Abstract
The efficiency of HIV infection is greatly enhanced when the virus is delivered at conjugates between CD4+ T cells and virus-bearing antigen-presenting cells such as macrophages or dendritic cells via specialized structures known as virological synapses. Using ion abrasion SEM, electron tomography, and superresolution light microscopy, we have analyzed the spatial architecture of cell-cell contacts and distribution of HIV virions at virological synapses formed between mature dendritic cells and T cells. We demonstrate the striking envelopment of T cells by sheet-like membrane extensions derived from mature dendritic cells, resulting in a shielded region for formation of virological synapses. Within the synapse, filopodial extensions emanating from CD4+ T cells make contact with HIV virions sequestered deep within a 3D network of surface-accessible compartments in the dendritic cell. Viruses are detected at the membrane surfaces of both dendritic cells and T cells, but virions are not released passively at the synapse; instead, virus transfer requires the engagement of T-cell CD4 receptors. The relative seclusion of T cells from the extracellular milieu, the burial of the site of HIV transfer, and the receptor-dependent initiation of virion transfer by T cells highlight unique aspects of cell-cell HIV transmission.
Keywords: 3D imaging, cell–cell contact, electron tomography, viral entry, neutralizing antibodies
As a pathogen with limited genomic coding capacity, HIV subverts many physiological processes of its host to facilitate key aspects of its own replication. Cell-cell interactions (1, 2) that play a critical role in normal immune system function are exploited by the virus to facilitate its transmission from antigen-presenting cells such as dendritic cells to susceptible target CD4+ T cells via a specialized structure designated a virological synapse (3, 4). Although virological synapses can also be formed between uninfected and infected T cells (5–7), such synapses appear to be less tightly structured than synapses between dendritic cells and CD4+ T cells. Importantly, the transmission of virus to CD4+ T cells is far more efficient via a virological synapse than via cell-free diffusion (8). In lymphoid tissues, the primary site of HIV replication in vivo, HIV can take advantage of physiological interactions between mature dendritic cells and CD4+ T cells to spread viral infection by cell-cell contact (9, 10).
Previous analyses of virological synapses formed between HIV-pulsed dendritic cells and T cells using light microscopy and conventional transmission electron microscopy (TEM) have demonstrated polarization of dendritic cell-internalized viruses at the cell-cell contact zone (3, 10–13) and have highlighted the importance of Env–CD4 interactions in achieving viral infection of the T cell (8). Although these and similar experiments with other kinds of cell–cell synapses (14, 15) have provided informative glimpses of the nature of cell-cell contact, imaging experiments using conventional 2D TEM provide only cross-sectional views. The focus of the experiments presented here is to describe the 3D architecture of virological synapses using two emerging technologies for 3D electron microscopy: ion abrasion (IA) SEM (16, 17) and electron tomography (18). Our experiments use these imaging approaches to reveal the 3D arrangement of virions and virion-containing compartments at the earliest stages of contact between HIV-pulsed mature dendritic cells and uninfected T cells when virological synapses have been established but before productive infection of either the dendritic cell or the T cell has occurred.
Results
Stimulated Emission Depletion Imaging of the Virological Synapse.
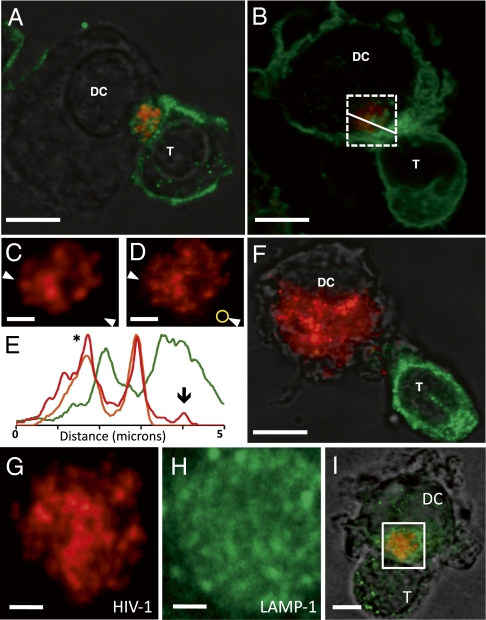
To establish the functional relevance of the synapses imaged using 3D electron microscopy, we first carried out light microscopy of cocultures of mature dendritic cells pulsed with HIV virions and uninfected autologous CD4+ T cells using cells prepared under the same conditions as those used for the electron microscopic experiments. With the use of stimulated emission depletion (STED) imaging (19), which allows imaging beyond the diffraction limit, the clustering of fluorescent viruses at the synapse could be easily visualized with single-virion resolution (Fig. 1 A–E). The HIV virions show strong colocalization with actin distribution in the dendritic cell as probed using fluorescent phalloidin, which binds actin (Fig. 1 B and E). Further, the higher resolution afforded with the STED imaging used here allows spatial separation of fluorescence signals arising from HIV virions and the closely situated cytosolic actin-associated fluorescent phalloidin (Fig. 1E). No colocalization of HIV was observed with markers of lysosome-associated membrane protein (LAMP-1; Figs. 1G–I) or early endosomal antigen 1 (EEA-1; Fig. S1). The requirement (13) of actin for HIV clustering at the synapse is also highlighted both by the extensive phalloidin staining of membranes at cell-cell contact regions (Fig. 1B) and in the loss of virion clustering and localization in the presence of the actin polymerization inhibitor cytochalasin D (Fig. 1F). Together, these experiments establish that the synapses used for the 3D electron microscopic analyses (Figs. 2 and 3) display the functional characteristics previously established for contacts between mature dendritic cells and CD4+ T cells (3, 10–13).
