Abstract
A chronic proinflammatory state precedes pathological change in arterial endothelial cells located within regions of susceptibility to atherosclerosis. The potential contributions of regulatory microRNAs to this disequilibrium were investigated by artery site-specific profiling in normal adult swine. Expression of endothelial microRNA10a (miR-10a) was lower in the athero-susceptible regions of the inner aortic arch and aorto-renal branches than elsewhere. Expression of Homeobox A1 (HOXA1), a known miR-10a target, was up-regulated in the same locations. Endothelial transcriptome microarray analysis of miR-10a knockdown in cultured human aortic endothelial cells (HAEC) identified IκB/NF-κB–mediated inflammation as the top category of up-regulated biological processes. Phosphorylation of IκBα, a prerequisite for IκBα proteolysis and NF-κB activation, was significantly up-regulated in miR-10a knockdown HAEC and was accompanied by increased nuclear expression of NF-κB p65. The inflammatory biomarkers monocyte chemotactic protein 1 (MCP-1), IL-6, IL-8, vascular cell adhesion molecule 1 (VCAM-1), and E-selectin were elevated following miR-10a knockdown. Conversely, knockin of miR-10a (a conservative 25-fold increase) inhibited the basal expression of VCAM-1 and E-selectin in HAEC. Two key regulators of IκBα degradation—mitogen-activated kinase kinase kinase 7 (MAP3K7; TAK1) and β-transducin repeat-containing gene (βTRC)—contain a highly conserved miR-10a binding site in the 3′ UTR. Both molecules were up-regulated by miR-10a knockdown and suppressed by miR-10a knockin, and evidence of direct miR-10a binding to the 3′ UTR was demonstrated by luciferase assay. Comparative expression studies of endothelium located in athero-susceptible aortic arch and athero-protected descending thoracic aorta identified significantly up-regulated MAP3K7, βTRC, phopho-IκBα, and nuclear p65 expression suggesting that the differential expression of miR-10a contributes to the regulation of proinflammatory endothelial phenotypes in athero-susceptible regions in vivo.
Keywords: β-transducing repeat-containing gene, hemodynamics, mitogen-activated kinase kinase kinase 7, NF-κB
Atherosclerosis, a local chronic inflammatory disease of arteries, originates and develops preferentially at sites of curvature, branching, and bifurcation in large elastic and muscular distributing arteries where complex hemodynamic conditions are prominent (1, 2). The endothelium of prelesional athero-susceptible regions is subtly different from that located at nearby athero-resistant sites; differential transcriptional, translational, and posttranslational phenotypes have been identified, including sensitization of inflammatory regulators, coagulation, redox balance, endoplasmic reticulum stress, and lipid balance (3–9). Complementary experiments using cultured endothelial cells subjected to various arterial flow waveforms modeled from regions of athero-protection and susceptibility show several differentially expressed transcription factors that regulate downstream pathways important in endothelial function relevant to atherogenesis. Notably, athero-protective Krüppel-like factors promote an antiinflammatory and anticoagulant endothelial phenotype, whereas activator protein-1 and NF-κB induce a proinflammatory and procoagulant endothelium (10–13). Additional levels of regulatory control by small RNAs are now emerging in the cardiovascular system (14, 15). MicroRNAs (miRNAs), highly conserved noncoding small RNAs of 19–26 nucleotides, are key posttranscriptional gene regulators that contribute to the maintenance of differentiated cell phenotypes (16); however, the identities of miRNAs mediating endothelial phenotypes in relation to athero-susceptibility in vivo are unknown.
Mammalian miRNAs usually bind to the 3′ UTR of target mRNAs, promoting mRNA degradation and/or inhibiting translation of the protein-coding genes (16). Evolutionarily conserved Watson–Crick pairing between cognate mRNA 3′ UTR and miRNA 5′ regions centered on seed nucleotides (nucleotides 2–7) primarily determines miRNA target selection (17). Emerging evidence suggests that individual miRNAs fine-tune the synthesis of many genes and that miRNA-mediated proteomes typically are mirrored by transcriptomes (18, 19). Given the widespread scope but modest repression of transcriptomes/proteomes by individual miRNAs, it is proposed that phenotypical consequences can be achieved by coordinated actions on multiple targets by single miRNAs or that multiple miRNAs have an integrated regulatory effect on key pathways (20, 21). Disruption of the tightly regulated spatial and temporal expression of miRNAs is implicated in human diseases such as cancer, infectious diseases, and cardiovascular diseases. In the vascular system, endothelial miRNA-mediated angiogenesis has been demonstrated by mutation or disruption of Dicer (22, 23), the rate-limiting enzyme involved in miRNA maturation, and individual miRNAs associated with proangiogenic or antiangiogenic endothelial phenotypes have been identified (24, 25). Furthermore, endothelial miR-126 has been shown to suppress vascular cell adhesion molecule 1 (VCAM-1) expression in vitro, implicating miRNAs in the control of endothelial responses to vascular perturbation (26).
