Abstract
Obesity and metabolic syndrome result from excess calorie intake and genetic predisposition and are mechanistically linked to type II diabetes and accelerated body aging; abnormal nutrient and insulin signaling participate in this pathologic process, yet the underlying molecular mechanisms are incompletely understood. Mice lacking the p66 kDa isoform of the Shc adaptor molecule live longer and are leaner than wild-type animals, suggesting that this molecule may have a role in metabolic derangement and premature senescence by overnutrition. We found that p66 deficiency exerts a modest but significant protective effect on fat accumulation and premature death in lepOb/Ob mice, an established genetic model of obesity and insulin resistance; strikingly, however, p66 inactivation improved glucose tolerance in these animals, without affecting (hyper)insulinaemia and independent of body weight. Protection from insulin resistance was cell autonomous, because isolated p66KO preadipocytes were relatively resistant to insulin desensitization by free fatty acids in vitro. Biochemical studies revealed that p66shc promotes the signal-inhibitory phosphorylation of the major insulin transducer IRS-1, by bridging IRS-1 and the mTOR effector p70S6 kinase, a molecule previously linked to obesity-induced insulin resistance. Importantly, IRS-1 was strongly up-regulated in the adipose tissue of p66KO lepOb/Ob mice, confirming that effects of p66 on tissue responsiveness to insulin are largely mediated by this molecule. Taken together, these findings identify p66shc as a major mediator of insulin resistance by excess nutrients, and by extension, as a potential molecular target against the spreading epidemic of obesity and type II diabetes.
Keywords: diabetes, longevity, mTOR/S6K, p66shc, nutrients
Obesity and metabolic syndrome represent ramping public health issues in the Western world, due to overnutrition and reduced physical activity, coupled with genetic susceptibility (1). Although a correct lifestyle remains the mainstream solution to this problem, pharmacological strategies are also being actively seeked; to this end, a better knowledge of the molecular players and biochemical mechanisms linking excess body fat to glucose intolerance and an increased cardiovascular risk is critically needed.
Genetic and diet-induced disturbances in insulin and nutrient signaling have been compellingly linked to diabetes, obesity, and accelerated aging (2–4). In particular, deregulated activity of the nutrient-sensitive kinase mTOR (mammalian target of rapamycin) and of its downstream effector S6 kinase (S6K) by high-fat diet or leptin deficiency promotes fat accumulation, induces insulin resistance, and shortens mouse lifespan (5, 6). On the other hand, calorie restriction and hypomorphic mutations or pharmacological blockade within the insulin and mTOR/S6 kinase signaling cascades increase longevity both in lower model organisms and in mammals (7–11), and adipose-specific deletion of the insulin receptor (IR) extends lifespan of FIRKO mice (12).
Mice lacking the 66-kDa isoform of the adaptor Shc (Src homology and collagen) are also long lived and resistant to a number of age-related pathologies mostly associated with tissue oxidative damage (13, 14). In a current model in fact, p66shc translocates to the mitochondria of stress-challenged cells where it catalyzes the formation of hydrogen peroxide (15, 16). However, only a marginal fraction of cellular p66shc is associated with the organelle, suggesting that additional mechanisms may contribute to the connection between loss of p66shc and organismal longevity and health.
Along parallel lines of investigation, p66shc has been shown to participate in insulin signaling and in particular to promote insulin-induced fat accumulation (17); thus, because excess adiposity limits animal lifespan and causes feedback desensitization of the insulin receptor cascade and insulin resistance (2, 12, 18), effects of p66shc on mouse lifespan and on age-related pathologies including vascular dysfunction (19) may be a consequence of the role of p66shc in nutrient-related signaling rather than of the redox properties of this molecule.
Prompted by this possibility we investigated the molecular interaction of p66shc with signaling cascades triggered by insulin and nutrients and evaluated the consequence of p66 deletion in an established mouse model of metabolic dysfunction by nutrient overload (leptin-deficient LepOb/Ob mice), with the goal of verifying whether p66shc may participate in obesity-induced insulin desensitization and, by extension, in animal susceptibility to metabolic syndrome and type 2 diabetes.
