Abstract
The microsomal antiestrogen binding site (AEBS) is a high-affinity target for the antitumor drug tamoxifen and its cognate ligands that mediate breast cancer cell differentiation and apoptosis. The AEBS, a hetero-oligomeric complex composed of 3β-hydroxysterol-Δ8-Δ7-isomerase (D8D7I) and 3β-hydroxysterol-Δ7-reductase (DHCR7), binds different structural classes of ligands, including ring B oxysterols. These oxysterols are inhibitors of cholesterol-5,6-epoxide hydrolase (ChEH), a microsomal epoxide hydrolase that has yet to be molecularly identified. We hypothesized that the AEBS and ChEH might be related entities. We show that the substrates of ChEH, cholestan-5α,6α-epoxy-3β-ol (α-CE) and cholestan-5β,6β-epoxy-3β-ol (β-CE), and its product, cholestane-3β,5α,6β-triol (CT), are competitive ligands of tamoxifen binding to the AEBS. Conversely, we show that each AEBS ligand is an inhibitor of ChEH activity, and that there is a positive correlation between these ligands’ affinity for the AEBS and their potency to inhibit ChEH (r2 = 0.95; n = 39; P < 0.0001). The single expression of D8D7I or DHCR7 in COS-7 cells slightly increased ChEH activity (1.8- and 2.6-fold), whereas their coexpression fully reconstituted ChEH, suggesting that the formation of a dimer is required for ChEH activity. Similarly, the single knockdown of D8D7I or DHCR7 using siRNA partially inhibited ChEH in MCF-7 cells, whereas the knockdown of both D8D7I and DHCR7 abolished ChEH activity by 92%. Taken together, our findings strongly suggest that the AEBS carries out ChEH activity and establish that ChEH is a new target for drugs of clinical interest, polyunsaturated fatty acids and ring B oxysterols.
Keywords: breast cancer, chemoprevention, cholesterol metabolism, oxysterol, docosahexaenoic acid
Tamoxifen (Tam) is one of the most commonly used drugs worldwide for hormonal treatment and chemoprevention of estrogen receptor (ER)-positive breast cancers (1). In this application, Tam activity is mediated through the modulation of gene expression under the control of ERs. Tam's pharmacology is complex, however; it exerts nongenomic effects through other targets at therapeutic doses (2). After the ER, the microsomal antiestrogen binding site (AEBS) is the target of highest affinity for Tam. The AEBS has no affinity for estrogens, but binds selective ER modulators (SERMs); these ligands contain a hydrophobic core that mimics the steroid backbone of estrogens, grafted to a dialkylaminoalkyl side chain. Thus, in addition to Tam, SERMs, such as raloxifene, 4-OH-Tam, and RU-39411, are ligands of the AEBS (3–5). The AEBS selectively binds diphenylmethane derivatives of Tam, including (4-benzyl-phenoxy)-ethyl-N-pyrrolidine (PBPE) and tesmilifene (6) and it also binds σ-receptor ligands and inhibitors of cholesterol biosynthesis, such as triparanol and AY-9944 (5, 7). All of these classes of synthetic AEBS ligands have a protonable dialkylaminoalkyl chain in common that is necessary for high-affinity binding to the AEBS (6). In the quest for natural AEBS ligands, several unsaturated fatty acids and ring B oxysterols have been identified (8, 9). Oxysterols that bind to the AEBS are cholesterol-oxidized on ring B of the cholesterol backbone and include 5-cholesten-3β-ol-7-one (7-ketocholesterol), 5α-cholestan-3β-ol-7-one (7-ketocholestanol), and 5α-cholestan-3β-ol-6-one (6-ketocholestanol) (8).
In previous work, we established that the coexpression of 3β-hydroxysterol-Δ8-Δ7-isomerase (D8D7I) and 3β-hydroxysterol-Δ7-reductase (DHCR7) in mammalian cells is necessary and sufficient to reconstitute the high-affinity binding site for [3H]Tam (i.e., the AEBS) (5). D8D7I and DHCR7 are two enzymes involved in specific catalytic steps in the postlanosterol biosynthesis of cholesterol. Consistent with the fact that the AEBS binds σ-receptor ligands (7), D8D7I (an AEBS subunit) carries a binding site for σ-receptor ligands (10). AEBS expression is ubiquitous, and AEBS is highly expressed in proliferative cells, such as tumor cells, and in cholesterogenic tissues, such as the liver and brain, in accordance with its relationship with cholesterol metabolism (5, 10, 11). We recently established that AEBS ligands induce breast cancer cell differentiation and apoptosis through a mechanism involving the production of sterol autoxidation products (3, 4, 12). Cholestan-5α,6α-epoxy-3β-ol (α-CE), cholestan-5β,6β-epoxy-3β-ol (β-CE), and 7-ketocholesterol are among the major autoxidation products of cholesterol (13). CEs (α-CE and β-CE) are the only known substrates of cholesterol epoxide hydrolase (ChEH), and 7-ketocholesterol is an inhibitor of ChEH (14, 15). ChEH (EC 3.3.2.11) catalyzes the hydrolysis of α-CE and β-CE into a unique geminal trans-diol, cholestane-3β,5α,6β-triol (CT) (16) (SI Appendix, Fig. S1).
We have observed that the AEBS and ChEH share similar characteristics and pharmacological properties. Indeed, ring B oxysterols, such as 6-ketocholestanol, 7-ketocholestanol, and 7-ketocholesterol, inhibit ChEH (14) and bind to the AEBS (8) with the same order of potency. In addition, ChEH is inhibited by an autoxidation product of 7-dehydrocholesterol (17), and 7-dehydrocholesterol is the substrate of DHCR7, an AEBS subunit (5). Moreover, as with the AEBS, ChEH is located in the endoplasmic reticulum of cells and is found in most mammalian tissues, with the liver being the richest source of both the AEBS and ChEH (18, 19). ChEH is the last member of the epoxide hydrolase family with an unidentified coding gene. The results from these studies, along with the relationship that we established between sterol autoxidation products and the functions of the AEBS (3, 4, 12), suggested to us a pharmacological and structural link between the AEBS and ChEH, which we investigated in the present study. Our results indicate that the enzymes that form the AEBS are involved in the catalytic activity of ChEH.
Results
α-CE, β-CE, and CT Are Competitive Ligands of the AEBS.