Fig. 1.
Visualization of T-cell–dendritic cell (T-DC) virological synapses by superresolution fluorescence microscopy. (A and B) STED microscopy of conjugates of dendritic cells pulsed with ATTO-647N–labeled HIV (red) and autologous CD4+ T cells labeled with anti-CD3 antibody (A) or with fluorescent phalloidin (B), which is a marker for actin distribution (green). (C and D) Expanded view of the boxed region in B showing comparison of images recorded using conventional confocal microscopy (C) or STED microscopy (D). (E) Fluorescence intensity profiles across synapse (arrows in C and D) show that the spatial resolution of STED imaging (red line, peak marked with asterisk) is better than that of confocal imaging (orange line for HIV, green line for phalloidin) and is adequate to detect single virions (marked by the downward arrow in E and by the yellow circle in D) not resolved with conventional confocal imaging of the same field. (F) STED image of conjugates labeled as in A but in the presence of cytochalasin D, added during synapse formation, illustrating that viruses are no longer clustered or localized at the synapse under these conditions (HIV, red; anti-CD3 antibody, green). Simultaneous imaging of ATTO-647N–labeled HIV-1 (G; red) and lysosomal marker LAMP-1 (H, green) shows that that they display nonoverlapping distributions (I). Pearson's correlation coefficient between fluorescent signals derived from HIV and LAMP-1 dropped to 0.29 with STED imaging as compared with 0.34 with confocal imaging of the same region. (Scale bars: A, B, and F, 3 μm; C, D, G, and H, 1 μm.)
Fig. 2.
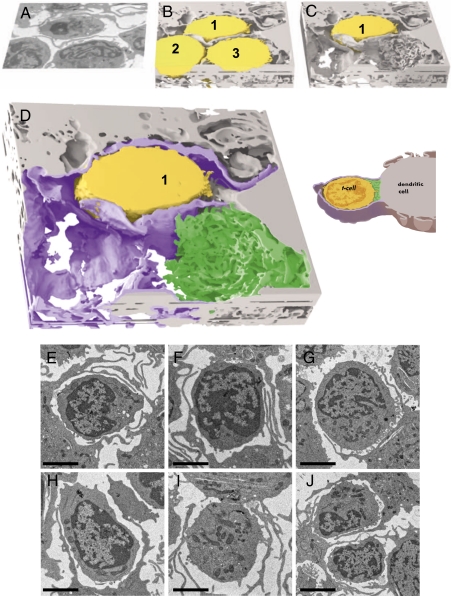
3D contacts between a dendritic cell and T cell at the synapse visualized with IA-SEM imaging. (A–C) Envelopment of the T cell by sheet-like membrane protrusions from the dendritic cell (Movies S1 and S2). The T-cells (numbered 1, 2, and 3) are colored yellow, and the dendritic cell is shown in light gray. Single-slice (A) and 3D-volume slabs before (B) and after (C) computational removal of the T cells labeled 2 and 3 from the imaged volume are shown. (D) Expanded view of C distinguishing the extensive membrane sheet encasement (magenta) and the localized filopodial interdigitations (green) at the region of cell-cell contact emanating from the dendritic cell. A schematic version of the contact is shown to the right. (E–J) Projection electron microscopic images from thin sections illustrating that every T cell is encased by membranes originating from the dendritic cell, with synapses visible in some instances. (Scale bars: 3 μm.)
Fig. 3.