We report differential endothelial miRNA expression in vivo at athero-susceptible and athero-protected regions of aorta and renal arteries in normal adult swine in the absence of known systemic risk factors. Significant low endothelial expression of microRNA10a (miR-10a) detected in regions of athero-susceptibility was investigated by a combination of genomic profiling, microRNA manipulations, and molecular analyses in freshly isolated arterial endothelium and in cultured cells.
Results
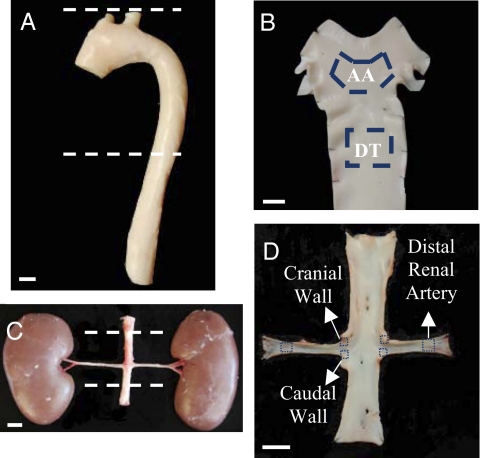
Endothelial cells were harvested from swine aortas and aorto-renal bifurcations from (i) the inner curvature of the aortic arch (AA) and nearby descending thoracic aorta (DT), and (ii) the cranial wall and caudal wall of the aorto-renal branches and the distal renal artery (Fig. 1).
Fig. 1.
Arterial regions of endothelial isolation. Endothelial cells were scraped gently from the inner curvature of AA, the DT, and the distal renal artery (A and B) as well as from the caudal and cranial regions of the aorto-renal bifurcations (C and D). (Scale bars, 1 cm.)
Low Expression of Endothelial miR-10a/b at Athero-Susceptible Arterial Sites in Vivo.
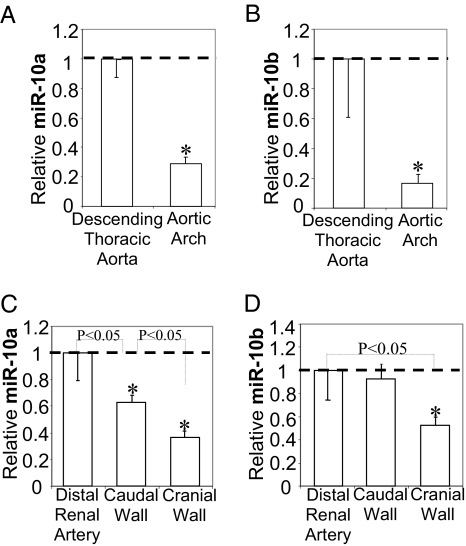
Differential expression analysis of multispecies miRNA microarrays identified seven down-regulated and twenty-seven up-regulated miRNAs in athero-susceptible AA endothelia (Fig. S1) when compared with athero-protected dorsal DT. The lowest relative expression was that of miR-10a/b. Real-time PCR demonstrated 71 ± 13% and 83 ± 15% lower expression of endothelial miR-10a and miR-10b, respectively, in the athero-susceptible AA (Fig. 2 A and B). A high incidence of human atherosclerosis at the cranial wall of the renal branch and low incidence in distal renal artery has been reported, whereas the caudal site of the renal branches exhibits modest susceptibility (27). Endothelial miR-10a and miR-10b expression were 63 ± 14% and 48 ± 25% lower, respectively, at the athero-susceptible cranial wall of the renal branches than at the protected distal renal artery (Figs. 2 C and D), whereas miR-10a expression at the caudal wall of the aorto-renal bifurcation was between the levels of the other two sites (Fig. 2C). The data demonstrate site-specific low expression of endothelial miR-10a/b in regions susceptible to atherosclerosis in vivo.
Fig. 2.
Low expression of endothelial miR-10a/b at athero-susceptible arterial sites. (A and B) Suppression of endothelial miR-10a and miR-10b at athero-susceptible AA compared with DT (n = 10). (C and D) Suppression of endothelial miR-10a and miR-10b at athero-susceptible cranial and caudal walls of the aorto-renal bifurcations compared with distal renal artery (n = 3 paired samples; each paired sample was pooled from the same four animals). Data represent mean ± SEM. *P < 0.05.
Endothelial Specificity and Relative Expression of miR-10a/b.
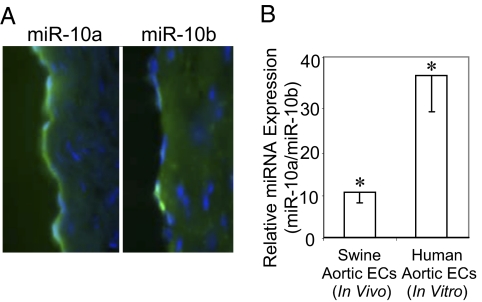
Locked nucleic acid (LNA)-FISH (28) of frozen sections of freshly isolated swine aorta showed miR-10a/b expression largely confined to the endothelium with stronger miR-10a staining than miR-10b staining (Fig. 3A and Fig. S2). The ratio of miR-10a/miR-10b expression in freshly isolated swine aortic endothelium and in cultured human aortic endothelial cells (HAEC), determined by quantitative real-time-PCR, showed endothelial miR-10a expression to be considerably higher than miR-10b (>10-fold and 38-fold respectively; Fig. 3B) in agreement with the FISH images. Low expression of miR-10a therefore was likely to be of greater consequence, and this isoform was investigated further.