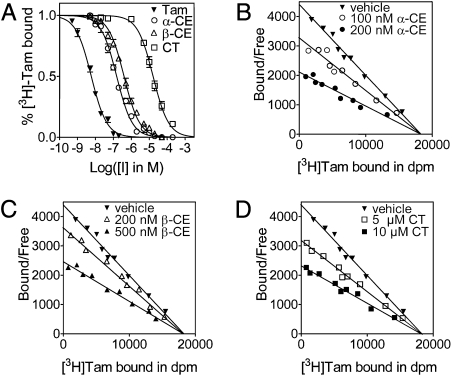
We began by investigating whether the substrates of ChEH (α-CE and β-CE) and CT, the product of ChEH activity, are ligands of the AEBS. We performed assays on rat liver microsomes, the richest source of the AEBS (6). Competitive binding assays showed that increasing concentrations of unlabeled Tam, α-CE, β-CE, or CT inhibited the binding of 2.5 nM [3H]Tam to the AEBS in a concentration-dependent manner (Fig. 1A). The IC50 was 5.9 ± 0.5 nM for Tam, 153 ± 0.5 nM for α-CE, 394 ± 1 nM for β-CE, and 16.3 ± 0.6 μM for CT. To study the modality of the inhibition by α-CE, β-CE, and CT, we performed Scatchard analyses of [3H]Tam binding in the absence or presence of two concentrations of α-CE (Fig. 1B), β-CE (Fig. 1C), and CT (Fig. 1D). For each experiment, a diminished slope of the lines indicated decreasing [3H]Tam affinity, whereas the Bmax remained unchanged. These data demonstrate that the substrates (α-CE and β-CE) and the product (CT) of ChEH are competitive ligands of the AEBS, with apparent Ki values of 66.5 ± 0.2 nM for α-CE, 171.3 ± 1.4 nM for β-CE, and 7.1 ± 0.6 μM for CT.
Fig. 1.
Competition for [3H]Tam binding to the microsomal AEBS by Tam, α-CE, β-CE, and CT. (A) Competition assays with increasing concentrations of unlabeled Tam (▼), α-CE (●), β-CE (▲), and CT (▪) were performed on rat liver microsomes using 2.5 nM [3H]Tam. (B) Scatchard plots of [3H]Tam binding to the microsomal AEBS in the absence (▼) or presence of 100 nM α-CE (○) or 200 nM α-CE (●). (C) Scatchard plots of [3H]Tam binding to the microsomal AEBS in the absence (▼) or presence of 200 nM β-CE (△) or 500 nM β-CE (▲). (D) Scatchard plots of [3H]Tam binding to the microsomal AEBS in the absence (▼) or presence of 5 μM CT (□) or 10 μM CT (▪). The lines intercept on the x-axis, indicating that α-CE, β-CE, and CT are competitive ligands of the AEBS with respect to Tam binding. Measurements were made in triplicate for at least three separate experiments. Data are presented as the mean ± SEM.
The Prototypical AEBS Ligands Tam and PBPE Are Inhibitors of ChEH.
To test the effects of AEBS ligands on ChEH, we first measured ChEH activity by incubating rat liver microsomes with 20 μM [14C]α-CE and separated it from CT by TLC. The migration of CE and CT was validated by reference to commercially available standards. As shown in the TLC autoradiogram (SI Appendix, Fig. S2A, Left), the conversion of α-CE to CT was time-dependent. Steady-state kinetics were apparent for the first 10 min of incubation (SI Appendix, Fig. S2A, Right). Consequently, all subsequent experiments were performed using a 9-min incubation period. The Km was 7.4 ± 0.5 μM, and the Vmax was 0.62 ± 0.01 nmol CT/mg protein/min (SI Appendix, Fig. S2B), consistent with data reported in the literature (14, 15). We then assessed ChEH activity, measured using [14C]α-CE or [14C]β-CE as the substrate, in the presence of known inhibitors (6-ketocholestanol and 7-ketocholestanol) (14), as well as in the presence of prototypical AEBS ligands (Tam and PBPE). The conversion of α-CE or β-CE to CT was inhibited by 10 μM 6-ketocholestanol or 7-ketocholestanol and by 1 μM Tam or PBPE (SI Appendix, Fig. S2C). Taken together, these results indicate that Tam and PBPE inhibit ChEH activity.
Tam and PBPE Are Competitive Inhibitors of ChEH.
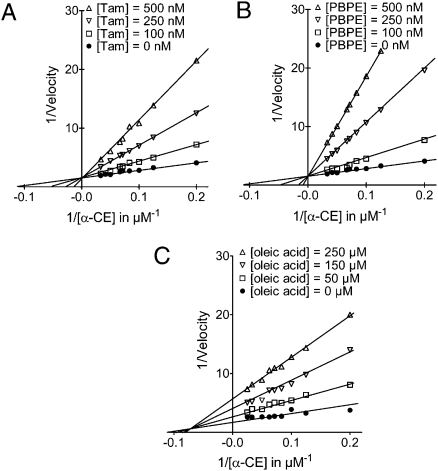
We next carried out experiments to determine the modality of ChEH inhibition by Tam and PBPE. Tam inhibited ChEH in a concentration-dependent manner, as shown by a double-reciprocal plot (Lineweaver-Burk) of the inhibition of [14C]α-CE hydration by Tam in Fig. 2A. The x-intercept (i.e., the 1/Km value) decreased, whereas the y-intercept (i.e., the Vmax value) was not affected; these changes are characteristic of competitive inhibition. This was confirmed through a Dixon plot of the inverse of the velocity as a function of increasing Tam concentration in the presence of two concentrations of [14C]α-CE (SI Appendix, Fig. S3A). The x-intercept gave a Ki value of 34 ± 8 nM for Tam. The same experiments carried out with PBPE, a selective AEBS ligand, showed competitive inhibition of ChEH, with a Ki value of 27 ± 6 nM (Fig. 2B and SI Appendix, Fig. S3B). These results indicate that Tam and PBPE are potent competitive inhibitors of ChEH activity.
Fig. 2.
Inhibition of ChEH by Tam, PBPE, and oleic acid. The relationship between the conversion rates of α-CE to CT and inhibitor concentrations is shown using 10 and 20 μM Tam and PBPE with rat liver microsomal ChEH. Shown are double reciprocal plots of Tam (A), PBPE (B), and oleic acid (C) versus [14C]α-CE.
Synthetic AEBS Ligands Are Inhibitors of ChEH.