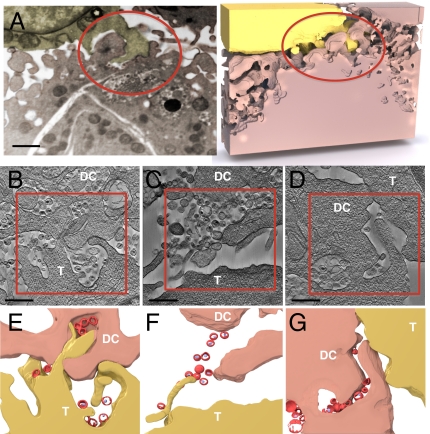
IA-SEM and electron tomography of contact regions between dendritic cells and T cells illustrating HIV-1 distribution within the virological synapse. (A) Interdigitation of membrane protrusions from the T cell (shown in yellow) into the dendritic cell (shown in pink) surface and connection of virion-rich compartments in the interior of the dendritic cell to the synapse via HIV-containing channels (red circle; Movie S3). (B–D) Tomographic slices obtained from ~200-nm thick sections of cocultures of HIV-1–pulsed dendritic cells (DC) mixed with CD4+ T cells (T) illustrating different structural aspects of the cell-cell contacts that define the synapse. (E–G) Segmentation of regions boxed in B–D to indicate membrane contact and virus (red) location. (Scale bar: A, 1 μm; B–D, 0.4 μm.)
IA-SEM Imaging of the Virological Synapse.
To define the 3D architecture of the cell–cell contact regions, we used IA-SEM, an approach for 3D structural analysis of whole cells at a resolution of ~20 nm (16, 17, 20, 21). With this imaging strategy, cells or tissues are exposed to a scanning gallium ion beam that iteratively abrades the specimen surface to remove a layer of defined thickness (typically ~15–20 nm) for each pass. Each newly exposed surface is imaged with a scanning electron beam, and successive slices are combined to generate a 3D reconstruction of the abraded volume (Movie S1). Visualization of the contact between dendritic cells and T cells using IA-SEM reveals that the membrane processes emerging from the dendritic cell envelop the T cell in large “sheets” of contact (Fig. 2 A and B and Movie S2); these can be mistaken for thin spaghetti-like filopodia when 2D images of single sections are examined. The presence of these sheets encasing the T-cell surface contact zone implies that the T-cell membrane is largely protected from the extracellular milieu. The striking 3D aspect of these interactions can be appreciated by the cut-away view of the contact zone (Fig. 2 C and D), in which two of the three T cells have been computationally removed to reveal the extensive wrapping of virtually the entire surface of the T cell by the membrane processes emanating from the dendritic cell. Examination of the ultrastructure of ~500 T cells in projection electron micrographs showed that >95% of the cells were surrounded by membrane processes (representative 2D images shown in Fig. 2 E–J). The presence of the membranes provides a shielded region for formation of the virological synapse in a localized region, where there is close contact between virion-rich regions of the mature dendritic cell and the T cell (schematic in Fig. 2D), with close interdigitation of filopodial extensions from both the dendritic cell and the T cell. Note that the membrane extensions are visible around almost every T cell seen in the thin-section images but that the localized contact sites representing the synapse are not always observed because only a small fraction of the cell volume is captured in each thin section.
IA-SEM imaging also shows that HIV taken up by the dendritic cells is located within large compartments that are connected to the cell surface via deep channels that can be many microns long and as narrow as the width of a virus in some regions (Fig. 3A, Fig. S2, and Movie S3), similar to those recently described in HIV-infected macrophages (22, 23). At the synapse, cell-cell contact involves extensive interdigitation of the respective cell membranes, with filopodial extensions from the T cell penetrating into the recesses of the virion-rich surface folds of the dendritic cell. Filopodial extensions from the dendritic cell are also apparent. The simultaneous presence of the two different modes of contact via the sheets and filopodial extensions can be seen clearly in the view after computational removal of the T cells (Fig. 2D). Tips of membrane protrusions from the T cell are present at the mouths of these virus-filled channels, which can extend deep into the dendritic cell (Fig. 2B and Movie S3). The dendritic and T-cell membranes are closely apposed at the tips of the protrusions, thus effectively separating the HIV in these compartments from the bulk medium. The combination of the membrane encasement, the deep virion channels, and the interdigitation between the donor and target cell membranes serves to ensure that HIV transfer to the T cell occurs in a highly secluded environment.
Electron Tomography of Cell–Cell Contacts at the Synapse.
