Keywords: yeast display, MHC peptide-binding interactions, MHC engineering, anchor specificity, directed evolution
Class II major histocompatibility complex (MHC-II)-restricted T cell responses are related to a great number of diseases including autoimmunity, graft rejection, and atypical immune response. MHC-II proteins are heterodimeric transmembrane proteins consisting of α and β chains containing two domains each (1), and these proteins capture antigenic peptides processed inside professional antigen-presenting cells (APCs) and present them on the APC surface for recognition by CD4+ T cells to initiate adaptive immunity (2, 3). The peptide-binding sites of MHC-II formed by the α1 and β1 domains contain several pockets that prefer to accommodate specific side chains of “anchor” residues on peptide antigens (4, 5). Thus, MHC-II are semipromiscuous binders capable of presenting numerous different peptides, but anchor pockets constrain the milieu of peptides presented by a given MHC-II. In depth characterization of peptide binding by MHC-II is therefore critical to understanding issues in vaccine design (6), autoimmune disease (7), infectious disease progression (8), and transplantation rejection (9, 10), but the lack of a rapid, efficient, robust, and quantitative methodology for characterizing the peptide-binding specificity and promiscuity of MHC alleles remains a bottleneck.
MHCs are the most polymorphic glycoproteins known in nature (11), and many polymorphisms impact peptide recognition; thus, investigation of peptide-binding properties of MHC-II is a challenging problem that requires high-throughput approaches. A number of studies have assessed peptide-binding to MHC-II on the surface of intact APCs (12, 13), whereas a routinely used in vitro approach entails purifying soluble recombinant MHC-II molecules from different expressing systems such as B cell lines (14), insect cells (15, 16), yeast (17), or Escherichia coli (18–20) and then characterizing binding of these molecules to different peptides generated either chemically by solid phase synthesis or genetically by phage display (4, 21). The labor-intensive preparation of soluble proteins, lengthy binding assays, or nonquantitative data generated (e.g., peptide abundance) limits the efficiency and throughput of these methods for mapping MHC-II binding specificities across the large number of existing alleles. Alternatively, surface display technologies commonly used for directed evolution could be applied to express MHC-II alleles for peptide-binding assays on the cell surface. As a single-cell eukaryotic system presenting advantages of simple molecular cloning and eukaryotic posttranslational modification for protein expression (22), yeast serves as a powerful platform for development of such an approach.
Here, we report a quantitative, high-throughput methodology, yeast codisplay (Fig. 1), for characterizing and engineering peptide-binding specificity of MHC-II. As a model system, the extracellular domains of the MHC-II human leukocyte antigen HLA-DR1 were expressed as a secreted heterodimer in Saccharomyces cerevisiae. Simultaneously, an antigenic peptide known to bind to HLA-DR1 was surface-displayed. Intracellular binding between the MHC-II and the peptide antigen thus anchored the soluble MHC-II to the cell surface upon secretion, allowing detection by immunofluorescence. The relative abundance of MHC-II compared to peptide on the cell surface depended on the strength of binding between these species, as assessed by studies of a panel of peptides mutated at a key anchor position governing interactions with HLA-DR1. Furthermore, HLA-DR1 mutants binding to peptides not recognized by wild-type HLA-DR1 were isolated from a combinatorial library, demonstrating the potential of this system for engineering MHC-II antigen presentation. A unique hyper-promiscuous MHC-II mutant was discovered with this tool. This method thus presents intriguing potential to impact the understanding of MHC-II/peptide recognition and related therapeutic applications benefiting from alterations in antigen presentation, such as mapping peptide specificity of MHC-II alleles (23), identification of small molecule agents that stabilize presentation of specific peptides (24), and construction of artificial antigen-presenting cells for immunotherapy (25).
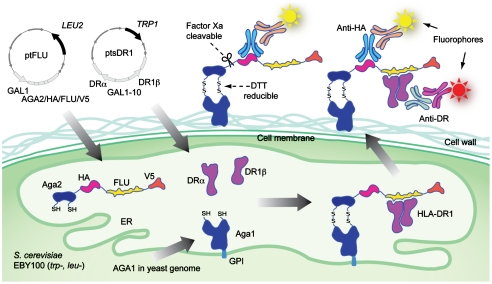
Fig. 1.
Design of the yeast codisplay system for HLA-DR1. Aga2p-fused FLU peptide (PKYVKQNTLKLAT) is expressed flanked by the HA and V5 epitope tags, enabling detection of peptide levels by immunofluorescent staining with antibodies specific for either tag. The HLA-DRα and HLA-DR1β chain extracellular domains are expressed from separate cassettes. The FLU peptide is anchored to the cell surface via native processing and secretion of the assembled a-agglutinin protein (composed of the Aga1p and Aga2p subunits) as described (22), and the HLA-DR1 heterodimer is anchored by noncovalent binding to FLU. Relative fluorescence levels of different fluorophores coupled to antitag and anti-DR reagents indicate the level of saturation of available peptides by bound HLA-DR1. GPI: glycosylphosphatidylinositol.