We tested the potency of AEBS ligands of different pharmacological classes to inhibit ChEH. All of the selective AEBS ligands (compounds 1–8; SI Appendix, Fig. S4A) inhibited ChEH activity (Table 1). The most potent inhibitors were PBPE, 1-{2-[4-(2-phenylpropan-2-yl) phenoxy]ethyl}pyrrolidine (PCPE), tesmilifene, 4-[2-(4-benzylphenoxy)ethyl]morpholine (MBPE), and 4-{2-[4-(2-phenylpropan-2-yl)phenoxy]ethyl}morpholine (MCPE) (5), with potencies in the nanomolar range, whereas 1-[4-(2-morpholino ethoxy)phenyl]-2-phenylethanone (MCOCH2PE) was the least potent, with a Ki value in the micromolar range for ChEH (Table 1). In the subsequent series, we found that SERMs (compounds 9–14; SI Appendix, Fig. S4B) inhibited ChEH in the following order of potency: clomiphene > nitromiphene > Tam ≥ raloxifene > 4OH-tamoxifen ≥ RU 39,411 (Table 1). ER ligands with no affinity for the AEBS, such as 17β-estradiol, diethylstilbestrol, ICI-164,384, RU-56668, and ICI-182,780 (SI Appendix, Fig. S7), did not inhibit the ChEH at concentrations up to 10 μM (SI Appendix, Table S1). Of the σ-receptor ligands (SI Appendix, Fig. S5A), compounds 15–22 were both ligands of the AEBS and inhibitors of ChEH with the following order of potency: SR-31747A > BD-1008 > trifluoroperazine > amiodarone > ibogaine ≥ rimcazole > AC-915 > haloperidol (Table 1). SR-31747A had the highest affinity for the AEBS and was the most potent ChEH inhibitor of the compounds tested, with a Ki value of 1.2 ± 0.1 nM for the inhibition of Tam binding to the AEBS and a Ki value of 6 ± 2 nM for ChEH inhibition. Other σ-receptor ligands (compounds S6–S10; SI Appendix, Fig. S7), including ditolyl guanidine (DTG), (+)-pentazocine, (+)-3PPP, PRE-084, and progesterone, failed to bind to the AEBS and inhibit ChEH, even at concentrations up to 1,000 μM (SI Appendix, Table S1). In the last series of synthetic compounds, inhibitors of cholesterol biosynthesis already reported to be AEBS ligands (5) (compounds 23–28; SI Appendix, Fig. S5A), including U-18666A, triparanol, AY-9944, and SKF-525A, and newly identified AEBS ligands, such as terbinafine and Ro 48–8071, were inhibitors of ChEH (Table 1). Together, these results establish that every tested drug that bound to the AEBS was an inhibitor of ChEH.
Table 1.
Inhibition of [3H]Tam binding to the AEBS and catalytic activity of ChEH by drugs
| Compound | Ki AEBS, nM | Ki ChEH, nM | |
| Selective AEBS ligands | |||
| PBPE | 1 | 9 ± 1 | 27 ± 6 |
| PCPE | 2 | 10 ± 1 | 35 ± 8 |
| Tesmilifene | 3 | 56 ± 2 | 62 ± 3 |
| MBPE | 4 | 18 ± 1 | 27 ± 6 |
| MCPE | 5 | 48 ± 2 | 57 ± 8 |
| PCOPE | 6 | 64 ± 4 | 203 ± 11 |
| MCOPE | 7 | 102 ± 16 | 241 ± 7 |
| MCOCH2PE | 8 | 850 ± 12 | 902 ± 13 |
| SERMs | |||
| Tamoxifen | 9 | 2.5 ± 0.2 | 34 ± 8 |
| 4OH-Tamoxifen | 10 | 11 ± 1 | 145 ± 4 |
| Raloxifene | 11 | 6 ± 1 | 36 ± 4 |
| Nitromiphene | 12 | 2.4 ± 0.3 | 18 ± 6 |
| Clomiphene | 13 | 1.5 ± 0.2 | 9 ± 2 |
| RU 39,411 | 14 | 38 ± 1 | 155 ± 8 |
| σ receptor ligands | |||
| BD-1008 | 15 | 83 ± 1 | 99 ± 9 |
| Haloperidol | 16 | 5,322 ± 9 | 18,067 ± 14 |
| SR-31747A | 17 | 1.2 ± 0.1 | 6 ± 2 |
| Ibogaine | 18 | 920 ± 12 | 2,150 ± 11 |
| AC-915 | 19 | 1,120 ± 8 | 3,527 ± 9 |
| Rimcazole | 20 | 640 ± 5 | 2,325 ± 8 |
| Amiodarone | 21 | 432 ± 22 | 733 ± 9 |
| Trifluoroperazine | 22 | 14 ± 2 | 135 ± 7 |
| Cholesterol biosynthesis inhibitors | |||
| Ro 48–8071 | 23 | 110 ± 4 | 89 ± 5 |
| U-18666A | 24 | 84 ± 2 | 90 ± 5 |
| AY-9944 | 25 | 358 ± 12 | 649 ± 6 |
| Triparanol | 26 | 17 ± 2 | 39 ± 3 |
| Terbinafine | 27 | 3,720 ± 16 | 9,105 ± 33 |
| SKF-525A | 28 | 897 ± 10 | 1,904 ± 11 |
| Ring B oxysterols | |||
| 6-Ketocholestanol | 29 | 1,122 ± 30 | 2,251 ± 21 |
| 7-Ketocholestanol | 30 | 580 ± 12 | 864 ± 22 |
| 7-Ketocholesterol | 31 | 1,223 ± 31 | 4,212 ± 32 |
| 7α-Hydroxycholesterol | 32 | 2,252 ± 32 | 6,151 ± 22 |
| 7β-Hydroxycholesterol | 33 | 4,471 ± 22 | 6,941 ± 21 |
| 6-Keto-5α-hydroxycholestanol | 34 | 5,320 ± 22 | 8,522 ± 12 |
| Cholestane-3β,5α,6β-triol | 35 | 7,131 ± 28 | 9,744 ± 11 |
| Fatty acids | |||
| Oleic acid | 36 | 48,144 ± 19 | 54,235 ± 38 |
| α-Linolenic acid | 37 | 38,232 ± 41 | 36,341 ± 42 |
| ARA | 38 | 26,171 ± 17 | 24,094 ± 18 |
| DHA | 39 | 18,284 ± 19 | 12,111 ± 16 |
Rat liver microsomes were incubated with a single concentration of 2.5 nM [3H]Tam and increasing concentrations of inhibitors ranging from 0.1 nM to 1,000 μM under the conditions described in SI Appendix, SI Materials and Methods. IC50 values were determined using the iterative curve-fitting program GraphPad Prism version 4 (GraphPad Software). For the AEBS, the apparent Ki was expressed as Ki = [IC50]/(1 + ([[3H]Tam])/Kd), using 2.5 nM [3H]Tam and a Kd of 2 nM. The Ki values of drugs for ChEH inhibition were determined using 150 μg of rat liver microsomal protein and 10 and 20 μM of [14C]α-CE with increasing concentrations of inhibitors ranging from 0.01 to 1000 μM, under the conditions described in SI Appendix, SI Materials and Methods. Ki was measured as the projection on the x-axis of the intersection of the lines obtained from 1/V versus [inhibitor] plots for ChEH. Values are the average of three experiments ± SEM, each carried out in duplicate.
