Abstract
The most remarkable outcome of a gene duplication event is the evolution of a novel function. Little information exists on how the rise of a novel function affects the evolution of its paralogous sister gene copy, however. We studied the evolution of the feminizer (fem) gene from which the gene complementary sex determiner (csd) recently derived by tandem duplication within the honey bee (Apis) lineage. Previous studies showed that fem retained its sex determination function, whereas the rise of csd established a new primary signal of sex determination. We observed a specific reduction of nonsynonymous to synonymous substitution ratios in Apis to non-Apis fem. We found a contrasting pattern at two other genetically linked genes, suggesting that hitchhiking effects to csd, the locus under balancing selection, is not the cause of this evolutionary pattern. We also excluded higher synonymous substitution rates by relative rate testing. These results imply that stronger purifying selection is operating at the fem gene in the presence of csd. We propose that csd's new function interferes with the function of Fem protein, resulting in molecular constraints and limited evolvability of fem in the Apis lineage. Elevated silent nucleotide polymorphism in fem relative to the genome-wide average suggests that genetic linkage to the csd gene maintained more nucleotide variation in today's population. Our findings provide evidence that csd functionally and genetically interferes with fem, suggesting that a newly evolved gene and its functions can limit the evolutionary capability of other genes in the genome.
Keywords: sex-determining gene, molecular constraint, balancing selection, honey bee
An important question in genome evolution is how the rise of a gene with a novel function influences the genomic region around that gene and the evolution of other genes in the genome. Under neofunctionalization, the newly arisen gene gains a function not present in the progenitor gene, whereas the original copy retains its function (1). Thus, duplicated genes that have evolved under a model of neofunctionalization provide an interesting case for exploring this question. Although the rise of novel gene functions has been studied in great detail in some examples of duplicated genes (2–4), how selection and the rise of a new function in the duplicated copy affects the evolution of the sister copy remains unclear. This evolutionary interference has not yet been studied in paralogous genes. This process differs from coevolutionary effects that implicate bidirectional interference between the tandem duplicated genes. Coevolution has been intensively studied in interacting protein systems, such as the cellular signaling pathway of protein ligands and their receptors (5, 6).
By examining the paralogous genes feminizer (fem) and complementary sex determiner (csd), which control sex determination in honey bees (7, 8), we can explore whether evolutionary interference plays a role in the evolution of tandemly arranged genes. We previously showed that after the duplication event in the honey bee lineage, fem preserved its ancestral function, whereas csd, following the model of neofunctionalization, achieved a new mode of protein activation by allelic combination (8). Both genes belong to the SR-type proteins, harboring an arginine/serine-rich domain, but only csd is required to induce the female pathway, whereas fem maintains this decision throughout development through a positive feedback splicing loop (9). We showed that the function of fem in Apis and its ortholog transformer (tra) in other insects (10, 11) is ancestral among holometabolous insects for the control of sexual development and regulation of doublesex (dsx) alternative splicing (9). The fem gene evolved under purifying selection and is the conserved progenitor from which csd arose after the split of stingless bees, bumble bees, and honey bees (∼70 mya) (12) but before honey bee divergence (∼10 mya). The csd locus evolved under a balancing mode of selection with a strong heterozygote advantage (7). Thus, rare alleles have a selective advantage and are maintained over extended periods compared with neutral polymorphism (13).
In the present study, we tested the hypothesis that the origin of csd affected the evolution of the fem gene within the Apis lineage. We raised the question whether the evolution of the fem protein and nucleotide sequence was affected by the rise of the csd gene and its new function. We then explored whether the observed evolutionary pattern is consistent with an evolutionary interference because of similar protein functions (functional interference) or hitchhiking effects of genetically linked genes (genetic interference). We examined the divergence of fem nucleotide sequences before and after the origin of the csd gene by comparing three Apis species and three non-Apis bees (stingless bees, bumble bees, and orchid bees). We used two other closely linked genes (GB13727 and GB11211) (8) as a contrast to explore the effect of linkage to csd and its strong balancing selection regime on sequence evolution. Finally, we analyzed nucleotide polymorphism of A. mellifera populations to gain insights into recent evolutionary forces acting on fem. Our findings provide evidence that the origin of a new regulatory function in csd constrains the molecular evolution of its sister copy gene fem, from which this novel function ancestrally derived.
Results
Evolutionary History of the fem and csd Genes.