Results
Peptide-Binding-Dependent Cell Surface Display of HLA-DR1.
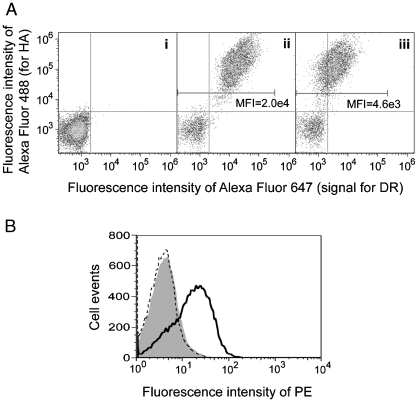
To verify that wild-type MHC-II heterodimers can be expressed in a functional form competent for binding to soluble peptides, the extracellular domain of HLA-DR1 both with and without a covalently fused antigen peptide was surface displayed by fusion to the N-terminus of the endogenous yeast adhesion receptor subunit Aga2p, as described previously for HLA-DR4 (with peptide linked to the β chain N-terminus) (26) and for other MHC-II proteins using a single-chain format and fusion to the C-terminus of Aga2p (27, 28). The antigen used in these studies is a 13 amino acid peptide (designated FLU) from a naturally HLA-DR1-presented fragment of influenza hemagglutinin (HA306-318), a well-studied peptide known to bind tightly to HLA-DR1 (5). Gene constructs directing expression of noncovalent extracellular domain heterodimers were transformed into yeast strain EBY100, and the surface display of peptide-fused or “empty” HLA-DR1 was assessed by simultaneous immunofluorescent labeling with anti-HLA-DRα antibody and antibodies specific for the HA epitope tag included at the Aga2p C-terminus (Fig. 2). The expression level of FLU-fused HLA-DR1 (Fig. 2A, ii) is significantly higher than that of the empty form (Fig. 2A, iii), whereas the expression level of Aga2p in these two scaffolds are similar, as represented by anti-HA signals. Enhanced expression of peptide-fused HLA-DR1 suggests that specific peptides might stabilize MHC-II folding in yeast similarly to in natural APCs (3), whereas the majority of the empty HLA-DR1 fails to express (approximately 5-fold reduction in cellular fluorescence in Fig. 2A), consistent with a prior study (28) and putatively due to degradation by the secretory quality control machinery (29). To validate the functionality of the yeast-displayed empty HLA-DR1, yeast were incubated with synthetic biotinylated peptides and stained with streptavidin-PE before detection by flow cytometry (Fig. 2B). Synthetic FLU peptide bound to HLA-DR1-displaying yeast, but not to a control strain displaying an irrelevant protein, whereas a control peptide derived from β2 microglobulin demonstrated no binding to either yeast. Similar results were obtained whether binding proceeded in citrate buffer at pH 5.0 or several other buffers or growth media at pH 5.0–7.4 (Fig. S1), in agreement with the demonstrated pH-independence of peptide recognition by HLA-DR1 (30). Whereas these results validate the function of yeast-secreted MHC-II, the dynamic range of the peptide-binding signal obtained with surface-displayed HLA-DR1 is low, motivating further development of an approach to characterize binding to peptides across a range of affinities and to enable simple and economical combinatorial variation of peptides in a high-throughput approach.
Fig. 2.
Expression of functional HLA-DR1 heterodimer in yeast by surface display. (A). Aga2p and HLA-DR1 levels of yeast expressing (i) an irrelevant soluble protein, (ii) Aga2p-fused HLA-DR1 with FLU-covalently linked to the β chain N-terminus, or (iii) empty Aga2p-fused HLA-DR1. Cells were double-labeled with reagents specific for the HA epitope tag (Aga2p level) and HLA-DRα. Mean fluorescence intensity (MFI) of DR signal for HA positive cell population was indicated. (B) Peptide-binding capability of yeast-displayed HLA-DR1. Yeast displaying empty HLA-DR1 (solid curve) or an irrelevant protein (shaded) were incubated with 100 μM biotinylated FLU at pH 5.0; HLA-DR1-displaying yeast were also incubated with 100 μM biotinylated control peptide (dashed curve). All samples were stained with streptavidin-PE and analyzed by flow cytometry.