Unesterified Ring B Oxysterols Are Inhibitors of ChEH.
We next evaluated a set of oxysterols (SI Appendix, SI Appendix,Figs. S6 and S8). Ring B oxysterols (compounds 29–34; SI Appendix, Fig. S6) inhibited ChEH according to the following order of potency: 7-ketocholestanol > 6-ketocholestanol > 7-ketocholesterol > 7α-hydroxycholesterol > 7β-hydroxycholesterol > 6-keto-5α-hydroxycholestanol > CT (Table 1). In contrast, side-chain oxysterols (compounds S13–S16; SI Appendix, Fig. S8) did not inhibit ChEH activity or bind to the AEBS (SI Appendix, Table S1). Ring B oxysterols were previously shown to be competitive inhibitors of ChEH (14) as well as of Tam binding to the AEBS (8). In addition, the sulfate ester α-CE (S17) and the stearic acid ester of CE (S18) had no affinity for the AEBS and were not inhibitors of ChEH (SI Appendix, Table S1). Thus, unlike α-CE, esterified forms of α-CE are not substrates of ChEH. Our data indicate that unesterified ring B oxysterols are both inhibitors of ChEH and ligands of the AEBS, whereas side-chain oxysterols and esterified ring B oxysterols are not.
Unsaturated Fatty Acids That Are AEBS Ligands Are Inhibitors of ChEH.
Because oleic acid is a noncompetitive ligand of the AEBS (20), we next studied whether oleic acid can inhibit ChEH activity, and analyzed the modality of its inhibition. Using Lineweaver-Burk analysis (Fig. 2C) and Dixon analysis (SI Appendix, Fig. S3C), we found that oleic acid is a noncompetitive inhibitor of ChEH, with a Ki value of 54 μM (Table 1). We extended this study by testing other fatty acids [compounds 36–39 (SI Appendix, Fig. S6) and S19–S21 (SI Appendix, Fig. S8)]. Unsaturated fatty acids, such as docosahexaenoic acid (DHA), α-linoleic acid, and arachidonic acid (ARA), are inhibitors of ChEH activity, whereas the saturated fatty acids stearic acid and palmitic acid and the methyl ester of oleic acid are not (SI Appendix, Table S1). These data indicate that unsaturated fatty acids are inhibitors of ChEH, and that oleic acid is a noncompetitive inhibitor.
Ligands’ Affinity for the AEBS Positively Correlates with Their Inhibition of ChEH.
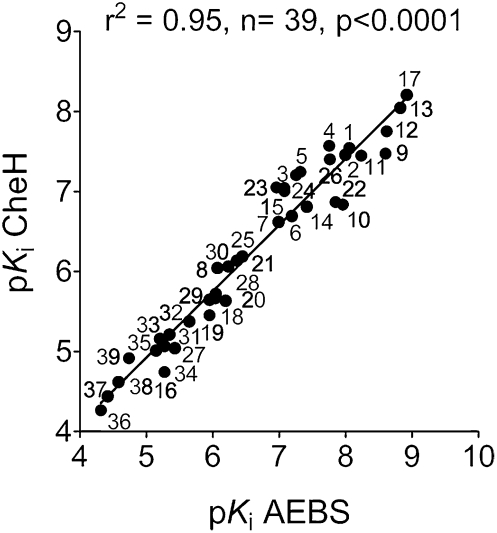
Plotting the pKi [-log(Ki)] of compounds that bound to the AEBS as a function of their pKi values for their inhibitory potency of ChEH activity yielded a positive linear correlation between both parameters, with an r2 value of 0.95 (n = 39; P < 0.0001) (Fig. 3). This demonstrates a clear correlation between the affinity for the AEBS and ChEH inhibition for the different classes of molecules.
Fig. 3.
Correlation between affinity of AEBS ligands for the AEBS and their potency to inhibit ChEH. Graph of the pKi for 39 compounds tested for the inhibition of [3H]Tam binding as a function of pKi on ChEH activity. The drug numbers and the corresponding pKi values [−log(Ki)] are listed in Table 1. Here r is the correlation coefficient between pKi values calculated for the inhibition of Tam binding and ChEH activity. The r2 value of 0.95 and significance of correlation (P < 0.0001) are given for all structural classes of compounds (n = 39).
D8D7I and DHCR7 Coexpression Allows the Reconstitution of ChEH.