To explore the evolutionary history of fem before and after the origin of csd, we compared fem nucleotide coding sequences of three Apis species (mellifera, cerana, and dorsata) and three related corbiculate non-Apis species (Bombus terrestris, Melipona compressipes, and Euglossa hemichlora), as well as csd nucleotide coding sequences of the three Apis species. We inferred differences in ratios of nonsynonymous to synonymous changes along branches of the fem genealogy and used these as indicators for differences in evolutionary forces that shaped fem nucleotide sequences. By comparing these ratios in bees that are increasingly distantly related, we can retrace differences in nucleotide evolution back in time to glean information on increased differences in selection before and after the origin of csd in the bee lineage. We inferred the ancestral gene sequence on all interior branch nodes of the genealogy of csd and fem coding sequences, relying on the phylogenetic tree of corbiculate bees [Apis species, B. terrestris, M. compressipes, and E. hemichlora (12)] using the orchid bee sequence E. hemichlora as an outgroup (Fig. 1A and Fig. S1A). Genealogies of csd and fem sequences using maximum likelihood and minimum evolution methods (see Materials and Methods) demonstrated the same relationship of the corbiculate bees as proposed in the phylogeny (Fig. S1B). We tested alternative phylogenetic relationships among corbiculate bees and found that they did not affect the outcome of our analysis. The dn/ds ratios of the fem gene were 0–0.52 in all of the tree branches, substantially less than 1 (P < 0.05, two-tailed Fisher's exact test) (Fig. 1B), suggesting that fem evolved under purifying selection in these species (8).
Fig. 1.
Nucleotide sequence evolution of the fem and csd genes in corbiculate bees. (A) Evolutionary tree of the fem and csd genes of corbiculate bees. Evolutionary time windows, as shown above the figure, correspond to the rise of csd before Apis species divergence, the preservation of csd within Apis species, and current populations. The absolute numbers of nonsynonymous (n) and synonymous (s) changes are shown along each branch. The summed numbers of these changes for Apis and non-Apis species (Melipona/Bombus) in the second evolutionary time window are shown in bold in the gray boxes. (B) Diagram of the ratios of nonsynonymous and synonymous changes per site as calculated from the estimates obtained from the evolutionary tree in A. The ratios of n/s substitutions per site (dn/ds) are shown for the different evolutionary time windows assigned in A. The ratios of the average pairwise nonsynonymous and synonymous diversity per site (πn/πs) were calculated from population sequence samples of A. mellifera (nA.m. csd = 28; nA.m. fem = 30) and B. terrestris (nB.t.fem = 23). Asterisks above columns indicate that n/s substitution ratios differ from 1 (two-tailed Fisher's exact test, performed on absolute numbers given in A; **P < 0.01; *P < 0.05). The striped column indicates a dn/ds ratio that has a denominator of 0.
Ratio of Nonsynonymous to Synonymous Substitutions in the fem Gene Was Reduced After the Rise of the csd Gene in the Honey Bee Lineage.
We first compared the ratio of nonsynonymous (n) to synonymous (s) substitutions in the two evolutionary time windows of the fem gene in the Apis lineage: (i) fem after the rise of csd but before Apis species divergence and (ii) preservation of csd within the Apis species. The ratios did not differ (n/s = 1/10 vs. n/s = 8/29, respectively; P = 0.43, two-tailed Fisher's exact test), providing no evidence that different strengths of purifying selection operated after divergence of fem and csd in the various Apis species.
We compared substitution ratios in Apis and other corbiculate bees representing lineages in which csd is absent (Fig. 1A). The n/s substitution ratios in three tree branches of the fem genealogy that present lineages lacking csd in the genome were all significantly elevated compared with the two evolutionary time windows of Apis (P < 0.05, two-tailed Fisher's exact test), but not significantly elevated compared with single branches representing the stingless (M. compressipes) and orchid bee (E. hemichlora) species (P = 0.087–0.099) (Fig. 1A and Table 1). We tested for heterogeneity among the n/s ratios of the non-Apis species derived from the different comparisons given in Table 1 by applying the replicated G test of goodness of fit. The heterogeneity G value was significant (G = 30.5; P < 0.001), indicating that the n/s ratios in the non-Apis species are significantly different from one another and cannot be viewed as a homogeneous group, suggesting different evolutionary and mutational rates in non-Apis species. This finding does not affect the finding of a reduced n/s ratio in the Apis lineages, however.
Table 1.
Comparison of n/s ratios of the fem and csd genes
| MB fem (20/12) | fem MB (64/73) | M. compressipes (28/45) | B. terrestris (36/28) | fem anc (67/26) | E. hemichlora (36/59) | |
| A fem (1/10) | P < 0.004 | P < 0.023 | P = 0.087 | P < 0.007 | P < 0.0001 | P = 0.092 |
| fem Apis (8/29) | P < 0.001 | P < 0.008 | P = 0.088 | P < 0.001 | P < 0.0001 | P = 0.099 |
Estimated n/s substitution ratios for each branch of the tree in Fig. 1A are given in parentheses. Differences between ratios were examined using the two-tailed Fisher's exact test. The n/s ratios of non-Apis species are heterogeneously distributed, as indicated by the G-test of heterogeneity (G = 30.5; P < 0.001).