Results and Discussion
Mice homozygous for the LepOb mutation are hyperphagic due to complete deficiency of leptin. These mice, fed ad libitum, develop overt obesity, hyperinsulinemia, and glucose intolerance, thereby modeling human metabolic syndrome. To address the impact of p66shc deficiency on the dismetabolic phenotype of Ob/Ob mice, double mutant animals (Ob/Ob-p66KO and Ob/Ob-p66WT littermates) were generated by sequencial crossbreeding.
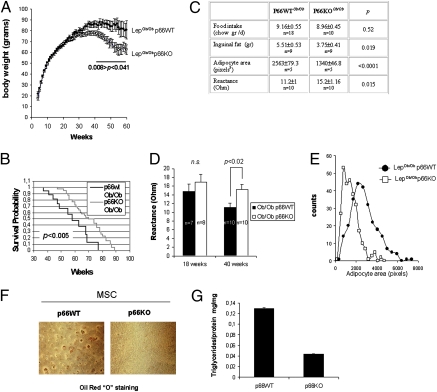
Ob/Ob p66KO mice and their p66WT controls (both in a B6/129 mixed background) were monitored over time and weight gain curves recorded. Although both strains developed severe obesity, the two growth curves diverged by week 34: the p66KO group displayed a lower plateau and the age-associated weight loss, expected in the Ob/Ob model (20), started earlier with respect to p66-proficient animals (Fig. 1A). These differences in total body weight between the two strains occurred independent of food consumption (Fig. 1C) and were most likely accounted for by a difference in fat mass, as confirmed by evaluation of inguinal fat weight and mean adipocyte size and by total body impedenziometric analysis (21) of representative animals showing decreased conductivity (and therefore a higher fat/water ratio) in older mice proficient for p66shc. (Fig. 1 C, D, and E). Of note, these findings, although obtained in a mixed B6/129 genetic background, are consistent with the previously reported (17) resistance to diet-induced obesity in pure sv129 p66-deficient mice.
Fig. 1.
Effect of p66shc deletion on fat accumulation and longevity of obese mice. (A) Attenuated pathologic obesity in LepOb/Ob mice lacking p66shc. Littermate male and female mice of the two genotypes (P66wt LepOb/Ob and p66KO LepOb/Ob) were caged individually, fed ad libitum, and monitored for weight gain over time (56 wk). Values are mean body weight ± SEM. Between 34- and 54-wk weight differences among the two strains were significant by at least P < 0.05 (two-tailed Student's t-test). (B) Kaplan-Meyer survival analysis of obese (p66WT, n = 20) and obese/p66-deficient (p66KO, n = 47) mice. Survival median was 58 wk for p66WT and 72 for p66-deficient animals. Extended survival of p66KO mice was significant (P < 0.0005) by the log-rank (Cox-Mantel) test. (C) Table displaying values for food intake and obesity-related parameters obtained from randomly chosen p66WT and KO mice in the 34- to 54-wk age range. Total body reactance (a correlate of the lean/fat mass ratio) was measured by bioelectrical impedance analysis (BIA). Data are mean ± SEM of n animals. P values, calculated by unpaired t-test, are indicated. (D) Histogram displaying total body conductivity values as a function of mouse age and genotype. Group sizes are indicated. P was calculated by unpaired t-test. (E) Distribution of adipocyte areas (values in pixels) in sections of inguinal white fat from p66WT and KO obese mice. A total of 50 cells per mouse (5 mice/group) were measured. (F) In vitro differentiation of primary preadipocytes (MSC) from p66WT and p66KO obese mice; lipid accumulation was revealed by Oil Red O staining. (G) Triglycerides were quantified and normalized for protein content.
In keeping with a cell-autonomous role for p66shc in fat accumulation/storage, primary preadipocytes (mesenchymal stem cells, MSC) purified from the fat of obese p66KO mice displayed much reduced triglyceride accumulation in response to an insulin + dexamethasone mix, as revealed by Oil Red O staining, compared with precursors derived from p66+/+ animals (Fig. 1 F and G).