Fig. 3.
Endothelial specificity of miR-10a/b. (A) In situ detection of endothelial miR-10a and miR-10b in frozen sections of freshly isolated swine DT by LNA-FISH. (B) Relative expression of endothelial miR-10a/b in freshly isolated swine aortic endothelial cells (n = 22, 11 AA and 11 DT) and in cultured HAEC (n = 8). Data represent mean ± SEM. *P < 0.05.
Validation of miR-10a Regulation of a Known Target Transcription Factor Homeobox A1.
Homeobox A1 (HOXA1) is an experimentally validated downstream target of miR-10a, as shown by direct HOXA1 3′ UTR-dependent suppression during megakaryocytopoiesis (29). Expression of HOXA1 therefore was examined in miR-10a knockdown HAEC in vitro and then the investigation was extended in vivo to swine endothelia at arterial sites exhibiting expression of different levels of miR-10a. In HAEC, miR-10a inhibitors suppressed endogenous expression by 81 ± 5% (Fig. S3A) while significantly up-regulating HOXA1 expression at the transcript and protein levels (Fig. S3B). The degree of inhibition of miR-10a obtained in vitro is comparable to the relatively low expression noted in AA in vivo (29% of DT). Similarly, HOXA1 was expressed at significantly higher levels in athero-susceptible AA (Fig. S3C) and at the cranial wall of aorto-renal bifurcations (Fig. S3D) in a reciprocal relationship to the down-regulation of miR-10a in these regions and consistent with miR-10a modulation of HOXA1 mRNA.
Activation of NF-κB Signal Transduction in miR-10a Knockdown HAEC.
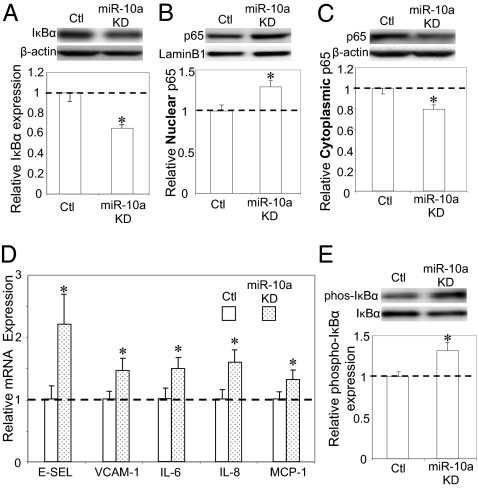
The effects of miR-10a knockdown on the endothelial transcriptome were determined in cultured HAEC by whole-genome microarray analyses. Differential transcript expression profiles identified 1,081 up-regulated and 794 down-regulated genes in miR-10a knockdown cells compared with control cells. The widespread impact on the endothelial transcriptome is consistent with recent studies demonstrating that a single miRNA can fine-tune the expression of many genes by direct or indirect effects (19, 20). Gene set enrichment analysis (GSEA), a knowledge-based approach for interpreting genome-wide expression profiles (30), identified IκB/NF-κB–mediated inflammation as the top biological processes up-regulated in miR-10a knockdown cells (Table 1). In nonactivated endothelial cells, NF-κB is bound to IκBs and is retained in the cytoplasm. Activation of NF-κB (nuclear translocation of p65) is contingent upon phosphorylation, polyubiquitination, and subsequent proteolysis of IκBs. In agreement with the GSEA analysis, nonphosphorylated IκBα was significantly decreased in miR-10a knockdown cells (Fig. 4A), whereas nuclear p65 increased at the expense of cytoplasmic p65 (Fig. 4 B and C). Confirmation of NF-κB activation in miR-10a knockdown HAEC was provided by significant up-regulation of the proinflammatory biomarkers monocyte chemotactic protein 1 (MCP-1), IL-6, IL-8, VCAM-1, and E-selectin (Fig. 4D). Endogenous phosphorylation of IκBα, a prerequisite for IκBα proteolysis, was not detectable, probably because of high turnover; however, when stimulated by a 10-min exposure to TNFα, phospho-IκBα was significantly elevated by miR-10a knockdown compared with control cells exposed to the cytokine (Fig. 4E). Therefore, inhibition of endogenous miR-10a led to changes in the endothelial transcriptome that promoted NF-κB activation, increased IκBα degradation, and augmented p65 nuclear translocation, resulting in up-regulation of inflammatory biomarkers.
Table 1.