The finding of specifically reduced n/s substitution ratio in the Apis lineage may suggest an increased strength of purifying selection after gene duplication and a rise in the paralog csd gene in the honey bees. Alternatively, this reduced ratio could be due to an increased synonymous substitution rate in the lineage of Apis fem, possibly resulting from an increased mutation rate. To examine these two hypotheses, we applied the relative rate test to identify an acceleration of synonymous substitution rates in the Apis tree branches as an indicator for increased mutation rates. We used synonymous changes because they are more or less neutral and immune to selection (except in the case of weak selection on codon usage in certain organisms). The relative substitution rates between Apis and non-Apis species did not differ (P > 0.5; Table 2), suggesting no specific increase in the mutation rate in the Apis branches of the genealogy. The reduced n/s ratios thus imply that purifying selection operated more strongly on the fem gene after the csd gene originated by gene duplication.
Table 2.
Tests of the relative evolutionary rate of fem and csd Apis sequences
| Lineage 1 | Lineage 2 | Ks1 | Ks2 | dKs | SD | P Ks |
| fem Apis | fem Melipona/Bombus | 0.46 | 0.42 | 0.04 | 0.06 | 0.55 |
| csd Apis | fem Apis | 0.48 | 0.46 | 0.02 | 0.02 | 0.52 |
Test of differences in relative evolutionary rate of fem and csd coding sequences based on synonymous substitutions (Ks) using Euglossa fem as an outgroup sequence. dK presents the difference between K1 and K2 (K1 − K2) with their SDs.
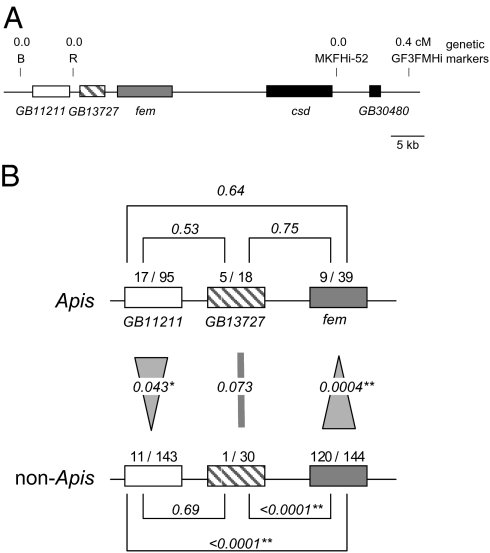
We compared divergence data of two other genetically linked genes, GB13727 and GB11211 (Fig. 2A), to contrast the general impact of genetic linkage with the csd gene and its balancing mode of selection on nucleotide sequence evolution. If genetic linkage to csd caused the reduced n/s ratio, then we would expect to find a similar pattern at the other two genes as well. We found no reduction in n/s ratio for GB13727 and even an increase in n/s ratio for GB11211 in Apis species compared with non-Apis species (Fig. 2B). This result suggests that the specific reduction in n/s ratio in Apis fem cannot be explained by general hitchhiking effects to the csd gene. A comparison of n/s ratios across Apis and non-Apis genes supports our conclusion. In non-Apis, fem shows relaxed constraints compared with the other two genes (Fig. 2B). This is consistent with the rapid evolutionary rate reported for fem orthologs in other dipteran species (14, 15). In contrast, fem in Apis does not show this pattern (Fig. 2B).
Fig. 2.
Genomic relationships of genes GB13727, GB11211, fem, and csd in the honey bee A. mellifera and their nucleotide sequence evolution in Apis and other corbiculate bees. (A) The genomic and genetic map of SDL (sex determination locus). GB13727 lies 3.5 kb and GB11211 lies 10.3 kb upstream of fem’s translational start codon. Neither gene is involved in sex determination. (B) Accumulated number of nonsynonymous (n) and synonymous (s) changes in the evolution of GB11211, GB13727, and fem genes in Apis and non-Apis species. The n/s ratio estimates are shown above the boxes, and the gene names are given below the boxes. The n/s ratios were compared by two-tailed Fisher's exact test, and the probabilities are given in italics above the connection lines denoting the comparison (**P < 0.01; *P < 0.05).
To further identify possible target regions of stronger purifying selection within the fem gene, we tested for evolutionary heterogeneity across the gene. Heterogeneity in the evolution of the fem orthologous gene transformer has been documented in several Drosophila species, in which the RS domain involved in protein binding evolves faster than other parts of the protein (14, 15). We compared the n/s ratios across the fem gene corresponding to the N-terminal (N-term) region, the arginine/serine-enriched (RS) domain, and the proline-rich (P) domain. We found no evidence of heterogeneity in the branches of the Apis and non-Apis trees (P > 0.2, two-tailed Fisher's exact test) (Table S1).