To investigate the 3D distribution of HIV within the synapse in greater detail, we performed electron tomography of thick sections containing the cell–cell contact regions. Tomographic studies reveal the presence of two distinct types of contacts at virological synapses with a similar frequency of occurrence (from a dataset of 81 individual synapses studied by electron tomography), which are distinguishable by differences in the location of HIV relative to the cell–cell interface (Fig. 3 B–G). In one type of contact (Fig. 3 B, C, E, and F), viruses evident at both cell surfaces and protrusions originating from the T-cell surface that penetrate into the virion-rich folds of the dendritic cell are apparent. HIV virions are found in contact with the T cell both at the tips (Fig. 3E) and along the length (Fig. 3F) of the protrusions, consistent with previous proposals of viral “surfing” along filopodia (24). STED imaging provides independent confirmation of the presence of individual virions that lie on actin-rich protrusions located at regions of cell-cell contact (Fig. S3), with the spatial resolution afforded by STED imaging demonstrating that the virions on the exterior surface of the cell membrane are distinct from the intracellular actin fluorescence profile. In a second type of contact (Fig. 3 D and G), the T-cell and dendritic cell make contact along their respective cell boundaries, where viruses are clearly visible in the vicinity of the contact zone. However, despite the close proximity to the T-cell membrane, the viruses are seen to be either predominantly within or in contact only with the dendritic cell.
Antibody Blocking of HIV Transfer Across the Synapse.
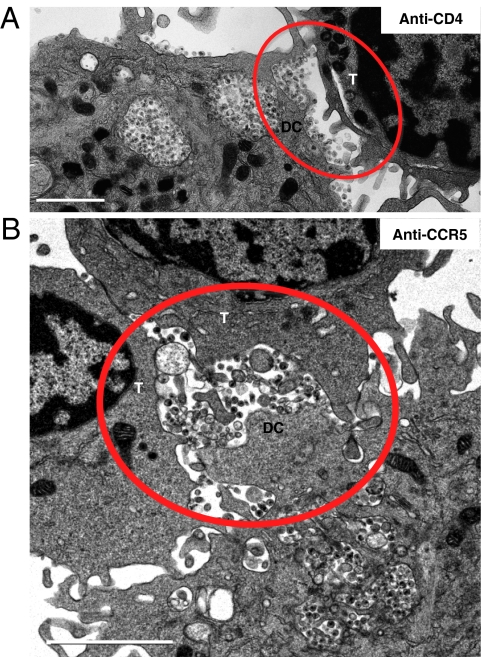
To define the requirements for effecting viral transfer across the narrow gap between the cell membranes in mature dendritic cell–T-cell conjugates (typically <500 nm, only three times the diameter of a single virion), we analyzed virus distribution in the synapse after preincubating CD4+ T cells with antibodies directed against CD4 or CCR5. The presence of anti-CD4 antibodies did not block synapse formation (Fig. 4A) but led to a >20-fold reduction in the number of viruses detected on the T-cell surface and a corresponding increase in viruses bound on the dendritic cell surface. In contrast, treatment with anti-CCR5 antibodies (Fig. 4B) resulted in synapses in which viruses were observed on both cell surfaces and the distribution of viruses was very similar (<10% difference) to that observed with untreated cells (Fig. S4). Infectivity assays confirmed that addition of either anti-CD4 or anti-CCR5 antibodies blocked infection, with p24 levels reduced by 65% and 90%, respectively, compared with controls in which no T cells were added to HIV-treated dendritic cells. No inhibition was observed with the addition of control antibody of the same isotype, whereas ~75% inhibition was obtained using the CCR5-binding entry inhibitor maraviroc. The CD4 blocking experiments thus demonstrate that HIV is only transferred to those T cells in which the first step in productive infection (i.e., gp120-CD4 binding) is ensured. This was further confirmed with control experiments in which the dendritic cells were incubated with the monoclonal antibody 2G12, which binds gp120 and blocks interaction with CD4, or with the monoclonal antibody 17b, which binds gp120 to a much greater extent after CD4 is bound but is sterically blocked from binding gp120 in the context of the virus-cell interface (25). As shown in Figs. S5 and S6, treatment with 2G12 resulted in retention of virions on the dendritic cell, whereas treatment with 17b, which binds tightly to gp120 only in the presence of CD4, resulted in synapses in which viruses were observed on both dendritic and T-cell surfaces. As expected, no T cell–associated HIV was observed when dendritic cells were treated with cytochalasin D either before HIV incubation or after HIV incubation but before addition of T cells (Fig. S7). Together, these results imply that neither virological synapse formation per se nor the mere presence of HIV virions at the synapse is sufficient for transfer of virus to T cells.
Fig. 4.
Projection TEM images of synapses formed between virus-pulsed dendritic cells (DC) and T cells (T) preincubated with blocking antibodies. (A) Treatment with anti-CD4 antibody M-T477 results in virions polarized toward the contact region with the T cell but with viruses predominantly positioned on the dendritic cell and only rarely on the T cell. (B) Treatment with anti-CCR5 antibody 3A9 results in virus distribution similar to that seen in untreated cells (Fig. S4), with viruses present on both cell surfaces at the synapse (red circles). (Scale bars: 2 μm.)