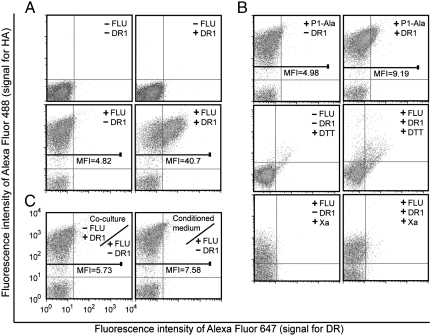
The yeast display approach was modified such that the MHC-II and target peptide are expressed from separate yeast shuttle vectors. One vector directs expression of the peptide as a fusion to Aga2p, following the classical yeast surface display approach (22). The second vector directs expression of both chains of the MHC-II heterodimer as separate cassettes from a bidirectional transcriptional promoter (Fig. 1). MHC-II heterodimers that bind the Aga2p-fused target peptide are thus anchored to the cell surface via Aga2p, whereas MHC-II that do not bind the target peptide or bind instead to endogenous peptides present in the secretory pathway will be either secreted as soluble species or potentially degraded by the secretory quality control machinery, as suggested by our surface display results. Plasmids directing expression of HLA-DR1 or Aga2p-FLU were transformed into the S. cerevisiae strain EBY100 (22). Yeast expressing combinations of these proteins were immunofluorescently costained to detect the level of FLU peptide and HLA-DR1 on the surface and analyzed by flow cytometry (Fig. 3). Yeast expressing only the soluble HLA-DR1 ectodomain show no detectable staining, whereas those coexpressing soluble HLA-DR1 and surface-displayed FLU demonstrate correlated signals for both (Fig. 3A). Based on the known peptide-binding motif of HLA-DR1 (21, 31), Ala was substituted for Tyr308 at the P1 major anchor position of the FLU peptide (peptide P1-Ala; PKAVKQNTLKLAT; see Fig. S2). As anticipated, this substitution abolished detectable HLA-DR1 on the cell surface (Fig. 3B); furthermore, treatment of yeast with reducing agents or Factor Xa protease, which respectively remove the surface-displayed Aga2p-FLU or FLU peptide (see Fig. 1), also abrogated surface display of HLA-DR1 (Fig. 3B), confirming the specific anchoring of HLA-DR1 to the surface via interaction with FLU.
Fig. 3.
Yeast codisplay and surface detection of FLU peptide and FLU-bound soluble HLA-DR1. (A) The yeast parent strain or yeast expressing HLA-DR1, Aga2p-FLU, or both were double-labeled with anti-HA and anti-DR1 antibodies and analyzed by flow cytometry. (B) The yeast parent strain and yeast expressing Aga2p-P1-Ala alone or with HLA-DR1 or yeast expressing Aga2p-FLU with or without coexpressed HLA-DR1 treated with DTT or Factor Xa protease, as indicated, were double-labeled and analyzed by flow cytometry. (C) Yeast expressing Aga2p-FLU were cocultured at 1∶1 ratio with yeast expressing soluble HLA-DR1 or cultured in medium preconditioned by growing codisplaying yeast prior to labeling and analysis.
To extend the codisplay approach to library screening, the genotype-phenotype linkage between plasmids and displayed proteins must be maintained; therefore, experiments were performed to determine whether HLA-DR1: (i) bound to FLU intracellularly and was subsequently exported to the surface, (ii) was secreted empty into the culture medium (or with weakly bound endogenous peptides) and then bound to FLU, or (iii) a combination of these mechanisms. Separate yeast strains expressing surface-displayed FLU or soluble HLA-DR1 ectodomain were cocultured in equal numbers; in addition, yeast displaying FLU were incubated in conditioned medium from a culture of soluble HLA-DR1-expressing yeast. In both cases, no significant binding of HLA-DR1 to the surface of any yeast was observed (Fig. 3C), indicating that resorting of secreted HLA-DR1 between different cells in the same culture fails to occur and suggesting that HLA-DR1/FLU binding occurs intracellularly within the secretory pathway.
Quantitative Mapping of HLA-DR1 P1 Anchor Specificity.