We used a sliding window method to identify and narrow down possible protein regions that were strongly targeted by purifying selection. We calculated the fem dn/ds ratios between A. mellifera and M. compressipes, between A. mellifera and B. terrestris, and between M. compressipes and B. terrestris in consecutive 30-bp windows. A region in fem showing a low dn/ds ratio in both the Apis–Melipona and Apis–Bombus comparisons but a high dn/ds ratio in the Melipona–Bombus comparison would be an indication of strong selective constraints of this region in the Apis lineage. We detected one region of 18 amino acids with a low dn/ds ratio between Apis and non-Apis (dn/ds Apis/Melipona = 0.25, dn/ds Apis/Bombus = 0.65) and high dn/ds in non-Apis (dn/ds Melipona/Bombus = 1.79; Z > 1.96; P < 0.05, two-tailed Z test) comparisons (Fig. S2). This region is a specific target of increased purifying selection operating specifically in the honey bee lineage. The region encodes part of the RS domain that has protein-binding abilities.
Genetic Linkage to csd Increases Nucleotide Diversity at fem in A. mellifera Populations.
We also studied the evolution of fem in today's honey bee population by analyzing nucleotide polymorphism. Balancing selection acting at the csd gene maintains 16 times more nucleotide diversity than the genome-wide average (13, 16). Alleles of csd are seldom lost through the process of genetic drift and are maintained over extended periods, during which they accumulate numerous synonymous mutations (17). We have previously shown that the nucleotide diversity of neutral linked loci decreases gradually with physical distance of 1.5–45 kb to csd (16). We compared the nucleotide diversity (π) of fem, csd, and the genome-wide average in A. mellifera to examine whether genetic linkage to csd could elevate the level of nucleotide diversity of fem. The synonymous diversity was substantially lower (∼8-fold) in fem compared with csd (π femsyn = 0.0084 ± 0.0036 vs. π csdsyn = 0.0646 ± 0.0142; Z = 3.8; P < 0.001, two-tailed Z test) (Table 3), consistent with the expectation that recombination would decrease linkage and nucleotide diversity. Comparing π femsyn with the genome-wide average data (16) (πgenome = 0.0055 ± 0.0012) revealed no differences (Z = 0.76; P > 0.8, two-tailed Z test). To further confirm our findings, we included fem intron sequences 8–10 into the comparison. The intron diversity of fem (fem πintron = 0.0151 ± 0.0026) exceeded the genome-wide average diversity (Z = 3.2; P < 0.001, two-tailed Z test). The distinct difference from the genome-wide average suggests that linkage to csd could have resulted in increased nucleotide diversity in fem; however, we failed to observe this difference for the few synonymous positions in exons 8–11 of fem that might lack statistical power.
Table 3.
Exon and intron polymorphism analysis of fem and csd genes
| A. mellifera fem | B. terrestris fem | A. mellifera csd | |
| Exon | |||
| Number of sequences | 30 | 23 | 28 |
| Number of sites (gaps excluded) | 439 | 492 | 330 |
| Number of polymorphic sites | 15 | 6 | 93 |
| Number of syn substitutions | 7 | 3 | 22 |
| Number of nonsyn substitutions | 7 | 3 | 56 |
| πtotal | 0.0034 ± 0.001 | 0.0036 ± 0.0016 | 0.0536 ± 0.0066 |
| πsyn | 0.0084 ± 0.0036 | 0.0059 ± 0.0036 | 0.0646 ± 0.0142 |
| πnonsyn | 0.0014 ± 0.0005 | 0.0029 ± 0.0016 | 0.0508 ± 0.0087 |
| πnonsyn/πsyn | 0.16 | 0.49 | 0.79 |
| Dtotal | −2.06* | 0.26 | −1.47 |
| Dsyn | −1.48 | −0.39 | −1.35 |
| Dnonsyn | −2.17** | 0.82 | −1.45 |
| Intron | |||
| Number of sites (gaps excluded) | 539 | 262 | 162 |
| Number of polymorphic sites | 48 | 5 | 39 |
| π | 0.0151 ± 0.0026 | 0.0023 ± 0.0017 | 0.038 ± 0.0072 |
| D | −1.3 | 0.28 | −1.71 |
| Genomewide average diversity | 0.0055 ± 0.0012 | ND | |
ND, not determined. Nucleotide diversity of fem and csd genes from the same Apis haplotypes (± SE) was calculated from the genomic region of exons 8–11 (fem) and of exons 6–9 (csd). Significance of Tajima's D: *P < 0.05; **P < 0.01.