The 3D imaging experiments presented here thus establish both the broader context of cell-cell interactions that define the virological synapse, including 3D envelopment of T cells by the mature dendritic cell, and the nature of local contacts mediated by membrane protrusions associated with HIV transfer to the T cell. Earlier studies have anticipated the importance of sheet-like “veils” that can be formed by antigen-presenting cells (26–29) as well as the enrichment and clustering of CD4, CCR5, and CXCR4 in filopodial protrusions from T cells, making them likely primary sites of contact with the virus (30). The contributions of our work are to place both types of membrane extensions in the context of the dendritic cell–T-cell virological synapse, to determine the 3D structure at the contact zone, and to show that interactions between gp120 on the virus and CD4 on the T cell are required for HIV transfer across the narrow gap between the cells.
Discussion
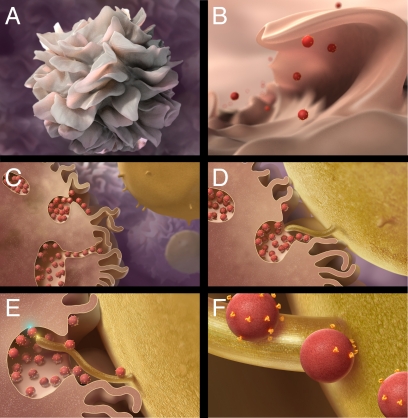
In Fig. 5, we present a plausible mechanism for transfer of HIV to the T cell at the virological synapse. We suggest that as part of the physiological process by which mature dendritic cells seek to capture antigens present in their environment, sheet-like protrusions of surface membrane that wrap around T cells in the vicinity are generated. They also result in trapping of the contents of the adjacent aqueous environment and material bound to the cell surface, including virions when present, in compartments that are essentially surface membrane invaginations. At the regions of local contact between dendritic cells and T cells, virion-rich compartments are predominantly localized toward the T-cell interface, presumably in an actin-dependent process (31), with cell-cell contact mediated by cell surface adhesion molecules (32). This model is also consistent with the evidence that HIV-1 alters endolysosomal traffic in dendritic cells (33), although virions themselves appear not to be present in endolysosomes. Contact of virions localized within surface-accessible membrane invaginations in the dendritic cell with microvillar/filopodial extensions from the T cell that are likely enriched for CD4 and CCR5/CXCR4 (30) initiates transfer of viruses onto the T-cell surface, leading to formation of entry claw structures (34) and membrane fusion. In the absence of CD4 availability on the T cell or when gp120 on the viral surface is prevented from binding CD4, the viruses remain associated with the dendritic cell, perhaps via interactions with dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN) (35, 36) (other receptors are possible) until the formation of productive synapses with T cells that are capable of supporting gp120-CD4 interaction. We note that virological synapses between T cells alone have also been described (7, 32, 37), and the available data suggest that these synapses may be less complex and devoid of the large membrane encasement reported here with mature dendritic cells.
Fig. 5.
Model for virus internalization, formation of virion channels, and transfer of virions from antigen-presenting cells to T cells at the virological synapse. (A and B) Sheet-like processes on the surfaces of dendritic cells fold back onto the membrane surface, entrapping viruses in the vicinity into compartments that retain continuity with the extracellular milieu. (C and D) Formation of conjugates between dendritic cells and T cells triggers actin-dependent polarization of virion conduits toward the contact zone with the T cell, resulting in contact of dendritic cell-associated viruses with CD4-rich filopodia/microvilli originating from the T cell. (E and F) Virus transfer from virion conduits to the T cell and diffusion along the surface of the T-cell membrane, likely driven by the active participation of cytoskeletal elements. The color burst in E schematically marks the proposed contacts at the dendritic cell-virus (cyan) and T-cell–virus (yellow) interface.
Our findings show that invaginations in the mature dendritic cell retain the virus in spaces that communicate with the external medium, ready for transfer to T cells at functional virological synapses. The electron microscopic experiments were designed to obtain structural snapshots at the earliest stages of cell–cell contact, formed under in vitro conditions similar to those in which most of the earlier mechanistic studies on dendritic cell–T-cell and T-cell–T-cell virological synapses have been carried out. Although the physiological relevance of HIV transmission by cell-to-cell spread in vivo is not fully established, this mode of transmission is plausible, given the strong evidence from in vitro studies. Even if virus transfer to the T cell can occur to some extent outside the context of these contacts, the finding that ~50% of cell contacts at the synapse appear to involve filopodial insertions into the dendritic cell membrane (Fig. 3 B and C) suggests that viral transfer in these secluded regions will be an important factor. A possible consequence of the wrapping of the T cell by the sheet-like envelopes is a reduction in the effective concentration of exogenously added reagents at the site of virus transfer, offering an explanation for the relative inability of neutralizing antibodies to block viral infection at dendritic cell–T-cell synapses in some instances (38). Inhibition of viral entry into CD4+ T cells by therapeutical intervention may thus require higher drug concentrations and local targeting, emphasizing the importance of pursuing complementary strategies aimed at neutralizing viruses at earlier stages in the process of infection such as virus uptake and delivery to T cells.