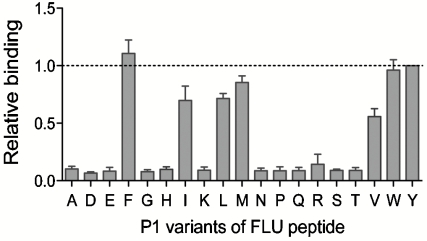
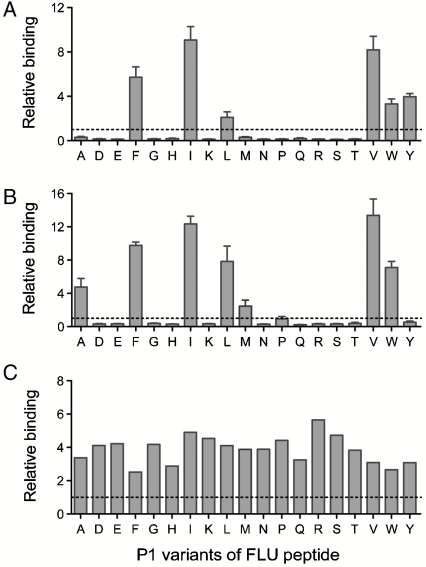
An obvious application of the yeast codisplay system is to determine the binding motif of a given MHC-II allele or to predict promiscuous MHC-II ligands by characterizing the relative peptide binding of MHC-II proteins to various peptides. Because the binding motif of HLA-DR1 has been carefully studied, and P1 (Fig. S2) has been determined to be the dominant anchor (21), we focused on this peptide position to validate the ability of the yeast codisplay approach to quantitatively assess peptide side-chain preferences in the context of FLU. A set of yeast surface-linked FLU analogues containing all natural amino acids except Cys at the P1 position were coexpressed with the soluble HLA-DR1 ectodomain; levels of codisplayed HLA-DR1 were measured by flow cytometry (Fig. S3) and relative binding between HLA-DR1 and the peptide variants was calculated based on fluorescence intensity (Fig. 4 and Fig. S4); costaining of these yeast strains with antibodies specific for the HA and V5 epitopes flanking the surface-displayed peptides verified that levels of these epitopes were equivalent (Fig. S5), indicating that mutations at the P1 residue did not influence display levels of full-length target peptides. Relative display levels of HLA-DR1 with the different peptides, presumably reflecting relative binding affinity between the MHC-II protein and peptide, follow the order Phe ≈ Tyr ≈ Trp > Met > Leu ≈ Ile > Val, whereas all other amino acids tested yield very low binding to HLA-DR1. These results show striking agreement with binding preferences previously determined using quantitative in vitro peptide-binding assays (21), qualitative phage display and bioinformatics methods (23, 31), and structure-based computational prediction approaches (32). Most importantly, the yeast codisplay method appears to demonstrate quantitative ability paralleling that of competitive binding assays with purified proteins and soluble peptides (21) while retaining the ease of rapid recombinant expression without component purification and compatibility with high-throughput screening approaches.
Fig. 4.
Peptide P1 anchor residue specificity of HLA-DR1 analyzed by yeast codisplay. Cells codisplaying HLA-DR1 and FLU peptides with the indicated P1 residue substitutions were analyzed by flow cytometry (Fig. S3) and normalized relative binding levels were calculated from fluorescence intensities (Materials and Methods). The dashed line represents binding to the wild-type FLU peptide with Tyr at P1. Error bars represent standard error of the mean determined from four independent experiments.
Engineering HLA-DR1 Specificity by Directed Evolution.
In addition to enabling rapid characterization of binding specificities of MHC-II, the codisplay system also presents the potential for straightforward application of directed evolution methods to generate MHC-II mutants that bind and present altered antigenic epitopes for recognition by T cells. Such redesign of MHC-II specificity would provide further resources for structure-function studies aimed at understanding constraints on peptide presentation and the related issue of immunodominance. Furthermore, such molecules might find use with artificial APC approaches to T cell activation as a means to functionally present altered antigenic epitopes, considering the ability of different MHC-II alleles to present different antigens is linked to infectious disease susceptibility and resistance (33).
Here, the codisplay system was applied to a combinatorial library of HLA-DR1 mutants to select molecules capable of binding tightly to several P1 variants of FLU. Six residues forming the HLA-DR1 pocket into which the P1 residue side chain extends were selected for saturation mutagenesis using degenerate PCR primers: Phe-α24, Ile-α31, Phe-α32, Gly-β86, Phe-β89, and Thr-β90 (Fig. S2). The resulting library was individually coexpressed with P1-mutated FLU target peptides P1-Val, P1-Ala, and P1-Glu, and mutants binding to these peptides were isolated by fluorescence-activated cell sorting. Binding motifs of a representative clone from each group were characterized (Fig. 5), and the data demonstrate that the P1 anchor position amino acid specificity is substantially altered in each case. In particular, the P1-Glu-binding mutant S4E1.3 appears to be a promiscuous binder, and all three mutants exhibit markedly enhanced binding to those peptides for which they are specific (approximately 3–14-fold increased relative binding signals). Yeast expressing only the S4E1.3 mutant but no target peptide or coexpressing an irrelevant peptide display no MHC-II on the surface (Fig. S6), confirming that S4E1.3 is anchored via interaction with the target peptides. Compared to wild-type HLA-DR1, the mutant selected for binding to P1-Val (S4V1.5) prefers the smaller hydrophobic P1 side chains of Val and Ile, but binds more tightly to peptides with any hydrophobic residue at P1 except Met (including wild-type FLU). The P1-Ala-binding mutant S4A1.9, on the other hand, shows greatly increased binding to all hydrophobic P1 residues with the exception of the Tyr present in wild-type FLU.
Fig. 5.
Peptide P1 anchor residue specificity of HLA-DR1 mutants generated by directed evolution. Relative binding to the P1 substituted FLU peptide panel were determined for (A) clone S4V1.5 selected against P1-Val, (B) clone S4A.19 selected against P1-Ala, and (C) S4E1.3 selected against P1-Glu. Dashed lines indicate the relative binding signal for wild-type HLA-DR1 codisplayed with wild-type FLU. Error bars represent standard error of the mean determined from four independent experiments.