Enriched GO biological processes up-regulated in miR-10a knockdown cells, identified by GSEA at 25% false discovery rate (FDR)
| GO biological process | Process gene set | FDR |
| GO:0043123 | Positive regulation of I-κB kinase/NF-κB cascade | 0.033 |
| GO:0043122 | Regulation of I-κB kinase/NF-κB cascade | 0.039 |
| GO:0007249 | I-κB kinase/NF-κB cascade | 0.058 |
| GO:0006955 | Immune response | 0.173 |
| GO:0006954 | Inflammatory response | 0.198 |
Fig. 4.
Activation of NF-κB signal transduction in miR-10a knockdown (KD) HAEC. (A) Suppression of total IκBα in miR-10a knockdown HAECs (n = 3). β-actin was used as an internal control (Ctl). (B and C) Increased nuclear expression and decreased cytoplasmic expression of NF-κB p65 in miR-10a knockdown cells. Nuclear (n = 4) and cytoplasmic (n = 3) expression of p65 was normalized to laminB1 and β-actin, respectively. (D) Up-regulation of E-selectin (E-SEL), VCAM-1, IL-6, IL-8, and MCP-1 in miR-10a knockdown HAEC. mRNAs of the inflammatory biomarkers were normalized to ubiquitin expression (n = 6 or 7). (E) Increased phosphorylated IκBα in miR-10a knockdown cells (n = 3). IκBα phosphorylation was stimulated by 5 ng/mL TNF-α for 10 min, and phospho-IκBα expression was normalized to total IκBα. Data represent mean ± SEM. *P < 0.05.
Suppressed Endothelial miR-10a Enhances Expression of MAP Kinase Kinase Kinase 7and β-Transducin Repeat-Containing Gene in Vitro.
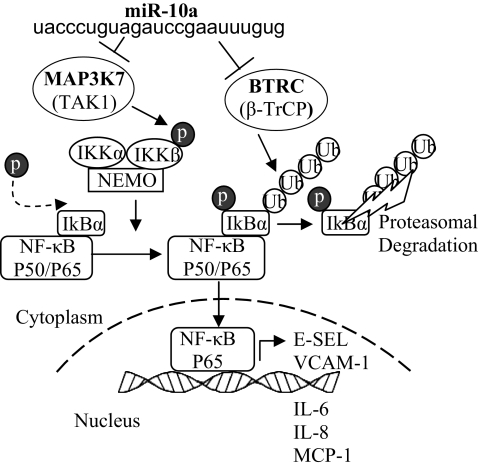
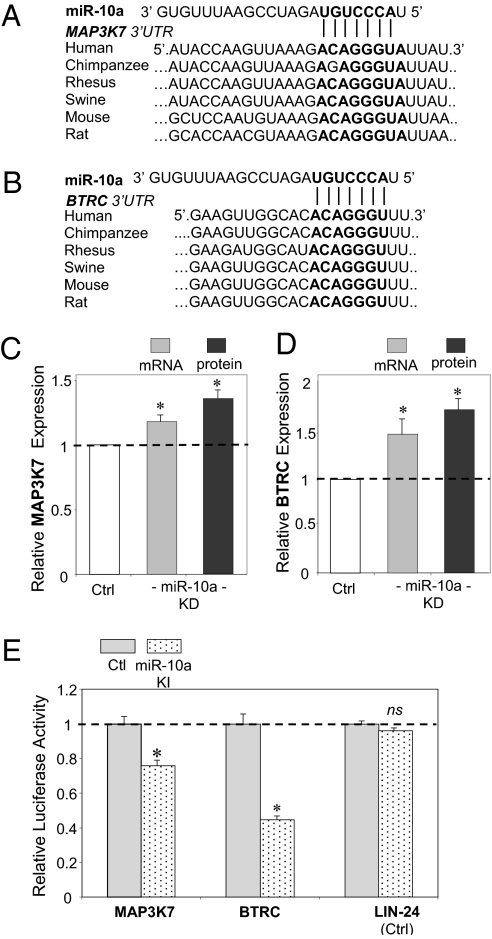
To investigate the molecular mechanisms underlying miR-10a regulation of proinflammatory molecules, genes in the canonical NF-κB pathway (Fig. S4) were interrogated with miR-10a putative downstream targets generated by TargetScan 5.1 that considers evolutionary conservation of miR seed sites (31). The in silico analyses identified two molecules: the mitogen-activated protein kinase kinase kinase 7 (MAP3K7; also known as TAK1) gene, and the β-transducin repeat-containing gene (βTRC; also known as β-TrCP). Both promote proteasomal degradation of IκBα and p65 nuclear translocation (Fig. 5) and contain evolutionarily conserved miR-10a binding sites in the 3′ UTRs (Fig. 6 A and B). MAP3K7 protein directly phosphorylates and activates IκB kinase β (IKKβ), stimulating phosphorylation of IκBs. βTRC recognizes phosphorylated IκBs and mediates phosphorylation-dependent ubiquitination leading to IκB proteolysis. Knockdown of endothelial miR-10a in HAEC significantly up-regulated MAP3K7 and βTRC expression as demonstrated by real-time PCR and Western blot (Fig. 6 C and D). The up-regulation of MAP3K7 and βTRC by knockdown of miR-10a, predicted by in silico analyses, is consistent with the activation of NF-κB signal transduction in miR-10a knockdown HAEC.