Methods
Cells and Viruses.
Mature dendritic cells were prepared from CD14+ peripheral blood monocytes isolated by magnetic bead immunoaffinity positive selection cultured in RPMI 1640 with 10% (vol/vol) human serum, IL-4 (100 IU/mL; R&D Systems), and GM-CSF (1,000 IU/mL; R&D Systems) for 6 d. On days 2 and 4 of dendritic cell generation, fresh media and cytokines were added to cultures, and on day 6, they were matured in RPMI containing 2% human antibody serum, TNF-α (20 ng/mL; R&D Systems), prostaglandin E2 (20 μM; Sigma–Aldrich), IL-6 (20 ng/mL; R&D Systems), and IL-1β (20 ng/mL; R&D Systems) and cultured for 1 d. Before being used in coculture experiments, cells were inspected by light microscopy to ensure the appropriate maturation phenotype and morphology. For electron microscopic studies, cells were pulsed with infectious HIV-1 BaL (1–2 μg HIVp24 capsid per 5 × 105 cells) for 1 h at 37 °C. HIV-1–pulsed dendritic cells were washed twice with culture medium and cocultured with autologous positively selected CD4+ T cells that had been maintained at 37 °C in RPMI 1640 media supplemented with IL-2 (25 IU/mL, obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health; human recombinant IL-2 obtained from Maurice Gately, Hoffmann–La Roche, Inc.) for the duration of the dendritic cell generation and maturation period. For fluorescence microscopic studies, the same procedures were used except that HIV-1 BaL labeled with ATTO-647N (ATTO-Tec, GmbH) was used. For blocking studies, CD4+ T cells were preincubated with HIV-1 blocking CD4 antibody (M-T477; Becton Dickinson) or anti-CCR5 antibody (3A9) at 10 μg of antibody per 106 T cells for 30 min (or for 1 h in some instances) at 4 °C. Antibodies were maintained at this concentration throughout the coculture period with dendritic cells. HIV-1–pulsed dendritic cell–T-cell mixtures were centrifuged at 500 × g for 1 min to facilitate conjugate formation and cultured in duplicate for 1 h at 37 °C with or without 6 μM cytochalasin D (Sigma–Aldrich). Following incubation for 1 h, replicate samples for electron microscopy studies were centrifuged at 200 × g for 5 min; after removal of the supernatant, they were fixed in glutaraldehyde/cacodylate buffer. The remaining replicate samples were gently mixed to resuspend the conjugates, and 20 μL was spotted onto poly-l-lysine-charged no. 1.7 Zeiss coverslips. Spotted cells were either air-dried and stored for subsequent analysis or immediately prepared for immunofluorescence analysis. For measurements of infectivity, the experiments were carried out with cell conjugates that either received no treatment (negative control) or were treated with maraviroc at a concentration of 1 μg/mL (positive control), control IgG (isotype control), anti-CCR5 (clone 3A9), or anti-CD4 (clone M-T477) for 1 h at 4 °C. The antibody concentration was maintained at 20 μg/mL, and cells were cultured for 3 d in duplicate or triplicate at 37 °C. Viral production was determined by triplicate determinations of the levels of HIV-1 p24 by ELISA analysis. Fluorescence microscopic experiments and electron microscopic imaging studies were carried out on cells from three different donors in three separate sets of experiments, leading to the same conclusions in each case.
Fluorescence Microscopy.
For immunofluorescence microscopy, cells prepared as described above were fixed in 4% (vol/vol) paraformaldehyde for 20 min at room temperature, washed, permeabilized with 0.1% saponin in PBS for 10 min, stained with Alexa-488– or Alexa-555–labeled phalloidin (Invitrogen) and with primary and secondary antibodies, and mounted with Mowiol (Calbiochem) onto glass slides. Microscopy was performed on a TCS STED microscope (Leica Microsystems) equipped for operation in both superresolution and conventional confocal imaging modes. The width of fluorescence peaks at half-maximum values were ~0.3 μm in confocal mode and ~160–190 nm for the same image captured in STED mode, providing a measure of the increase in resolution with STED imaging. Primary and secondary antibodies used were mouse anti-CD3 mAb (clone F7.2.38; Dako), rabbit anti-CD3 mAb (clone SP7; Thermo Scientific), mouse anti-HIVp24 mAb (clone Kal-1; Dako), rabbit anti-actin mAb (clone 13E5; Cell Signaling Technology), rabbit anti-EEA-1 polyclonal antibody (Cell Signaling Technology), rabbit anti-LAMP-1 mAb (clone C54H11; Cell Signaling Technology), and Alexa555-labeled donkey anti-mouse IgG and donkey anti-rabbit IgG (Invitrogen).