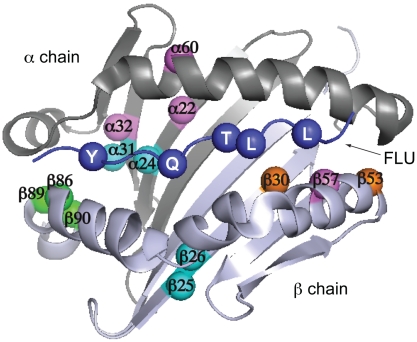
In all three clones, mutations occurred at P1 pocket positions designed in the library and at other positions presumably arising due to PCR-induced mutagenesis (Fig. 6). Both S4V1.5 and S4E1.3 contain mutations in both the α and β chains, highlighting the orthogonal nature of this in vitro directed evolution approach compared to natural evolution of MHC-II alleles, in which HLA-DR α chains are monomorphic. Furthermore, both polymorphic and nonpolymorphic positions in the β chain were mutated in these clones. These data suggest that the HLA-DR architecture is more tolerant to mutations than suggested by the existing database of natural HLA-DR molecules (11).
Fig. 6.
Mutation sites conferring altered P1 anchor specificity on HLA-DR1. Positions mutated in S4V1.5 (cyan), S4A1.9 (orange), S4E1.3 (magenta), or all three clones (green) are highlighted on a top view of the α1 and β1 domains, with the peptide anchor residue positions P1, P4, P6, P7, and P9 also highlighted (blue). Images generated from coordinates in PDB file 1DHL.
Discussion
Understanding the binding specificities of MHC-II alleles is critical to applications from transplantation to vaccine design. Yeast codisplay adds an important experimental tool for quantitatively determining MHC-II antigen-binding motifs with the potential to enable assessment of anchor residue context-dependence, significant information currently absent in scoring algorithms predicting peptide binding (34). Numerous MHC-II alleles could be examined for binding to specific target peptides via a library approach, or alternatively a single allele could be screened against a peptide library to rapidly identify binders using high-throughput sequencing technologies. Small libraries of important peptides could be codisplayed with MHC-II allele libraries, as well, to quickly assess the potential for antigen presentation. In any format, the high-throughput capability of yeast display and flow cytometry or FACS implies rapid and economical characterization is possible (e.g., assessment of target peptide binding by a library of MHC-II alleles could be accomplished in < 3 weeks) and suggests the possibility of quantitative peptide-binding information databases at the level of the impact of individual MHC-II polymorphisms. The codisplay system demonstrates measurable and varying signals among the weak binders studied here and thus appears particularly suited for quantitatively characterizing naturally existing weak interactions (such as the P1-Val peptide studied here), an important addition to the dataset toward further mechanistic understanding of MHC-peptide recognition.
Unlike common surface display systems such as phage display (23, 31), animal cell display (35), baculovirus/insect cell display (36), or even classical yeast display (28), which were developed for identification of T cell receptor ligands, the yeast codisplay method eliminates the limitations of expressing single-chain derivatives of multimeric proteins and covalently attaching MHC-II with peptides or other proteins. In the codisplay approach, MHC-II/peptide binding takes place intracellularly between nontethered species, better mimicking peptide loading of MHC-II in APCs (3), and yet yeast codisplay retains the advantage of creating a genotype-phenotype link for high-throughput screening and easy information retrieval. Adaptation of this approach for screening peptide libraries to identify T cell stimulatory ligands (28, 35, 36) should be straightforward. Furthermore, whereas we have focused on characterizing and engineering MHC-II/peptide recognition, the approach is amenable to extension to a host of other systems for quantitative characterization of protein–protein and protein–peptide-binding specificities, complementing other methods for studying such interactions (37, 38) and broadening the impact of the method.
To our knowledge, in vitro engineering of HLA-DR1 peptide binding specificity has not been previously demonstrated; here we have shown that HLA-DR1 can be engineered to accommodate peptides with substantially altered P1 anchor side chains, including isolation of an unprecedented HLA-DR mutant showing nonspecific P1 anchor binding. Introduction of counterselection steps in the directed evolution scheme could potentiate tighter control of specificity and restrict binding promiscuity, if desired. Furthermore, our approach might be directly extended to other major peptide anchor positions (e.g., P4, P6, P7, and P9 for HLA-DR1), enabling custom design of MHC-II reagents with tailored antigen-binding specificities. A subset of such molecules retaining the ability to interact with HLA-DR1-restricted T cells could find use in engineered cellular or artificial antigen-presenting cell vaccines (6, 25) and could shed additional light on the limitations of T cell antigen presentation. Furthermore, the methods demonstrated here could be simply adapted for facile quantitative screening of compound libraries to identify modulators of natural peptide antigen presentation (39).
Materials and Methods
Expression of Functional HLA-DR1 Heterodimers by Classical Yeast Display.