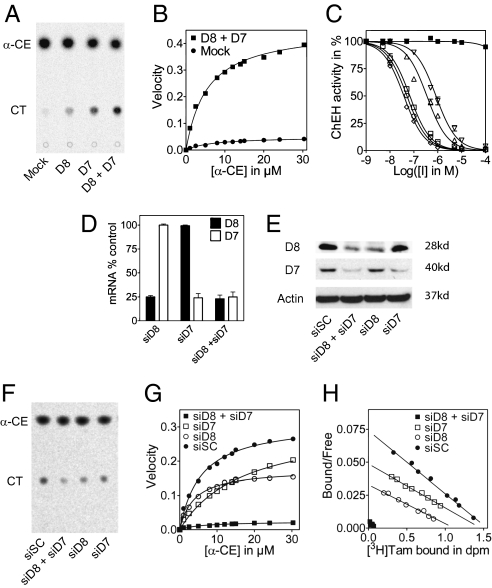
We previously reported that the coexpression of D8D7I and DHCR7 is necessary for reconstitution of the AEBS in mammalian COS-7 cells (5). We evaluated whether these two enzymes were involved in ChEH activity. As shown in Fig. 4A, the basal activity of ChEH was low in COS-7 cells transfected with the vector control (mock). The single expression of D8D7I (D8) or DHCR7 (D7) in COS-7 cells induced a slight increase in ChEH activity (1.8- and 2.6-fold, respectively) compared with the vector control. In contrast, the coexpression of D8D7I and DHCR7 (D8 + D7) in COS-7 cells potentiated ChEH activity by 8.5-fold (Fig. 4A). A Vmax value of 0.46 ± 0.04 nmol CT/mg protein/min and a Km value of 4.47 ± 0.05 μM was measured in the D8D7I and DHCR7 coexpression experiments, compared with a Vmax = 0.05 ± 0.002 nmol CT/mg protein/min and Km = 7.01 ± 0.05 μM for the vector-only transfected cells (Fig. 4B). These data indicate that coexpression of D8D7I and DHCR7 led to the reconstitution of robust ChEH activity in mammalian cells, indicating that ChEH activity requires both enzymes. We tested the inhibition of ChEH by various AEBS ligands on COS-7 cell lysates that coexpressed D8D7I and DHCR7 (Fig. 4C). The order of potency of AEBS ligands for inhibiting the reconstituted ChEH was as follows: clomiphene (Ki = 32.3 ± 0.5 nM) > PBPE (Ki = 43.3 ± 0.4 nM) > Tam (Ki = 57.3 ± 0.6 nM) > 4OH-tamoxifen (Ki = 295.8 ± 0.8 nM) > 7-ketocholestanol (Ki = 711.6 ± 0.9 nM). 17β-Estradiol, which does not bind to the AEBS (SI Appendix, Table S1), did not inhibit the reconstituted ChEH. These data establish that the pharmacological profile obtained with the ChEH is similar to that of the AEBS (5).
Fig. 4.
Expression and knockdown of D8D7I and DHCR7 in mammalian cells: Impact on ChEH and AEBS activities. (A) ChEH activity of microsomal extracts from COS-7 cells transfected with control vector (mock), D8, D7, and D8 + D7. (B) Michaelis-Menten plot of velocity versus α-CE in ChEH assays from COS-7 cells transfected with mock (●) or D8 + D7 (▪). (C) Inhibition of ChEH in microsomal extracts from COS-7 cells coexpressing human recombinant D8 and D7 with increasing concentrations of clomiphene (⋄), PBPE (○), Tam (□), 4OH-tamoxifen (△), 7-ketocholestanol (▽), or 17β-estradiol (▪). (D, E) Expression of D8 and D7 in MCF-7 cells transfected with siSC scrambled, siD8, siD7, or siD8 + siD7 at the mRNA level (D) and at the protein level (E). (F) Representative TLC autoradiogram showing ChEH activity in MCF-7 cells from three independent experiments. (G) Michaelis-Menten plot of velocity versus α-CE in ChEH assays from MCF-7 cells transfected with control scrambled siRNA (siSC; ●), siD8 (○), siD7 (□), or siD8 + siD7 (▪). (I) Scatchard plots of [3H]Tam binding to microsomal AEBS from MCF-7 cells transfected with siSC (●), siD8 (○), siD7 (□), or siD8 + siD7 (▪). Measurements were made in triplicate for at least three separate experiments. Data are presented as mean ± SEM.
Knockdown of D8D7I and DHCR7 Abolishes ChEH Activity.
To confirm that D8D7I and DHCR7 both contribute to ChEH, we conducted knockdown experiments using siRNA against D8D7I and DHCR7 in a human breast adenocarcinoma cell line, MCF-7 cells. The siRNA specificities were evaluated at the mRNA and at the protein levels for the expression of D8D7I and DHCR7. The impact of knockdowns on the kinetic parameters of ChEH and on the binding parameters of [3H]Tam to the AEBS was measured. MCF-7 cells expressed the AEBS (Kd = 5.2 ± 1.4 nM, Bmax = 1,553 ± 25 fmol/mg proteins) (4), and ChEH activity was found, with a Vmax value of 0.38 ± 0.07 nmol CT/mg protein/min and a Km value of 5.91 ± 0.06 μM (Fig. 4 G and H). Transfection of the cells with D8D7I siRNA, but not with scrambled siRNA, led to decreased D8D7I expression at the mRNA level (72%) (Fig. 4D) and protein level (60%) (Fig. 4E). Interestingly, it also reduced ChEH activity by 47% (Fig. 4F), with Vmax = 0.18 ± 0.09 nmol CT/mg protein/min, Km = 3.87 ± 0.07 μM (Fig. 4G), and a 42% decrease in the amount of AEBS (Kd = 6.1 ± 0.4 nM, Bmax = 897 ± 18 fmol/mg proteins) (Fig. 4H). Transfection of the cells with DHCR7 siRNA, but not with scrambled siRNA, decreased DHCR7 expression at the mRNA level (73%) (Fig. 4D) and protein level (64%) (Fig. 4E). Knockdown of DHCR7 increased the Km value of ChEH by 66% (Km = 17.22 μM) with no significant changes in the Vmax value (Fig. 4G), and reduced the affinity of Tam for the AEBS with no changes in the Bmax value (Kd = 7.2 ± 0.6 nM, Bmax = 1,445 ± 23 fmol/mg proteins) (Fig. 4H). Finally, transfection of the cells with D8D7I siRNA and DHCR7 siRNA produced a comparable decrease in D8D7I and DHCR7 expression as in the single knockdown experiments, associated with a drastic reduction (92%) in ChEH activity (Fig. 4F) that was more than additive (Vm = 0.023 ± 0.06 nmol CT/mg protein/min, with no change in the Km value (5.75 ± 0.07 μM) (Fig. 4G). A similar effect on the AEBS was observed, with a 93% decrease in the Bmax value (103.5 ± 29 fmol/mg proteins) with no changes in the Kd value (Fig. 4H). These decreases are greater than expected based on the reduction in protein expression and strongly suggest that D8D7I and DHCR7 cooperate in the ChEH activity. These data further demonstrate that D8D7I and DHCR7 are involved in the ChEH activity.
Discussion
The present study provides evidence that the AEBS and ChEH are pharmacologically and molecularly related. We have shown that substrates (CEs) and the product (CT) of ChEH are competitive inhibitors of Tam binding to the AEBS, and have established that ChEH is inhibited by all AEBS ligands tested, demonstrating that inhibition of ChEH is a hallmark of AEBS ligands. We found that different structural classes of AEBS ligands inhibit ChEH with the same modality as they inhibit [3H]Tam binding to the AEBS. We established the pharmacological similarities by showing a positive correlation between inhibition of ChEH activity and affinity of compounds for the AEBS. Importantly, compounds belonging to different pharmacological or biochemical classes that had no affinity for the AEBS did not inhibit ChEH. With the series of synthetic compounds, the arylaminoalkyl structure was not sufficient for ChEH inhibition; such compounds as (+)-3-PPP, (+)-pentazocine, PRE-084, and MCH3PE did not inhibit ChEH or compete with Tam for binding to the AEBS.