Fig. 5.
Role of miR-10a in canonical NF-κB signaling. MAP3K7 and βTRC contain an evolutionarily conserved miR-10a putative binding site, and both promote proteasomal degradation of IκBα and p65 nuclear translocation with induction of downstream inflammatory biomarkers.
Fig. 6.
miR-10a negatively regulates MAP3K7 and βTRC through their 3′ UTR binding site. (A and B) Evolutionarily conserved putative miR-10a binding site within the 3′ UTR of MAP3K7 and βTRC. (C and D) Increased expression of MAP3K7 in miR-10a knockdown HAEC. Gene (n = 7–10) and protein (n = 3) expressions were normalized to GAPDH and β-actin, respectively. (E) Reduced luciferase activity in HEK293 cells overexpressing miR-10a following insertion of miR-10a binding elements cloned from MAP3K7 and βTRC 3′ UTRs. Data represent mean ± SEM. ns, not significant. *P < 0.05.
miR-10a Inhibits MAP3K7 and βTRC Through 3′ UTR Binding.
A dual-luciferase reporter assay demonstrated a direct interaction of miR-10a and the 3′ UTR sequences of MAP3K7 and βTRC. Three tandem repeats of the predicted miR-10a recognition elements present in MAP3K7 or βTRC 3′ UTR were inserted into the 3′ UTR of Renilla luciferase (Fig. S5). Intracellular delivery of miR-10a biomimetics significantly repressed the Renilla luciferase activity, by 25% and 55%, respectively, in HEK293 cells expressing MAP3K7 3′ UTR- or βTRC 3′ UTR-containing luciferase transcripts (Fig. 6E). Renilla luciferase inserted with putative let-7b binding sites, cloned from LIN-24 3′ UTR, was not responsive to miR-10a knockin (Fig. 6E). It is notable that insertion of the miR-10a recognition element cloned from βTRC led to higher luciferase inhibition than seen with MAP3K7, even though both sequences contain an identical evolutionarily conserved binding site for the miR-10a seed region; this finding indicates that additional structural features of these miR-10a recognition elements may influence the miR-10a–mediated regulation (32). Consistent with this observation, inhibition of endogenous endothelial miR-10a led to higher regulation of βTRC than MAP3K7 at the transcriptional and translational levels (Fig. 6C).
Overexpression of Endothelial miR-10a Down-Regulates Inflammatory Molecules in Vitro.
Intracellular delivery of biomimetic miRs (typically at 25–50 nM) results in overexpression by several thousand-fold. We titrated miR-10a overexpression to a lower, more physiologically relevant range (∼25-fold at 0.01 nM; Fig. S6A) in cultured HAEC. This level of miR-10a up-regulation significantly inhibited basal transcript expression of MAP3K7, βTRC, and the downstream proinflammatory markers E-selectin and VCAM-1 (Fig. S6B).
MAP3K7, βTRC, IκBα, and p65 in Athero-Susceptible AA in Vivo.
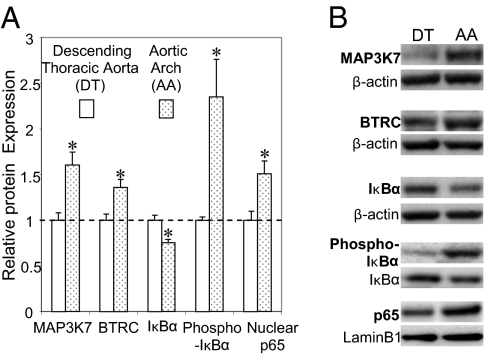
Finally, to investigate evidence for an in vivo equivalent of the in vitro knockdown and knockin experiments, endothelial MAP3K7 and βTRC expression were measured in athero-susceptible swine AA, a site of suppressed expression of miR-10a. Both genes were significantly up-regulated in AA compared with DT (Fig. 7). Furthermore, in AA endothelium, steady-state phospho-IκB was increased, but IκBα expression declined, indicating increased proteolysis of IκBα with a significant elevation of nuclear p65 in the same athero-susceptible region (Fig. 7). These in vivo data are consistent with a steady-state reciprocal relationship between miR-10a suppression and proinflammatory endothelial phenotype at athero-susceptible sites.
Fig. 7.
Endothelial protein expression in swine athero-susceptible AA andathero-protected DT. (A) Differential endothelial expression of MAP3K7, βTRC, IκBα, phosphorylated IκBα (n = 5), and nuclear p65 (n = 4; each sample was pooled from the seven animals) in paired comparisons of AA and DT. Data represent mean ± SEM. *P < 0.05. (B) Representative Western blots.