Plasmid pfluDR1 or pDR1 was constructed (SI Materials and Methods) and transformed into engineered S. cerevisiae strain EBY100 (a GAL1-AGA1:URA3 ura3-52 trp1 leu2Δ1 his3Δ200 pep4:HIS2 prb1Δ1.6R can1 GAL) (22) by electroporation using the MicroPulser Electroporation Apparatus (BioRad), for surface-displaying FLU-fused or empty HLA-DR1 using previously described method (26). Yeast displaying empty HLA-DR1 were incubated with biotinylated FLU peptide or β2m peptide (derived from human β2 microglobulin 52–64: SDLSFSKDWSFYL) (Abgent, Inc.) and stained by streptavidin-PE (Sigma–Aldrich) as described (SI Materials and Methods) for assessing the peptide-binding ability.
Construction and Transformation of Yeast Shuttle Vectors for Codisplay.
The entire expression cassette (GAL1-10//AGA2-HA//scFv 4-4-20//MFα Term.) was excised from pCT302 (22) by double digest using KpnI and SacI and subcloned into KpnI/SacI partially digested yeast shuttle vector pRS315 (40) to create a yeast surface display plasmid with a LEU2 selectable marker. Vector ptFLU was constructed for displaying Aga2-FLU fusion by inserting an oligonucleotide encoding the 13-residue FLU peptide followed by a short spacer (GGGS) and the V5 epitope tag (GKPIPNPLLGLDST) in place of the scFv in pCT302. To create vector ptsDR1 for expressing the soluble HLA-DR1 ectodomain heterodimers, the plasmid pDR1 was modified by replacing the AGA2 and HA-tag sequences downstream of the DRB1*010101 gene with an in-frame stop codon. ptFLU (LEU+) and ptsDR1 (TRP+) were transformed into EBY100 (URA+, trp-, leu-) by electroporation.
Preparation of Codisplaying Yeast for Flow Cytometry.
Two mL SD-SCAA minimal medium (2% (wt/vol) glucose, 0.67% (wt/vol) yeast nitrogen base without amino acids, 0.062% (wt/vol) Leu/Trp/Ura dropout supplement mixtures of amino acids (Clontech), 38 mM Na2HPO4, 62 mM NaH2PO4·2H2O, pH 6.0) was inoculated by a single colony and incubated in a 30 °C shaker until a density of 2.5–5.0 × 107 cells/mL (an OD600 of 2.5–5.0) was reached. To induce GAL1-10 promoted protein expression, 107 cells were collected by centrifugation and transferred to 2 mL SG-SCAA medium (glucose replaced by galactose). After 16–18 h induction at 30 °C, approximately 106 cells per sample were harvested by centrifugation for immunofluorescent labeling. When protein stripping was desired, the induced cells were first washed with 400 μL Tris-buffered saline (137 mM NaCl, 20 mM Tris-Cl, pH 7.6) 1–2× and incubated either in 20 μL of reducing buffer [50 mM Tris-Cl pH 8.0, 1 mM DTT (Sigma; added just before use)] at 4 °C for 24 h with gentle shaking or in 20 μL of Factor Xa buffer (100 mM NaCl, 2 mM CaCl2, 20 mM Tris-Cl, pH 8.0) with 20 μg/mL Factor Xa protease (New England Biolabs) at 23 °C for at least 48 h. Cells were pelleted and washed with 400 μL ice cold PBS + 1% BSA at least once before primary labeling. Cell pellets were resuspended in 25 μL PBS + 1% BSA with mouse anti-HLA-DR monoclonal antibody (mAb) L243 (1∶2.5 dilution; BD Biosciences) and rabbit anti-HA polyclonal antibody (1∶25; Sigma) and stained for 30 min at room temperature followed by 10 min on ice. After removal of primary reagents by centrifugation, cells were washed with ice cold PBS + 1% BSA again and resuspended in 40 μL PBS + 1% BSA containing highly cross-adsorbed secondary antibodies (Molecular Probes, Invitrogen): Alexa FLuor 647 goat antimouse IgG (H + L) (1∶80) and Alexa FLuor 488 goat antirabbit IgG (H + L (1∶80) for an 30 min incubation at room temperature followed by 10 min on ice. A final wash with ice cold PBS + 1% BSA was applied before resuspending cells in 500–700 μL PBS + 1% BSA for flow cytometry. For the simultaneous detection of HA-tag and V5-tag flanking FLU peptide, mouse anti-V5 mAb (Invitrogen) was used at a dilution of 1∶30 instead of the mAb L243 as the primary labeling reagent, whereas other labeling steps were unchanged.
Flow Cytometry and Quantitative Analysis for Relative Binding.