As shown previously for reconstitution of the AEBS (5), transient coexpression of D8D7I and DHCR7 in COS-7 cells led to the reconstitution of ChEH. We confirmed the involvement of D8D7I and DHCR7 in ChEH through knockdown experiments using siRNA directed against D8D7I and DHCR7. These data show that both D8D7I and DHCR7 are required for the dual reconstitution of ChEH and the AEBS and provide more evidence of the molecular nature of the ChEH. The fact that our single knockdown experiments showed that D8D7I and DHCR7 affect the kinetic parameters of ChEH differently suggests that D8D7I carries the catalytic activity, and that DHCR7 cooperates in the binding of substrates to ChEH (Fig. 5). We find it interesting that three epoxide hydrolases in three unrelated structural classes have such high structural complexity. The leukotriene A4 hydrolase/aminopeptidase (21) and the soluble epoxide hydrolase/phosphatase (22) are both bifunctional enzymes. Our data suggest even greater complexity with ChEH in being bifunctional and composed of two independent gene products that unite in the microsomes to create a functional protein.
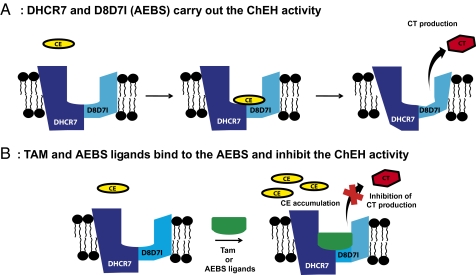
Fig. 5.
Functional relationship between the AEBS and ChEH. (A) The subunits of the AEBS that bind Tam (D8D7I and DHCR7) carry out the ChEH activity. (B) Ligands of the AEBS, such as cationic SERMs (Tam and raloxifene), diphenylmethane compounds (tesmilifene and PBPE), ring B oxysterols (7-ketocholesterol and 7-hydroxycholesterol) and polyunsaturated fatty acids (DHA), are inhibitors of ChEH, leading to the blockage of CT production and CE accumulation.
Here we have shown that anticancer drugs of clinical interest are inhibitors of ChEH at pharmacologically and therapeutically active concentrations. In vitro studies on breast cancer cells have previously used 1- to 40-μM concentrations of selective AEBS ligands, SERMS, or σ-receptor ligands (3, 4, 7) and 10–100 μM polyunsaturated fatty acids (23). The present study has demonstrated the existence of ChEH activity in MCF-7 cells, and thus ChEH inhibition and CE accumulation are likely to play a role in the mechanism of induction of breast cancer cell differentiation and apoptosis by AEBS ligands that require sterol autoxidation products. The therapeutic plasma concentration of Tam is 1–10 μM (24), and that of tesmilifene is 5 μM (25), whereas their respective Ki values for ChEH are 33 nM and 26 nM, indicating total inhibition of ChEH at therapeutic concentrations. The plasma concentration of nonesterified DHA is 2–10 μM in humans receiving a diet supplemented with 1.5 g DHA/d (26), whereas the DHA Ki value for the AEBS is 12.1 μM, indicating possible inhibition of ChEH. Thus, the accumulation of CEs is likely to be part of the pharmacologic action of these compounds and might be involved in the anticancer and chemopreventive actions of SERMs and DHA; this merits further investigation.
CEs are autoxidation products of cholesterol, and their production can be blocked by lipophilic antioxidants such as vitamin E (27). We recently established that AEBS ligands induce differentiation and apoptosis in breast cancer cells through a mechanism involving the production of sterol autoxidation products, and that vitamin E inhibits these effects (3, 4, 12). Consistent with these data, previous reports have shown a limited clinical outcome with Tam treatment when cholesterol metabolism enzymes (28) or antioxidant enzymes (29) are overexpressed in breast tumors. These data, along with our present findings, suggest that the modulation of CE metabolism resulting from ChEH inhibition by AEBS ligands might be involved in these effects. Further investigation is warranted. In particular, it will be interesting to study the relationship that may exist between CE metabolism and the sensitivity of and resistance to Tam of breast cancer cell lines and to define how accumulation of CEs or the products of their transformation might be involved in the effects of AEBS ligands.
The physiological function of ChEH has been proposed to be involved in the control of lipid metabolism (16) based on the biological properties of CT, not for detoxification as initially proposed (14). As opposed to toxic aliphatic and aromatic epoxides, which can spontaneously alkylate proteins and nucleic acids in association with their cytotoxicity or carcinogenicity, CEs are stable (30) and are less toxic than CT (13, 31) and nontumorigenic substances (32). CT's greater toxicity and the mutagenic nature of its oxidation products (33, 34) suggest that the inhibition of ChEH and the inhibition of CT production protects cells against cytotoxic insults.
Several other lines of evidence point to the existence of a dynamic metabolism centered on CEs. α-CE is the only epimer of CE found in the adrenal cortex, where it is produced by an as-yet-unidentified cytochrome p450 (35). α-CE can be esterified by Sult2B1b (36) to give a 3β-sulfated product that antagonizes liver X-receptor signaling (37, 38), or it can be transformed by glutathione transferase into 3β,5α-dihydroxycholestan-6β-yl-S-glutathione (39, 40). In addition, we have reported that the aminolysis of α-CE by biogenic amines under catalytic conditions is possible and generates powerful cell-differentiating alkylaminooxysterols (30). In contrast, β-CE is nonreactive even under catalytic conditions (30) and has been reported to accumulate in breast fluids (41) and in the plasma of endometrial cancer patients (42). Thus, it would be interesting to study whether β-CE can deregulate CE metabolism at the level of the epoxidation step or of CE metabolism, including ChEH, leading to the appearance of toxic CT.
In conclusion, these data shed light on the molecular nature and potential functions of ChEH and open up the existence of an active metabolic pathway at the level of the CEs.
Materials and Methods
Descriptions of chemical synthesis, AEBS binding, cell culture, and transfections are provided in SI Appendix, SI Materials and Methods.
ChEH Activity Assays.