To lend further support to the idea that linkage to the csd gene affects fem polymorphism, we analyzed the polymorphism frequency spectrum by applying Tajima's test. Using the total fem polymorphism, we found that Tajima's D significantly deviated from 0 (D = −2.06; P < 0.05) (Table 3), indicating an excess of variants of low frequency. We found the same excess when we confined our analysis to nonsynonymous polymorphism (Dnonsyn = −2.17; P < 0.01), indicating that purifying selection is operating. Tajima's D calculated on synonymous and intron polymorphism did not deviate from 0 (Dsyn = −1.48, Dintron = −1.3; P > 0.1 for both), consistent with a neutral evolutionary model. Although we found greater polymorphism than the genome-wide average, the frequency spectrum of fem polymorphism shows no indication that balancing selection is operating, implying that combined forces are operating at the fem locus.
Does the greater nucleotide sequence polymorphism at fem affect the rate of divergence and thus the n/s ratio estimates in the Apis lineage? Or is the the accumulation of more polymorphism counterbalanced by slower fixation rates and longer maintenance times, which would result in similar substitution rates when comparing balancing and nonbalancing loci? We compared the number of synonymous substitutions that have accumulated along the Apis csd (sAcsd = 33) and Apis fem (sAfem = 39) branches and the number of synonymous sites that have remained unchanged in csd (S = 170) and fem (S = 164) and found no difference (P > 0.5, two-tailed Fisher's exact test). These findings suggest a similar substitution rate in the two genes despite the substantial differences in strength of balancing selection, causing an ∼8-fold increase in csd over fem polymorphism. We conclude that csd affects silent polymorphism at linked loci, but not the silent substitution rates at these loci. This conclusion is consistent with our previous result obtained by the relative rate test suggesting similar silent substitution rates of Apis csd and fem genes (P > 0.5; Table 2).
The comparison supports our previous conclusion that the reduced n/s ratios result from stronger purifying selection, not from a higher silent substitution rate caused by the specific selection regime operating at the csd gene.
Discussion
The rise of csd within the Apis lineage gene has led not only to a gene harboring a new function, but also to a regulatory relationship with its sister copy gene fem. Together, the two genes initiate and implement sexual differentiation. Our findings imply that the evolution of fem, which has retained its ancestral function, was affected by the origin of csd’s new function. In previous studies, we obtained comprehensive information about the evolutionary forces acting on csd (13, 16, 17), whereas we had only limited information about the evolution of fem. In the present study, we followed the evolutionary fate of fem in Apis and non-Apis species over time and detected a lower ratio of nonsynonymous to synonymous substitutions (n/s) in fem of Apis species compared with fem of non-Apis species (Fig. 1 and Table 1).
The results of the relative rate test (Table 2) suggest that the synonymous substitution rate in fem of Apis species is not specifically increased and thus does not explain the lower observed n/s ratio. A genetic linkage between fem and csd, for which we found evidence in the intron nucleotide polymorphism of current populations (Table 3), led us to question whether balancing selection might affect the synonymous substitution rate of fem. The similar numbers of synonymous substitutions of fem and csd that have accumulated in Apis species along branches (P > 0.51) suggest that balancing selection does not affect the synonymous substitution rate, irrespective of the contrasting levels of standing nucleotide variation in the two genes. This could be taken as an indication that greater polymorphism due to the impact of balancing selection does not lead to a higher fixation rate of these variants. This finding has the potential to extend existing population genetics models on hitchhiking effects under balancing selection (18).
The closely genetically linked genes GB13727 and GB11211 showed a contrasting evolutionary pattern compared with fem. GB13727 and GB11211 demonstrated no specific reduction in n/s ratios in the Apis lineages, suggesting that genetic linkage to csd is not the cause of fem's substitution pattern (Fig. 2B). Based on our observation that the stronger purifying selection operating at the fem gene in the different Apis species lineages specifically coincides with the origin and maintenance of the csd gene, we conclude that the origin of csd's new function is the cause of stronger evolutionary constraints in Fem protein.