A clear tendency to extended survival (median survival, 72 wk versus 58 wk; P < 0.005; Fig. 1B) was also observed in obese p66KO mice compared with p66WT controls, in agreement with the reported lifespan-extending effect of p66 deletion in mice, and with the general notion that fat accumulation negatively affects longevity in mammals and in model organisms (12, 22). Thus, taken together, these findings reveal p66shc as contributing to excess fat accumulation and accelerated aging observed in LepOb/Ob mice.
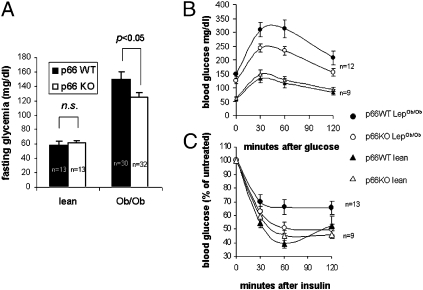
Obesity is mechanistically linked to hyperglycemia and glucose intolerance both in humans and laboratory animals. Because p66shc participates in insulin signaling (17), we reasoned that its deletion may affect insulin desensitization in obese animals. In 20-wk-old Ob/Ob mice lacking p66shc, fasting glycemia values were significantly lower than in p66shc+/+, weight-matched controls (Fig. 2A, two-way ANOVA, P < 0.05). Moreover, a glucose tolerance test (GTT) revealed glucose intolerance to be less severe, although not completely prevented, in obese, p66-deficient mice (Fig. 2B). Of note, at the indicated age, body weight and composition did not differ between the two strains (Fig. 1 A and D), and weight-matched animals were used for comparisons, which points to a direct signaling effect of p66shc in the observed differences in glucose homeostasis. Interestingly, p66 deficiency did not significantly affect fasting glycemia in lean mice, nor their response under GTT (Fig. 2 A and B).
Fig. 2.
Absence of p66 partially prevents glucose intolerance in obese p66 KO mice. (A) Fasting glycemia was recorded in 20-wk-old lean and obese mice, either proficient (WT, black columns) or deficient (KO, white columns) for p66shc. Obese mice of different p66 genotype were weight matched to avoid the confounding effect of fat mass on insulin sensitivity. N values for each group are indicated. Values are mean ± SEM. Significance was determined by 2 × 2 ANOVA followed by Tukey HSD post hoc test. (B) Glucose tolerance test (GTT) was performed on the same animals as in A, by injecting 1 g /kg glucose in peritoneum after overnight starvation. Glycemia was recorded at the indicated times. Values are mean ± SEM. Curves were compared by two-way ANOVA with repeated measures on one factor. (C) Insulin tolerance test (ITT) after 16 h starvation mice were injected with 2 U/kg body weight insulin in peritoneum. Glycemia was recorded at the indicated time points. Data are expressed as percentage of basal glycemia. Values are mean ± SEM of n mice.
In parallel, peripheral response to insulin, as assessed by an insulin tolerance test in the same groups as above, was markedly decreased in p66WT obese mice (P = 0.0018 versus the corresponding lean population), whereas largely preserved in p66-deficient obese mice of the same weight, with a glycemic curve largely overlapping that of the corresponding lean controls (P = 0.4, Fig. 2C). Plasma insulin levels were markedly increased, as expected, in obese animals, but that was irrespective of the p66 status (Fig. S1); hence, collectively, these findings suggest that p66shc deletion attenuates insulin resistance of Ob/Ob mice independent of body weight and likely at the level or downstream of the insulin receptor.