Discussion
Despite exposure of the entire arterial tree to systemic risk factors, atherosclerosis preferentially develops in predisposed arterial regions of flow disturbance. In the prelesion stage of normal arteries, athero-susceptible regions exhibit low-grade chronic inflammation in the intima where enhanced NF-κB activity in endothelial cells may influence the regional sensitivity for atherogenesis (7, 33). In a mouse model, increased phosphorylation of IκBα in lesion-susceptible regions is consistent with preferential activation of endothelial NF-κB signaling under the systemic stimuli of hypercholesterolemia and lipopolysaccharide (7). Regional expression of kinases, small GTPases, and transcription factors is proposed to account, in part, for the predisposition of NF-κB activation in the lesion-prone regions (3, 6, 10, 34, 35). Here we addressed miRNA regulation by directly accessing athero-susceptible and athero-protected endothelial regional miRNA profiles to determine steady-state miRNA expression in vivo. In silico analyses and in vitro interventions demonstrated that miR-10a suppression in athero-susceptible regions probably contributes to endothelial proinflammatory NF-κB signaling in vivo.
Two phases of the study used unbiased genomics, first to identify regional miRNA expression in arterial endothelium and second to determine the target pathways most affected by miR-10a knockdown in HAEC. From this study emerged in silico putative miR-10a targets that are influential in the regulation of IkB. We prioritized miR-10 for study because it is the most highly modulated miRNA identified. When the results suggested by the in vitro HAEC manipulations were probed in vivo, a pattern consistent with miR-10a regulation of proinflammatory phenotype emerged.
The interplay of miRNAs with their targets is complex, and it is unlikely that any single miRNA or its targets are entirely responsible for spatially related differences in endothelial phenotype in arteries. As reported in Fig. S1, transcript profiling identified the differential expression of several other miRNAs in swine athero-susceptible regions; these miRNAs also are under investigation and may relate to the current findings. In that context, Zhou et al. (36) recently identified four miRNAs in inflammatory epithelial cells that are directly activated by the binding of p65 to their promoter regions: miR-21, miR-23b, miR-30b, and miR-125b. In our global profiling, all four miRNAs are modestly up-regulated in AA endothelial cells (Fig. S1), a site of suppressed miR-10a and elevated p65 nuclear expression. These data not only support a different equilibrium state of NF-κB activation in the athero-protective and athero-susceptible endothelial phenotypes in vivo but also support a putative indirect role of other miRNAs in endothelial miR-10a regulation.
Without preconceived expectations of NF-κB pathway regulation by miR-10a, GSEA identified up-regulated IκB/NF-κB signaling pathways and inflammatory response in miR-10a knockdown HAEC, and the mechanistic link was predicted when genes that direct or mediate the NF-κB canonical pathways were cross-referenced with the miR-10a putative downstream targets generated in silico by TargetScan 5.1. Evolutionarily conserved miR-10a binding sites were identified in the IκB/NF-κB regulators MAP3K7 and βTRC. Both genes have a well-established role in activating NF-κB signal transduction by provoking IκB degradation. MAP3K7 activates IKKβ that phosphorylates IκBs in response to inflammatory stimuli (37). βTRC constitutes the substrate recognition subunit of SCFb-TrCP E3 ubiquitin ligase that ubiquitinates the phosphorylated IκBs (38). Phosphorylation and subsequent ubiquitination of IκBs leads to a 26S proteome-mediated proteolysis of IκBs and consequent nuclear translocation of active NF-κB dimers. Inhibition of either MAP3K7 or βTRC increases the total expression of IκB and impairs NF-κB activation. Our data demonstrate miR-10a as a posttranscriptional regulator of the IκB/NF-κB signaling pathway by suppressing MAP3K7 and βTRC. It has been suggested that coordinated action on multiple target genes by a single miRNA could provide a powerful mechanism by which a single miRNA can impact a complex regulatory network and ultimately the physiological process or disease (21).
Members of the miR-10 family are regulators of HOX genes during development and are deregulated in several cancer forms (39). Garzon et al. (29) reported the inhibition of miR-10a during megakaryocytic differentiation and identified its direct downstream target HOXA1 (used for site-specific validation in our study). miR-10a knockdown in HAEC led to a widespread impact on the transcriptome (∼1,900 genes) but did not identify the HOX-mediated gene network as one of the top regulated biological processes in the present study. The global and moderate influence of miR-10a on endothelial mRNA expression is consistent with recent transcriptional analyses in individual RNA knockin/knockdown HeLa cells (19, 20) and is in agreement with the emerging view that a single miRNA represses numerous genes that mediate a wide variety of biological and molecular functions (20). Recently, miR-10a regulation of ribosomal proteins via a HOX-independent mechanism was reported (40).