What is the possible nature of this functional interference? Csd and Fem proteins share similar sequences (>70% identical amino acid residues) (8) and are both involved in the control of splicing, but they differ in their sex-specific mode of activation (8, 9). The Csd protein is activated by a heterozygous allelic combination of proteins, whereas the fem gene is activated by sex-specific splicing process of its transcript, producing Fem proteins only in females. We propose that under relaxed molecular constraints at Apis fem, more nonsynonymous substitutions could accumulate, possibly interfering with the novel molecular regulation mechanism that originated with the rise of the csd gene. This might be a nonspecific cross-reaction to proteins of the csd-determining pathway that would inhibit the function of fem. By analyzing networks of protein–protein interaction, Makino et al. (19) found a much slower evolutionary rate of duplicated genes in which proteins have more interacting partners compared with genes in which proteins have fewer interacting partners. It can be speculated that if fem is evolutionary more relaxed, then the Fem proteins can nonspecifically interact with more interaction partners that could interfere with Csd protein and putative cofactor functions. One candidate region for a possible strong affect of purifying selection is a portion of the first RS domain (Fig. S2) that reportedly has protein-binding abilities (20). Further functional studies may identify the molecular mechanisms involved and confirm the conclusions drawn from this evolutionary genetic survey (21). In the ortholog tra protein, the RS domain evolves more rapidly compared with other parts of the protein among distinct Drosophila species (15), providing additional support for the exceptional evolution of Fem in Apis. The fem(tra) regulatory part of the sex-determining pathway, including the downstream target gene doublesex (dsx), is ancestral and conserved over a broad range of holometabolous insect species (9), lending further support to our conclusion that csd’s new function caused the evolutionary constraints in Fem protein.
Along with functional interference, we also found evidence of genetic interference due to physical linkage (Table 3). Balancing selection at csd also increased the maintenance time of linked loci, resulting in an accumulation of more mutations, which we detected as an increase in polymorphism (16). The intron nucleotide diversity of fem in current A. mellifera populations is higher than the genome-wide average diversity (Table 3). This is consistent with a previous finding of increased nucleotide diversity of neutral linked loci at 12 kb to csd (16), the same genomic distance that we observed between csd and fem. We detected no signatures of balancing selection at the fem locus when we applied Tajima's D test on silent sites, suggesting that a composite of hitchhiking effects and purifying selection is operating at the fem locus.
The effect of balancing selection on the nucleotide diversity in adjacent genomic regions can vary considerably depending on the recombination rate, as has been reported at loci with similar selection regimes (22, 23). In the self-incompatibility locus of Arabidopsis lyrata, balancing selection affects the diversity of silent sites for as much as 100 kb. This is accompanied by a 10- to 100-fold reduction in recombination compared with the neighboring genomic region (24). Within the genomic region of the sex determination locus (SDL), recombination is substantially reduced compared with the adjacent genomic regions, which, conversely, show a significant increase in recombinational exchange (up to 70 cM/Mb) compared with the genome-wide average (16). A decline in silent polymorphism when moving toward the 5′ part of the csd gene (16) indicates that meiotic recombination is operating; however, this reflects a process over prolonged evolutionary time due to extended maintenance times of alleles. These rare recombination events within the SDL explain the reduced association between fem and csd polymorphism over 12 kb.
Overall, the present study has provided evidence that the csd gene functionally and genetically interferes with the fem gene; we term this evolutionary interference. The gene duplication in the Apis lineage that gave rise to csd’s new function had a profound impact on the evolution of Fem protein, which was subjected to stronger molecular constraints. The evolutionary origin of a new regulatory function limited the evolutionary capability of another gene. A genetic linkage to csd and the specific selection region shaped fem’s polymorphism in current A. mellifera population by genetic hitchhiking. These findings suggest a complex interplay between newly originated gene functions and selection regimes of other genes in the genome, which might play a more general role in shaping the genome's gene repertoire than considered thus far.
Materials and Methods
Sequence Data.