At least 10,000 cell events, gated by forward and side scatter, were collected per sample. Flow cytometers used included Accuri C6 (Accuri Cytometers Inc.), FACSCalibur, FACSVantage, and LSR II (BD Biosciences). Flow cytometric data for a codisplaying yeast strain was analyzed (Fig. S4) using Flowjo software (Tree Star Inc). The peptide (e.g., FLU) display level on the surface of yeast is proportional to the background-corrected mean fluorescence intensity value normalized to the background intensity
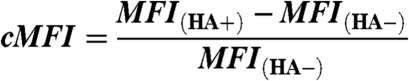 |
where MFI(HA-) and MFI(HA+) represent mean fluorescence intensity of HA-tag coupled Alexa Fluor 488 emission calculated using Flowjo for negative and positive cell populations, respectively. Analysis using anti-V5-tag staining resulted in equivalent cMFI values. The intracellular peptide-binding-dependent MHC-II display level was properly calculated using normalized, background-corrected fluorescence associated with both anti-DR1 staining and peptide display level (DR-ratio)
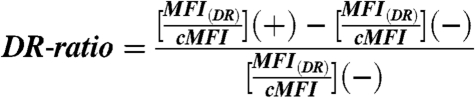 |
where (+) and (-) represent the codisplaying yeast and peptide-only-displaying yeast, respectively. MFI(DR) and cMFI represent the mean fluorescence intensity of HLA-DR1 coupled Alexa Fluor 647 emission and the FLU display level on corresponding yeast strain surface. Normalization minimizes variability between experiments due to laser power output, detector amplification, and other cytometer parameters. Relative binding of MHC-II peptide variants is determined as DR-ratio of the corresponding coexpressing yeast strain using the above equation and divided by the value for yeast codisplaying FLU/HLA-DR1 complexes.
Anchor Preference Assay via Yeast Codisplay.
The key anchor position (i.e., P1) of FLU peptide for HLA-DR1 binding in plasmid ptFLU was first substituted to all natural amino acids except for Cys by site directed mutagenesis using the QuikChange II kit (Stratagene) following the manufacturer’s recommended procedures. Yeast were transformed, cultured, induced for expression, and analyzed as described above to determine relative binding of MHC-II to each peptide variant.
Construction and Screening of HLA-DR1 Mutant Library.
Polymerase chain reactions primed by two degenerate oligonucleotides enabled saturation mutagenesis for the six selected residues—α24, α31, α32, β86, β89, and β90, forming the P1 pocket of HLA-DR1 peptide-binding site (SI Materials and Methods). HLA-DR1-Mutant-expressing plasmids were generated by homologous recombination in yeast coexpressing target P1-variant of FLU peptide. The constructed library was cultured, labeled, sorted and characterized as described (SI Materials and Methods).
Supplementary Material
Acknowledgments.
We thank L. J. Stern (University of Massachusetts School of Medicine) for providing plasmid pDLM1-drb1s. We also thank N. R. Neilsen, D. J. Trent, and S. Eda (University of Tennessee) for assistance with cell sorting and flow cytometry analysis, and D. Monos (Children’s Hospital of Philadelphia) and S. L. Diamond (University of Pennsylvania) for stimulating discussions. This work was funded by the US National Science Foundation (CAREER BES-0239099 to E.T.B.) and the US National Institutes of Health (R01 GM081444 to E.T.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006344107/-/DCSupplemental.
References
- 1.Brown JH, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 2.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watts C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat Immunol. 2004;5:685–692. doi: 10.1038/ni1088. [DOI] [PubMed] [Google Scholar]
- 4.Hammer J, et al. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993;74:197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- 5.Stern LJ, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 6.Sette A, Fikes J. Epitope-based vaccines: An update on epitope identification, vaccine design, and delivery. Curr Opin Immunol. 2003;15:461–470. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 7.Casares S, et al. Down-regulation of diabetogenic CD4+ T cells by a soluble dimeric peptide-MHC class II chimera. Nat Immunol. 2002;3:383–391. doi: 10.1038/ni770. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, et al. Suppression of major histocompatibility complex class II-associated invariant chain enhances the potency of an HIV gp120 DNA vaccine. Immunology. 2007;120:207–216. doi: 10.1111/j.1365-2567.2006.02492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevanovic S. Structural basis of immunogenicity. Transpl Immunol. 2002;10:133–136. doi: 10.1016/s0966-3274(02)00059-x. [DOI] [PubMed] [Google Scholar]
- 10.Hong JC, Kahan BD. Immunosuppressive agents in organ transplantation: Past, present, and future. Semin Nephrol. 2000;20:108–125. [PubMed] [Google Scholar]
- 11.Reche PA, Reinherz EL. Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J Mol Biol. 2003;331:623–641. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 12.Chicz RM, et al. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vacchino JF, McConnell HM. Peptide binding to active class II MHC protein on the cell surface. J Immunol. 2001;166:6680–6685. doi: 10.4049/jimmunol.166.11.6680. [DOI] [PubMed] [Google Scholar]