Rat liver microsomes were prepared as described previously (43), and ChEH activity was assayed as described previously (14). Drugs and [14C]α-CE were dissolved in acetonitrile for the biological tests. The concentration of [14C]α-CE in the test tubes was 10 or 20 μM for the Dixon analyses and 5, 10, 20, 30, or 40 μM for the Lineweaver-Burk analyses. The maximal velocity (Vmax) and Michaelis constant (Km) were determined by nonlinear regression analysis using GraphPad Prism version 4.01 for Windows (GraphPad Software). One-way ANOVA with Dunnett's multiple-comparison posttest was performed with vector-only cells as the control using GraphPad Prism 4.01.
Supplementary Material
Acknowledgments
We thank Dr. Michel Record for his critical reading of the manuscript. This work was funded by Institut National de la Santé et de la Recherche Médicale Conseil Regional Midi-Pyrénées, the Institut National du Cancer through the ResisTH network, the Ministère Français de la Recherche et de l'Enseignement Supérieur through the GenHomme project and a predoctoral fellowship (G.S.), Affichem, and European Commission FP6 Integrated Project EuroHear (LSHG CT-2004-512063A). S.S.-P. is in charge of research at the Centre National de la Recherche Scientifique.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002922107/-/DCSupplemental.
References
- 1.Jordan VC. Tamoxifen: A most unlikely pioneering medicine. Nat Rev Drug Discov. 2003;2:205–213. doi: 10.1038/nrd1031. [DOI] [PubMed] [Google Scholar]
- 2.de Médina P, Favre G, Poirot M. Multiple targeting by the antitumor drug tamoxifen: A structure-activity study. Curr Med Chem Anticancer Agents. 2004;4:491–508. doi: 10.2174/1568011043352696. [DOI] [PubMed] [Google Scholar]
- 3.de Medina P, et al. Ligands of the antiestrogen-binding site induce active cell death and autophagy in human breast cancer cells through the modulation of cholesterol metabolism. Cell Death Differ. 2009;16:1372–1384. doi: 10.1038/cdd.2009.62. [DOI] [PubMed] [Google Scholar]
- 4.Payré B, et al. Microsomal antiestrogen-binding site ligands induce growth control and differentiation of human breast cancer cells through the modulation of cholesterol metabolism. Mol Cancer Ther. 2008;7:3707–3718. doi: 10.1158/1535-7163.MCT-08-0507. [DOI] [PubMed] [Google Scholar]
- 5.Kedjouar B, et al. Molecular characterization of the microsomal tamoxifen binding site. J Biol Chem. 2004;279:34048–34061. doi: 10.1074/jbc.M405230200. [DOI] [PubMed] [Google Scholar]
- 6.Poirot M, et al. Synthesis, binding and structure-affinity studies of new ligands for the microsomal anti-estrogen binding site (AEBS) Bioorg Med Chem. 2000;8:2007–2016. doi: 10.1016/s0968-0896(00)00119-x. [DOI] [PubMed] [Google Scholar]
- 7.Kedjouar B, et al. Structural similitudes between cytotoxic antiestrogen-binding site (AEBS) ligands and cytotoxic sigma-receptor ligands: Evidence for a relationship between cytotoxicity and affinity for AEBS or sigma-2 receptor but not for sigma-1 receptor. Biochem Pharmacol. 1999;58:1927–1939. doi: 10.1016/s0006-2952(99)00285-3. [DOI] [PubMed] [Google Scholar]
- 8.Hwang PL, Matin A. Interactions of sterols with antiestrogen-binding sites: Structural requirements for high-affinity binding. J Lipid Res. 1989;30:239–245. [PubMed] [Google Scholar]
- 9.Hwang PL. Unsaturated fatty acids as endogenous inhibitors of tamoxifen binding to antioestrogen-binding sites. Biochem J. 1986;237:749–755. doi: 10.1042/bj2370749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moebius FF, et al. Pharmacological analysis of sterol delta8-delta7 isomerase proteins with [3H]ifenprodil. Mol Pharmacol. 1998;54:591–598. doi: 10.1124/mol.54.3.591. [DOI] [PubMed] [Google Scholar]
- 11.Gray JM, Ziemian L. Antiestrogen binding sites in brain and pituitary of ovariectomized rats. Brain Res. 1992;578:55–60. doi: 10.1016/0006-8993(92)90229-3. [DOI] [PubMed] [Google Scholar]
- 12.de Medina P, Silvente-Poirot S, Poirot M. Tamoxifen and AEBS ligands induced apoptosis and autophagy in breast cancer cells through the stimulation of sterol accumulation. Autophagy. 2009;5:1066–1067. doi: 10.4161/auto.5.7.9820. [DOI] [PubMed] [Google Scholar]
- 13.Schroepfer GJ., Jr. Oxysterols: Modulators of cholesterol metabolism and other processes. Physiol Rev. 2000;80:361–554. doi: 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- 14.Sevanian A, McLeod LL. Catalytic properties and inhibition of hepatic cholesterol-epoxide hydrolase. J Biol Chem. 1986;261:54–59. [PubMed] [Google Scholar]
- 15.Nashed NT, Michaud DP, Levin W, Jerina DM. Properties of liver microsomal cholesterol 5,6-oxide hydrolase. Arch Biochem Biophys. 1985;241:149–162. doi: 10.1016/0003-9861(85)90371-6. [DOI] [PubMed] [Google Scholar]
- 16.Newman JW, Morisseau C, Hammock BD. Epoxide hydrolases: Their roles and interactions with lipid metabolism. Prog Lipid Res. 2005;44:1–51. doi: 10.1016/j.plipres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Nashed NT, Michaud DP, Levin W, Jerina DM. 7-Dehydrocholesterol 5,6 beta-oxide as a mechanism-based inhibitor of microsomal cholesterol oxide hydrolase. J Biol Chem. 1986;261:2510–2513. [PubMed] [Google Scholar]
- 18.Watts CK, Murphy LC, Sutherland RL. Microsomal binding sites for nonsteroidal anti-estrogens in MCF 7 human mammary carcinoma cells: Demonstration of high affinity and narrow specificity for basic ether derivatives of triphenylethylene. J Biol Chem. 1984;259:4223–4229. [PubMed] [Google Scholar]