Preparation of Specimen Blocks for Electron Microscopy.
Dendritic cells were pulsed with HIV-1 BaL and incubated with autologous CD4+ T cells for between 1 and 2 h as described. At the designated harvest times, cells were fixed in 5% (vol/vol) glutaraldehyde in 200 mM sodium cacodylate buffer at a pH of 7.4 for 24–48 h. Cells were then rinsed three times (5 min each time) in 100 mM sodium cacodylate buffer and five times in water (1 min each time). For IA-SEM analysis, pellets were treated with 1% osmium tetroxide in water for 30 min, rinsed three times (5 min each time) with water, and then treated with 1% thiocarbohydrazide in water for 10 min. Pellets then went through two more cycles of osmium tetroxide and thiocarbohydrazide for a total of three treatments with osmium tetroxide and two treatments with thiocarbohydrazide. The cell pellet was then washed three times in water (5 min each time). Pellets went through dehydration in a graded ethanol series (50%, 70%, 90%, 95%, 100%) and were then embedded using Eponate-12 (Ted Pella, Inc.).
IA-SEM.
Resin blocks were imaged using a Nova 200 NanoLab dual-beam instrument (FEI) equipped with a gallium ion source for milling and a field emission gun scanning electron microscope with an in-lens secondary electron detector for imaging. Before focused ion beam milling and SEM imaging, the entire sample surface was coated with a platinum/palladium layer 1,000 nm thick using the gas injector system in the main specimen chamber. Secondary electron SEM images were typically recorded at accelerating voltages of 3 kV, a magnification of 10,000×, and a beam current of 68–270 pA in the immersion lens mode, with a 5-mm working distance and pixel size of ~3 nm. Material was removed in step sizes of ~15 nm using the focused ion beam. Image segmentation was carried out in the environment of Amira (Visage Imaging) and rendered with 3ds MAX software (Autodesk, Inc.).
Electron Tomography.
Fixed embedded cell specimens were cut into 200-nm thick sections using a Leica Ultracut T Microtome (Leica Microsystems), placed on carbon-coated copper EM grids, stained with lead citrate, coated with 10-nm gold beads (Nanoprobes), and imaged in a Tecnai 12 transmission electron microscope (FEI) at 120 kV, a magnification of ×52,000 (image pixel size = 0.44 nm), and −1-μm defocus. Tilt series were recorded using Xplore3D software (FEI), and 3D reconstructions were carried out using weighted-back projection.
Supplementary Material
Acknowledgments
This work was supported by funds from the intramural program of the National Cancer Institute and the IATAP program (to S.S.) and federal funds from the National Cancer Institute under Contract HHSN261200800001E (to J.D.L.). R.L.F. was supported in part by a Pharmacology Research Associate Fellowship from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003040107/-/DCSupplemental.
References
- 1.Grakoui A, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285:221–227. [PubMed] [Google Scholar]
- 2.Revy P, Sospedra M, Barbour B, Trautmann A. Functional antigen-independent synapses formed between T cells and dendritic cells. Nat Immunol. 2001;2:925–931. doi: 10.1038/ni713. [DOI] [PubMed] [Google Scholar]
- 3.McDonald D, et al. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 4.Sattentau Q. Avoiding the void: Cell-to-cell spread of human viruses. Nat Rev Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 5.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 2004;199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin N, Sattentau Q. Cell-to-cell HIV-1 spread and its implications for immune evasion. Current Opinion in HIV and AIDS. 2009;4:143–149. doi: 10.1097/COH.0b013e328322f94a. [DOI] [PubMed] [Google Scholar]
- 7.Rudnicka D, et al. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J Virol. 2009;83:6234–6246. doi: 10.1128/JVI.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P, Hübner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lekkerkerker AN, van Kooyk Y, Geijtenbeek TB. Viral piracy: HIV-1 targets dendritic cells for transmission. Curr HIV Res. 2006;4:169–176. doi: 10.2174/157016206776055020. [DOI] [PubMed] [Google Scholar]
- 10.Wang JH, Janas AM, Olson WJ, Wu L. Functionally distinct transmission of human immunodeficiency virus type 1 mediated by immature and mature dendritic cells. J Virol. 2007;81:8933–8943. doi: 10.1128/JVI.00878-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu HJ, Reuter MA, McDonald D. HIV traffics through a specialized, surface-accessible intracellular compartment during trans-infection of T cells by mature dendritic cells. PLoS Pathog. 2008;4:e1000134. doi: 10.1371/journal.ppat.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia E, et al. HIV-1 trafficking to the dendritic cell-T-cell infectious synapse uses a pathway of tetraspanin sorting to the immunological synapse. Traffic. 2005;6:488–501. doi: 10.1111/j.1600-0854.2005.00293.