In the cardiovascular system miRNAs are expressed in a temporal-spatial fashion. Emerging studies show them to be important modulators of cardiovascular development (41) and angiogenesis (25), and aberrant miRNA expression is associated with cardiac arrhythmia, cardiac hypertrophy, and arterial stenosis (42). They are implicated in arterial hyperplasia through regulation of proliferation and phenotype switching of vascular smooth muscle cells (43). Two recent studies in cultured endothelial cells have linked miR126 to TNF-induced endothelial VCAM-1 expression (26) and miR-31 and miR17-3p to E-selectin and intercellular adhesion molecule 1 expression, respectively (44). However, we did not identify differential expression of these miRNAs in atherosusceptible sites in vivo. A recent report (45) included miR-10a (among others) as a flow-responsive miR in vitro which may be related to a hemodynamic mechanism underlying our in vivo findings. By identifying miR-10a as a posttranscriptional regulator of NF-κB activation through MAP3K7 and βTRC, our study links miRNA regulation in arterial endothelium with endothelial athero-susceptibility/protection signatures in vivo. Furthermore, because inhibition of IκBα reduces atherosclerotic plaque formation in apoE−/− mice (46), noncoding miRNAs may be suitable therapeutic targets in vascular inflammation.
Methods
Expanded methods are provided in SI Methods.
Swine Sample Collection.
Arterial tissues were obtained from adult pigs (6-mo-old; ∼250 lb) immediately after pigs were killed at a local abattoir (Hatfield Industries). Endothelial cells were freshly harvested by gentle scraping of regions described in Fig. 1. Endothelial cell purity assessed by antibody staining for endothelium, smooth muscle cells, and leukocytes ranged between 96–100% (3, 9) and was not significantly different for AA and DT regions.
MicroRNA Microarray.
Endothelial cells were collected from the athero-susceptible AA and athero-protected DT of 35 swine. For each arterial site, endothelial samples from five animals were pooled to provide seven biological replicates for microarray experiments. miRNAs were isolated using the mirVana isolation kit (Ambion) and profiled using NCode multispecies miRNA microarrays (Invitrogen). The complete Minimum Information about a Microarray Experiment (MIAME)-compliant annotated study has been deposited in the public repository ArrayExpress (accession number E-CBIL-46). Detailed MIAME-compliant annotation and data for this study also are available for query at www.cbil.upenn.edu/RAD.
MicroRNA and cDNA Quantitative Real-Time PCR.
Expression of selected miRNAs was quantified by two-step quantitative real-time PCR (Applied Biosystems) and normalized to endogenous small nuclear U6B RNA. cDNA was quantified using LightCycler FastStart DNA Master SYBR Green I or Masterplus SYBR Green I (Roche). PCR primers for genes of interest are listed in Table S1.
In Situ Hybridization.
Expression of miR-10a/b in freshly isolated swine arterial tissues was determined by a modified FISH assay that employs LNA probes and tyramide signal amplification, described by Silahtaroglu et al. (28).
Human Whole-Genome Microarray.
The complete MIAME-compliant annotated study has been deposited in ArrayExpress (accession number E-CBIL-45), and annotation and data are available at www.cbil.upenn.edu/RAD. Whole-genome transcriptomes were determined in control and miR-10a knockdown cells by whole-human genome 4 × 44K microarrays (Agilent).
Transfection of miR-10a Mimics and Inhibitors.
HAECs (Lonza) were transfected with Homo sapiens miR-10a (hsa-miR-10a) mimics, miRNA mimic negative controls, hsa-miR-10a inhibitors, or miRNA inhibitor negative controls (Dharmacon) using Lipofectamine RNAiMAX transfection reagent.
Bioinformatics.
Data preprocessing of the microarray studies is described in SI Methods. Differential expression analysis was performed using Patterns of Gene Expression (47; PaGE v5.1.6). Genes/miRNAs with less than a 5% false discovery rate were considered to be differentially expressed. GSEA was used to identify enriched Gene Ontology (GO) gene sets.
Luciferase Reporter Assay.
Three tandem repeats of the miR-10a binding elements were synthesized and cloned into a pRL-TK vector (Promega). HEK 293 cells were cotransfected with pRL-TK vectors, control pGL3 vectors, and hsa-miR-10a mimics or miRNA mimic negative controls, followed by dual-luciferase assays (Promega).
Endothelial Protein Extraction and Western Blots.
Protocols are described in a previous study (9) and in SI Methods. Table S2 lists the primary antibodies used. Statistical significance was assessed for the gene and protein expression ratios using unpaired two-tailed Student's t test assuming experimental samples of unequal variance.
Supplementary Material
Acknowledgments
We thank Drs. Zissimos Mourelatos, Michael May, and Hun-Way Hwang for critical reading of the manuscript, Drs. Chris Stoeckert and Greg Grant for advice on experimental design, Mr. Junmin Liu for data submission to ArrayExpress, and Drs. Yohei Kirino, Zissimos Mourelatos, and Scott Diamond and Ms. Olga Lozynska for assistance with luciferase constructs and assays. This work was supported by National Institutes of Health Grant HL62250 (to P.F.D.) and an American Heart Association Postdoctoral Fellowship 0725385U (to Y.F.) and Predoctoral Fellowship 0315286U (to M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C. is a guest editor invited by the Editorial Board.
Data deposition: The complete MIAME compliant annotated study of the miRNA microarray experiment has been deposited in the public repository ArrayExpress (accession number E-CBIL-46). The complete MIAME compliant annotated study of the cDNA microarray experiment has been deposited into ArrayExpress (accession number E-CBIL-45).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002120107/-/DCSupplemental.