- 19.Aström A, et al. Subcellular and organ distribution of cholesterol epoxide hydrolase in the rat. Biochim Biophys Acta. 1986;882:359–366. doi: 10.1016/0304-4165(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 20.Hwang PL. Interaction of unsaturated fatty acids with antioestrogen-binding sites. Biochem J. 1987;243:359–364. doi: 10.1042/bj2430359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haeggström JZ. Leukotriene A4 hydrolase/aminopeptidase, the gatekeeper of chemotactic leukotriene B4 biosynthesis. J Biol Chem. 2004;279:50639–50642. doi: 10.1074/jbc.R400027200. [DOI] [PubMed] [Google Scholar]
- 22.Newman JW, Morisseau C, Harris TR, Hammock BD. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc Natl Acad Sci USA. 2003;100:1558–1563. doi: 10.1073/pnas.0437724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chamras H, Ardashian A, Heber D, Glaspy JA. Fatty acid modulation of MCF-7 human breast cancer cell proliferation, apoptosis and differentiation. J Nutr Biochem. 2002;13:711–716. doi: 10.1016/s0955-2863(02)00230-9. [DOI] [PubMed] [Google Scholar]
- 24.Trump DL, et al. High-dose oral tamoxifen, a potential multidrug-resistance–reversal agent: Phase I trial in combination with vinblastine. J Natl Cancer Inst. 1992;84:1811–1816. doi: 10.1093/jnci/84.23.1811. [DOI] [PubMed] [Google Scholar]
- 25.Brandes LJ, Bracken SP, Ramsey EW. N,N-diethyl-2-[4-(phenylmethyl)phenoxy]ethanamine in combination with cyclophosphamide: An active, low-toxicity regimen for metastatic hormonally unresponsive prostate cancer. J Clin Oncol. 1995;13:1398–1403. doi: 10.1200/JCO.1995.13.6.1398. [DOI] [PubMed] [Google Scholar]
- 26.Bougnoux P, Hajjaji N, Maheo K, Couet C, Chevalier S. Fatty acids and breast cancer: Sensitization to treatments and prevention of metastatic re-growth. Prog Lipid Res. 2010;49:76–86. doi: 10.1016/j.plipres.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Bergstrom S, Samuesson B. The autoxidation of cholesterol. In: Lundberg WO, editor. Autoxidation and Antioxidants. I. New York: Interscience; 1961. pp. 233–248. [Google Scholar]
- 28.Pitroda SP, Khodarev NN, Beckett MA, Kufe DW, Weichselbaum RR. MUC1-induced alterations in a lipid metabolic gene network predict response of human breast cancers to tamoxifen treatment. Proc Natl Acad Sci USA. 2009;106:5837–5841. doi: 10.1073/pnas.0812029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiff R, et al. Oxidative stress and AP-1 activity in tamoxifen-resistant breast tumors in vivo. J Natl Cancer Inst. 2000;92:1926–1934. doi: 10.1093/jnci/92.23.1926. [DOI] [PubMed] [Google Scholar]
- 30.de Medina P, Paillasse MR, Payré B, Silvente-Poirot S, Poirot M. Synthesis of new alkylaminooxysterols with potent cell differentiating activities: Identification of leads for the treatment of cancer and neurodegenerative diseases. J Med Chem. 2009;52:7765–7777. doi: 10.1021/jm901063e. [DOI] [PubMed] [Google Scholar]
- 31.Smith LL, Johnson BH. Biological activities of oxysterols. Free Radic Biol Med. 1989;7:285–332. doi: 10.1016/0891-5849(89)90136-6. [DOI] [PubMed] [Google Scholar]
- 32.el-Bayoumy K, et al. Lack of tumorigenicity of cholesterol epoxides and estrone-3,4-quinone in the rat mammary gland. Cancer Res. 1996;56:1970–1973. [PubMed] [Google Scholar]
- 33.Cheng YW, Kang JJ, Shih YL, Lo YL, Wang CF. Cholesterol-3-beta, 5-alpha, 6-beta-triol induced genotoxicity through reactive oxygen species formation. Food Chem Toxicol. 2005;43:617–622. doi: 10.1016/j.fct.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Black HS. Analysis and physiologic significance of cholesterol epoxide in animal tissues. Lipids. 1980;15:705–709. doi: 10.1007/BF02534024. [DOI] [PubMed] [Google Scholar]
- 35.Watabe T, Sawahata T. Biotransformation of cholesterol to cholestane-3beta,5alpha,6beta-triol via cholesterol alpha-epoxide (5alpha,6alpha-epoxycholestan-3beta-ol) in bovine adrenal cortex. J Biol Chem. 1979;254:3854–3860. [PubMed] [Google Scholar]
- 36.Fuda H, Javitt NB, Mitamura K, Ikegawa S, Strott CA. Oxysterols are substrates for cholesterol sulfotransferase. J Lipid Res. 2007;48:1343–1352. doi: 10.1194/jlr.M700018-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Song C, Hiipakka RA, Liao S. Auto-oxidized cholesterol sulfates are antagonistic ligands of liver X receptors: Implications for the development and treatment of atherosclerosis. Steroids. 2001;66:473–479. doi: 10.1016/s0039-128x(00)00239-7. [DOI] [PubMed] [Google Scholar]
- 38.Argmann CA, et al. Regulation of macrophage cholesterol efflux through hydroxymethylglutaryl-CoA reductase inhibition: A role for RhoA in ABCA1-mediated cholesterol efflux. J Biol Chem. 2005;280:22212–22221. doi: 10.1074/jbc.M502761200. [DOI] [PubMed] [Google Scholar]
- 39.Meyer DJ, Ketterer B. 5 alpha,6 alpha-epoxy-cholestan-3 beta-ol (cholesterol alpha-oxide): A specific substrate for rat liver glutathione transferase B. FEBS Lett. 1982;150:499–502. doi: 10.1016/0014-5793(82)80798-9. [DOI] [PubMed] [Google Scholar]
- 40.Watabe T, Sawahata T, Horie J. Evidence for the formation of a steroid S-glutathione conjugate from an epoxysteroid precursor. Biochem Biophys Res Commun. 1979;87:469–475. doi: 10.1016/0006-291x(79)91819-9. [DOI] [PubMed] [Google Scholar]
- 41.Wrensch MR, et al. Breast fluid cholesterol and cholesterol beta-epoxide concentrations in women with benign breast disease. Cancer Res. 1989;49:2168–2174. [PubMed] [Google Scholar]
- 42.Küçük O, et al. Increased plasma level of cholesterol-5 beta,6 beta-epoxide in endometrial cancer patients. Cancer Epidemiol Biomarkers Prev. 1994;3:571–574. [PubMed] [Google Scholar]
- 43.de Medina P, et al. Tamoxifen is a potent inhibitor of cholesterol esterification and prevents the formation of foam cells. J Pharmacol Exp Ther. 2004;308:1165–1173. doi: 10.1124/jpet.103.060426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.