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang JH, Wells C, Wu L. Macropinocytosis and cytoskeleton contribute to dendritic cell-mediated HIV-1 transmission to CD4+ T cells. Virology. 2008;381:143–154. doi: 10.1016/j.virol.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nobile C, et al. HIV-1 Nef inhibits ruffles, induces filopodia and modulates migration of infected lymphocytes. J Virol. 2009;84:2282–2293. doi: 10.1128/JVI.02230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sol-Foulon N, et al. ZAP-70 kinase regulates HIV cell-to-cell spread and virological synapse formation. EMBO J. 2007;26:516–526. doi: 10.1038/sj.emboj.7601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heymann JA, et al. 3D imaging of mammalian cells with ion-abrasion scanning electron microscopy. J Struct Biol. 2009;166:1–7. doi: 10.1016/j.jsb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildebrand M, Kim S, Shi D, Scott K, Subramaniam S. 3D imaging of diatoms with ion-abrasion scanning electron microscopy. J Struct Biol. 2009;166:316–328. doi: 10.1016/j.jsb.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne JL, Subramaniam S. Cryo-electron tomography of bacteria: Progress, challenges and future prospects. Nat Rev Microbiol. 2009;7:666–675. doi: 10.1038/nrmicro2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willig KI, et al. Nanoscale resolution in GFP-based microscopy. Nat Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 20.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heymann JA, et al. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J Struct Biol. 2006;155:63–73. doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett AE, et al. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 2009;5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia E, Nikolic DS, Piguet V. HIV-1 replication in dendritic cells occurs through a tetraspanin-containing compartment enriched in AP-3. Traffic. 2008;9:200–214. doi: 10.1111/j.1600-0854.2007.00678.x. [DOI] [PubMed] [Google Scholar]
- 24.Sherer NM, et al. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrijn AF, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher PJ, Bulur PA, Vuk-Pavlovic S, Prendergast FG, Dietz AB. Dendritic cell microvilli: A novel membrane structure associated with the multifocal synapse and T-cell clustering. Blood. 2008;112:5037–5045. doi: 10.1182/blood-2008-04-149526. [DOI] [PubMed] [Google Scholar]
- 27.Knight SC, et al. Non-adherent, low-density cells from human peripheral blood contain dendritic cells and monocytes, both with veiled morphology. Immunology. 1986;57:595–603. [PMC free article] [PubMed] [Google Scholar]
- 28.Mellman I, Steinman RM. Dendritic cells: Specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 29.Peters JH, Ruhl S, Friedrichs D. Veiled accessory cells deduced from monocytes. Immunobiology. 1987;176:154–166. doi: 10.1016/s0171-2985(87)80107-9. [DOI] [PubMed] [Google Scholar]
- 30.Singer II, et al. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. J Virol. 2001;75:3779–3790. doi: 10.1128/JVI.75.8.3779-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jolly C, Mitar I, Sattentau QJ. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J Virol. 2007;81:5547–5560. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolly C, Mitar I, Sattentau QJ. Adhesion molecule interactions facilitate human immunodeficiency virus type 1-induced virological synapse formation between T cells. J Virol. 2007;81:13916–13921. doi: 10.1128/JVI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turville SG, et al. Immunodeficiency virus uptake, turnover, and 2-phase transfer in human dendritic cells. Blood. 2004;103:2170–2179. doi: 10.1182/blood-2003-09-3129. [DOI] [PubMed] [Google Scholar]
- 34.Sougrat R, et al. Electron tomography of the contact between T cells and SIV/HIV-1: Implications for viral entry. PLoS Pathog. 2007;3:e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrighi JF, et al. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J Exp Med. 2004;200:1279–1288. doi: 10.1084/jem.20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arrighi JF, et al. Lentivirus-mediated RNA interference of DC-SIGN expression inhibits human immunodeficiency virus transmission from dendritic cells to T cells. J Virol. 2004;78:10848–10855. doi: 10.1128/JVI.78.20.10848-10855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin N, et al. Virological synapse-mediated spread of human immunodeficiency virus type-1 between T cells is sensitive to entry inhibition. J Virol. 84:3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganesh L, et al. Infection of specific dendritic cells by CCR5-tropic human immunodeficiency virus type 1 promotes cell-mediated transmission of virus resistant to broadly neutralizing antibodies. J Virol. 2004;78:11980–11987. doi: 10.1128/JVI.78.21.11980-11987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.