The data set comprises six csd cDNA sequences and one fem cDNA sequence, each from the honey bees A. cerana, A. dorsata, and A. mellifera (GenBank accession nos. EU100887, EU100888, EU100891, EU100894, EU100898, EU100899, EU100921, EU100923, EU100929, EU100932, EU100933, EU100935, EU100903, EU100905, EU100910, EU100913, EU100914, EU100916, and EU100936–EU100941) and of fem cDNA sequences from the bumble bee Bombus terrestris (EU288185) and stingless bee Melipona compressipes (EU139305). We also obtained sequence data from the orchid bee Euglossa hemichlora, which was isolated using primers developed for a 200-bp region corresponding to exons 2 and 3 of A. melifera fem (Euglfw4, 5′-ATGATACAAAAAGAGCGGGAACGA-3′; EuglRev4, 5′-TTGAAGTTCACCAGTTGT TGTTTTA- 3′). Primers were designed based on sequence information obtained from this fragment to perform rapid amplification of cDNA ends (RACE) experiments using the FirstChoice RLM-RACE kit (Ambion), following the manufacturer's protocol. All attempts to isolate the complete 5′ region failed, and thus the ORF of E. hemichlora fem lacks the first ≈45 amino acids. Primers for 3′ RACE were as follows: S-3 5′- CCGGGTAAAACAACAACTGGTGAAC-3′; S-4 5′-GATGGTACAACACCC TTATTTAAAGG-3′. The combination of primers Euglw4 and S-17 5′-CAAGTCTTCCATTAATACATTGGCCC-3′ was used to obtain a fragment with maximal sequence information of fem. The csd and fem sequences were aligned on the basis of the deduced amino acids with ClustalX version 2.0.4 (25) and were edited manually using Bioedit (26) to improve conformity with the ORF. All positions containing gaps were removed to exclude ambiguous sites. Samples of 30 and 23 chromosomes (individual haploid drones) for A. mellifera (18 different colonies from Texas, North Carolina, and Australia) and B. terrestris (Germany) were used to amplify fragments from genomic DNA of csd and fem spanning the same corresponding region (for csd, exons 6–9; for fem, exons 8–11) by PCR using standard protocols (27) and high-fidelity proofreading DNA-polymerase (Phusion; Finnzyme). Primers and their combinations were as follows: for csd A. mellifera: genoRfw 5′- AGACRATATGAAAAATTACACAATGA-3′/Conscsdrev 5′ TCA TCTCATWTTTCATTATTCAAT-3′; S-39 5′-TATAATGAAAAAGAAAAATTTTTAGAAG-3′/S-40 5′-ACTATGTGCATCAATATA AATTC-3′; fem A. mellifera: csdIIsiRNA714for 5′-AGATTCAAGACATGAAGACAG-3′/GenoRNAiIIRTrev 5′-TATCT GGAGGAATAAATCGTGGTG-3′; fem B. terrestris: 3race28I 5′- GCCAGGTATGAGGATACAAAATATG-3′/femBom_rev2 5′- CTATCGGCACCATTCCCTTCCAACT-3′. A fragment of the GB13727 gene was amplified from genomic DNA (for Apis) and cDNA (for non-Apis) using primer GB13727_fw 5′cgtttggtgccattttggatg and GB13727_Aug07_rv CATCAGTGCTACCTGATGATAG (for Apis) and S-119 5′GATAAATGTCCACCAAATTTTACAG and S-121 5′ CTATATTAACATATGATTGTG (for non-Apis). A fragment of the GB11211 gene was amplified from the same DNA as used for GB13727 with the primers S-57 CGTTACTTTTCGATCATAGTGACGATG and S-58 CCATTGTCGGCGTACACCTATTGG, except for M. compressipes, for which no PCR product could be obtained. PCR products obtained for fem, GB13727, and GB11211 were double-strand sequenced directly (MWG Biotech). csd PCR fragments were cloned into the pGEM-T vector (Promega), and the clones were subjected to double-strand sequencing.
Molecular Evolutionary Analysis of Nucleotide and Amino Acid Substitutions.
An evolutionary tree of csd and fem sequences was generated using genealogies obtained by the minimum evolution (ME) method and applying Poisson distances on deduced amino acids using MEGA version 4.0 (28) and the maximum likelihood method implemented in DNAML of PHYLIP version 3.5c (29). Potential protein domains were defined following the prediction of Prosite (http://www.expasy.org/tools/scanprosite/), and nucleotide polymorphism analyses limited to single domains of csd and fem were performed on the minimum spanning length of that domain in the alignment. Nucleotide sequence polymorphism of A. mellifera fem and csd and B. terrestris fem sequences were analyzed using DnaSP version 4.0 (30). Ancestral sequence construction at interior nodes of the tree was performed using the ANCESTOR program (31). The program first infers the amino acids by the distance-based Bayesian method, and then infers the underlying nucleotide sequences by fixing the inferred amino acids. The computed average posterior probability is > 98% for all ancestral sequences. This sequence information was used to calculate the number of synonymous (dS) and nonsynonymous (dN) substitutions per site for each branch and the absolute number of synonymous (s) and nonsynonymous (n) substitutions for each branch. Evolutionary analyses of GB13727 and GB11211 coding sequences were performed as described for csd and fem. Relative rate tests for substitutions rate of nucleotide differences were analyzed using RRtree version 1.1 (32). Tests of heterogeneity of n/s ratios was performed using the replicated G-test of goodness of fit (33).
Supplementary Material
Acknowledgments
We thank K. Aronstein (US Department of Agriculture, Texas), C. Grozinger (Penn State University, PA), D. Roubik (Smithsonian Tropical Research Institute, Panama) R. Malezska (Australian National University, Canberra, Australia), and M. Schindler (University Bonn, Germany) for providing bee samples and G. E. Heimpel and A. Zayed for their comments on the manuscript. This work was supported by a grant from the German funding association (DFG HA 5499/3-1; to M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HM584938–HM584947 and HM588917–HM588997).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005617107/-/DCSupplemental.