- 14.Gorga JC, Horejsi V, Johnson DR, Raghupathy R, Strominger JL. Purification and characterization of class II histocompatibility antigens from a homozygous human B cell line. J Biol Chem. 1987;262:16087–16094. [PubMed] [Google Scholar]
- 15.Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty alpha beta heterodimers in the absence of antigenic peptide. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 16.Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 17.Kalandadze A, Galleno M, Foncerrada L, Strominger JL, Wucherpfennig KW. Expression of recombinant HLA-DR2 molecules. Replacement of the hydrophobic transmembrane region by a leucine zipper dimerization motif allows the assembly and secretion of soluble DR alpha beta heterodimers. J Biol Chem. 1996;271:20156–20162. doi: 10.1074/jbc.271.33.20156. [DOI] [PubMed] [Google Scholar]
- 18.Joshi RV, Zarutskie JA, Stern LJ. A three-step kinetic mechanism for peptide binding to MHC class II proteins. Biochemistry. 2000;39:3751–3762. doi: 10.1021/bi9923656. [DOI] [PubMed] [Google Scholar]
- 19.Justesen S, Harndahl M, Lamberth K, Nielsen LL, Buus S. Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome Res. 2009;5:2. doi: 10.1186/1745-7580-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato AK, et al. Determinants of the peptide-induced conformational change in the human class II major histocompatibility complex protein HLA-DR1. J Biol Chem. 2000;275:2165–2173. doi: 10.1074/jbc.275.3.2165. [DOI] [PubMed] [Google Scholar]
- 21.Hammer J, et al. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci USA. 1994;91:4456–4460. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 23.Sturniolo T, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 24.Ghumman B, Bertram EM, Watts TH. Chemical chaperones enhance superantigen and conventional antigen presentation by HLA-DM-deficient as well as HLA-DM-sufficient antigen-presenting cells and enhance IgG2a production in vivo. J Immunol. 1998;161:3262–3270. [PubMed] [Google Scholar]
- 25.Oelke M, Krueger C, Giuntoli RL, II, Schneck JP. Artificial antigen-presenting cells: artificial solutions for real diseases. Trends Mol Med. 2005;11:412–420. doi: 10.1016/j.molmed.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Boder ET, Bill JR, Nields AW, Marrack PC, Kappler JW. Yeast surface display of a noncovalent MHC class II heterodimer complexed with antigenic peptide. Biotechnol Bioeng. 2005;92:485–491. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 27.Starwalt SE, Masteller EL, Bluestone JA, Kranz DM. Directed evolution of a single-chain class II MHC product by yeast display. Protein Eng. 2003;16:147–156. doi: 10.1093/proeng/gzg018. [DOI] [PubMed] [Google Scholar]
- 28.Wen F, Esteban O, Zhao H. Rapid identification of CD4+ T-cell epitopes using yeast displaying pathogen-derived peptide library. J Immunol Methods. 2008;336:37–44. doi: 10.1016/j.jim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sette A, et al. Effect of pH on MHC class II-peptide interactions. J Immunol. 1992;148:844–851. [PubMed] [Google Scholar]
- 31.Hammer J, Takacs B, Sinigaglia F. Identification of a motif for HLA-DR1 binding peptides using M13 display libraries. J Exp Med. 1992;176:1007–1013. doi: 10.1084/jem.176.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Androulakis IP, Nayak NN, Ierapetritou MG, Monos DS, Floudas CA. A predictive method for the evaluation of peptide binding in pocket 1 of HLA-DRB1 via global minimization of energy interactions. Proteins. 1997;29:87–102. [PubMed] [Google Scholar]
- 33.Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–192. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med Res Rev. 2002;22:168–203. doi: 10.1002/med.10006. [DOI] [PubMed] [Google Scholar]
- 35.Boen E, Crownover AR, McIlhaney M, Korman AJ, Bill J. Identification of T cell ligands in a library of peptides covalently attached to HLA-DR4. J Immunol. 2000;165:2040–2047. doi: 10.4049/jimmunol.165.4.2040. [DOI] [PubMed] [Google Scholar]
- 36.Crawford F, et al. Use of baculovirus MHC/peptide display libraries to characterize T-cell receptor ligands. Immunol Rev. 2006;210:156–170. doi: 10.1111/j.0105-2896.2006.00365.x. [DOI] [PubMed] [Google Scholar]
- 37.Hu X, Kang S, Chen X, Shoemaker CB, Jin MM. Yeast surface two-hybrid for quantitative in vivo detection of protein–protein interactions via the secretory pathway. J Biol Chem. 2009;284:16369–16376. doi: 10.1074/jbc.M109.001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fields S. Interactive learning: Lessons from two hybrids over two decades. Proteomics. 2009;9:5209–5213. doi: 10.1002/pmic.200900236. [DOI] [PubMed] [Google Scholar]
- 39.Marin-Esteban V, Falk K, Rotzschke O. “Chemical analogues” of HLA-DM can induce a peptide-receptive state in HLA-DR molecules. J Biol Chem. 2004;279:50684–50690. doi: 10.1074/jbc.M407598200. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.