References
- 1.Schwartz CJ, Mitchell JR. Observations on localization of arterial plaques. Circ Res. 1962;11:63–73. [PubMed] [Google Scholar]
- 2.Ku DN, Giddens DP, Zarins CK, Glagov S. Pulsatile flow and atherosclerosis in the human carotid bifurcation. Positive correlation between plaque location and low oscillating shear stress. Arteriosclerosis. 1985;5:293–302. doi: 10.1161/01.atv.5.3.293. [DOI] [PubMed] [Google Scholar]
- 3.Passerini AG, et al. Coexisting proinflammatory and antioxidative endothelial transcription profiles in a disturbed flow region of the adult porcine aorta. Proc Natl Acad Sci USA. 2004;101:2482–2487. doi: 10.1073/pnas.0305938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross PL, Aird WC. The endothelium and thrombosis. Semin Thromb Hemost. 2000;26:463–478. doi: 10.1055/s-2000-13202. [DOI] [PubMed] [Google Scholar]
- 5.Volger OL, et al. Distinctive expression of chemokines and transforming growth factor-beta signaling in human arterial endothelium during atherosclerosis. Am J Pathol. 2007;171:326–337. doi: 10.2353/ajpath.2007.061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magid R, Davies PF. Endothelial protein kinase C isoform identity and differential activity of PKCzeta in an athero-susceptible region of porcine aorta. Circ Res. 2005;97:443–449. doi: 10.1161/01.RES.0000179767.37838.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajra L, et al. The NF-kappa B signal transduction pathway in aortic endothelial cells is primed for activation in regions predisposed to atherosclerotic lesion formation. Proc Natl Acad Sci USA. 2000;97:9052–9057. doi: 10.1073/pnas.97.16.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai G, et al. Biomechanical forces in atherosclerosis-resistant vascular regions regulate endothelial redox balance via phosphoinositol 3-kinase/Akt-dependent activation of Nrf2. Circ Res. 2007;101:723–733. doi: 10.1161/CIRCRESAHA.107.152942. [DOI] [PubMed] [Google Scholar]
- 9.Civelek M, Manduchi E, Riley RJ, Stoeckert CJ, Jr, Davies PF. Chronic endoplasmic reticulum stress activates unfolded protein response in arterial endothelium in regions of susceptibility to atherosclerosis. Circ Res. 2009;105:453–461. doi: 10.1161/CIRCRESAHA.109.203711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekker RJ, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Krüppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 11.Parmar KM, et al. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkins GB, Jain MK. Role of Krüppel-like transcription factors in endothelial biology. Circ Res. 2007;100:1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- 13.Lan Q, Mercurius KO, Davies PF. Stimulation of transcription factors NF kappa B and AP1 in endothelial cells subjected to shear stress. Biochem Biophys Res Commun. 1994;201:950–956. doi: 10.1006/bbrc.1994.1794. [DOI] [PubMed] [Google Scholar]
- 14.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 15.van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 18.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang M. MicroRNA: A new entrance to the broad paradigm of systems molecular medicine. Physiol Genomics. 2009;38:113–115. doi: 10.1152/physiolgenomics.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 23.Suárez Y, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonauer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 25.Suárez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debakey ME, Morris GC, Jr, Morgen RO, Crawford ES, Cooley DA. Lesions of the Renal Artery. Surgical Technic and Results. Am J Surg. 1964;107:84–96. doi: 10.1016/0002-9610(64)90244-2. [DOI] [PubMed] [Google Scholar]
- 28.Silahtaroglu AN, et al. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 29.Garzon R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Mourelatos Z. Small RNAs: The seeds of silence. Nature. 2008;455:44–45. doi: 10.1038/455044a. [DOI] [PubMed] [Google Scholar]
- 33.Jongstra-Bilen J, et al. Low-grade chronic inflammation in regions of the normal mouse arterial intima predisposed to atherosclerosis. J Exp Med. 2006;203:2073–2083. doi: 10.1084/jem.20060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzima E, et al. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 35.Bhullar IS, et al. Fluid shear stress activation of IkappaB kinase is integrin-dependent. J Biol Chem. 1998;273:30544–30549. doi: 10.1074/jbc.273.46.30544. [DOI] [PubMed] [Google Scholar]
- 36.Zhou R, et al. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 38.Winston JT, et al. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund AH. miR-10 in development and cancer. Cell Death Differ. 2010;17:209–214. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 40.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 43.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suárez Y, Wang C, Manes TD, Pober JS. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: Feedback control of inflammation. J Immunol. 2010;184:21–25. doi: 10.4049/jimmunol.0902369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin X, et al. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci USA. 2010;107(7):3240–3244. doi: 10.1073/pnas.0914882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gareus R, et al. Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metab. 2008;8:372–383. doi: 10.1016/j.cmet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 47.Grant GR, et al. A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;21:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.