References
- 1.Goodman M, Moore GW, Matsuda G. Darwinian evolution in the genealogy of haemoglobin. Nature. 1975;253:603–608. doi: 10.1038/253603a0. [DOI] [PubMed] [Google Scholar]
- 2.Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Dean AM, Brunet F, Long M. Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci USA. 2004;101:16246–16250. doi: 10.1073/pnas.0407066101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbin CJ, et al. Paralogues of porcine aromatase cytochrome P450: A novel hydroxylase activity is associated with the survival of a duplicated gene. Endocrinology. 2004;145:2157–2164. doi: 10.1210/en.2003-1595. [DOI] [PubMed] [Google Scholar]
- 5.Moyle WR, et al. Co-evolution of ligand-receptor pairs. Nature. 1994;368:251–255. doi: 10.1038/368251a0. [DOI] [PubMed] [Google Scholar]
- 6.Pazos F, Helmer-Citterich M, Ausiello G, Valencia A. Correlated mutations contain information about protein-protein interaction. J Mol Biol. 1997;271:511–523. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- 7.Beye M, Hasselmann M, Fondrk MK, Page RE, Jr, Omholt SW. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114:419–429. doi: 10.1016/s0092-8674(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 8.Hasselmann M, et al. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature. 2008;454:519–522. doi: 10.1038/nature07052. [DOI] [PubMed] [Google Scholar]
- 9.Gempe T, et al. Sex determination in honeybees: Two separate mechanisms induce and maintain the female pathway. PLoS Biol. 2009;7:e1000222. doi: 10.1371/journal.pbio.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cline TW, Meyer BJ. Vive la différence: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 11.Pane A, Salvemini M, Delli Bovi P, Polito C, Saccone G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development. 2002;129:3715–3725. doi: 10.1242/dev.129.15.3715. [DOI] [PubMed] [Google Scholar]
- 12.Grimaldi D, Engel MS. Evolution of the Insects. Cambridge, UK: Cambridge University Press; 2005. [Google Scholar]
- 13.Hasselmann M, Beye M. Signatures of selection among sex-determining alleles of the honey bee. Proc Natl Acad Sci USA. 2004;101:4888–4893. doi: 10.1073/pnas.0307147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAllister BF, McVean GA. Neutral evolution of the sex-determining gene transformer in Drosophila. Genetics. 2000;154:1711–1720. doi: 10.1093/genetics/154.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulathinal RJ, Skwarek L, Morton RA, Singh RS. Rapid evolution of the sex-determining gene, transformer: Structural diversity and rate heterogeneity among sibling species of Drosophila. Mol Biol Evol. 2003;20:441–452. doi: 10.1093/molbev/msg053. [DOI] [PubMed] [Google Scholar]
- 16.Hasselmann M, Beye M. Pronounced differences of recombination activity at the sex determination locus of the honeybee, a locus under strong balancing selection. Genetics. 2006;174:1469–1480. doi: 10.1534/genetics.106.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasselmann M, et al. Evidence for convergent nucleotide evolution and high allelic turnover rates at the complementary sex-determiner gene of Western and Asian honeybees. Mol Biol Evol. 2008;25:696–708. doi: 10.1093/molbev/msn011. [DOI] [PubMed] [Google Scholar]
- 18.Slatkin M. Balancing selection at closely linked, overdominant loci in a finite population. Genetics. 2000;154:1367–1378. doi: 10.1093/genetics/154.3.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino T, Suzuki Y, Gojobori T. Differential evolutionary rates of duplicated genes in protein interaction network. Gene. 2006;385:57–63. doi: 10.1016/j.gene.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner A. How the global structure of protein interaction networks evolves. Proc Biol Sci. 2003;270:457–466. doi: 10.1098/rspb.2002.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schierup MH, Vekemans X. Genomic consequences of selection on self-incompatibility genes. Curr Opin Plant Biol. 2008;11:116–122. doi: 10.1016/j.pbi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Kamau E, Charlesworth D. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr Biol. 2005;15:1773–1778. doi: 10.1016/j.cub.2005.08.062. [DOI] [PubMed] [Google Scholar]
- 25.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 27.Hasselmann M, Fondrk MK, Page RE, Jr, Beye M. Fine-scale mapping in the sex locus region of the honey bee (Apis mellifera) Insect Mol Biol. 2001;10:605–608. doi: 10.1046/j.0962-1075.2001.00300.x. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 30.Rozas J, Rozas R. DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Nei M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol. 1997;44(Suppl 1):S139–S146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]
- 32.Robinson-Rechavi M, Huchon D. RRTree: Relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics. 2000;16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- 33.McDonald JH. Handbook of Biological Statistics. Baltimore, MD: Sparky House Publishing; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.