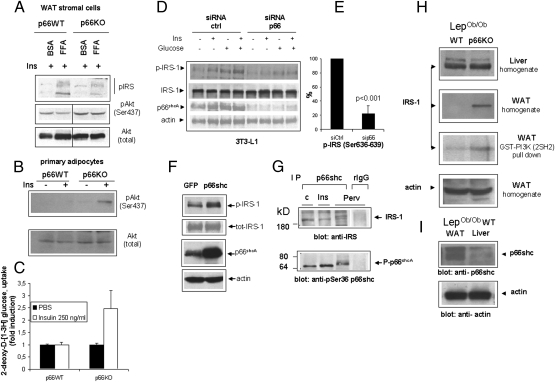
Isolated insulin-responsive cells exposed in vitro to excess free fatty acids (FFAs) or inflammatory cytokines recapitulate biochemical correlates of insulin resistance in vivo, including inhibitory phosphorylation of the insulin receptor substrate-1 (IRS-1) on serines 307 and 636–639, reduced recruitment of the PI3-kinase, and impaired downstream signaling through the Akt/PKB kinase (18). Serine phosphorylation of IRS-1 eventually leads to protein degradation (23), accounting in part for decreased IRS content in fat cells from type II diabetic patients (24). To further investigate p66shc effects on nutrient-elicited insulin desensitization, vascular stromal cells (which include vascular cells, macrophages, preadipocytes, and stem cells) were isolated from adipose tissue of p66+/+ and p66KO mice and exposed in vitro to FFA and high (25 mM) glucose for 16 h, before stimulation with insulin. In these conditions, insulin-induced phosphorylation of the Akt/PKB kinase on serine 473 was blunted in p66WT cells, but preserved in p66-deficient cells (Fig. 3 A and B), in parallel with a decreased inhibitory phosphorylation of IRS-1 (serine 307 + serine 636–639) (Fig. 3A). A similar difference in response to insulin, based on phosphorylation of Akt (Fig. 3B) and uptake of 2-deoxy-D-[1-3H]glucose (Fig. 3C), was also observed in mature adipocytes freshly isolated from p66WT and p66KO adipose tissue and immediately stimulated in vitro. Furthermore, siRNA-mediated knockdown of p66shc in 3T3L1 preadipocytes also significantly reduced IRS-1 serine phosphorylation in response to high glucose (4.5 g/L for 30 minutes) and glucose + insulin, two conditions mimicking nutrient replenishment (Fig. 3 D and E) without effects on total IRS expression level. Conversely, overexpression of p66shc led to a clear increase in IRS-1 phosphorylation at the above serine residues (Fig. 3F). Importantly, p66shc and IRS-1 could be coimmunoprecipitated, albeit at low stoichiometry, in this cell line, confirming that the two proteins can form a complex in vivo (25) (Fig. 3G and Fig. S2). This interaction is not significantly modulated by cell exposure to insulin or the potent insulinomimetic, pervanadate, although both stimuli induce significant Ser-36 phosphorylation of p66shc (Fig. 3G Lower).
Fig. 3.
p66shc regulates IRS-1 and insulin desensitization in vitro and in vivo. (A) Phospho-specific immunoblot analysis of protein lysates of undifferentiated preadipocytes (passages 1–3) from p66WT and p66KO mice exposed overnight to excess free fatty acids and restimulated with insulin (100 ng/mL). Phosphorylation of Akt on Ser-473 and of IRS-1 on serines 636–639 and 307 (pool of two distinct antisera) were investigated. Total Akt was used as a protein loading control (Lower). (B) Immunoblot analysis of Akt/PKB phosphorylation in insulin-stimulated adipocytes from the inguinal fat of p66WT and KO mice. The same filter was rehybridized with an anti-total Akt antiserum to confirm protein loading. Panel representative of two independent experiments. (C) Glucose uptake assay on the same cell populations as in B. Primary adipocytes isolated from mice after overnight fast were stimulated with 250 ng/mL (50 nM) insulin for 30 min and uptake of 2-deoxy-D-[1-3H] glucose was determined by liquid scintillation counting. Values are mean ± SD of triplicate samples. Picture representative of two independent experiments. (D) siRNA-mediated knockdown of p66shc in 3T3L1 preadipocytes reduces IRS-1 serine phosphorylation by glucose and insulin. Immunoblot analysis of total lysates reveals reduced phosphorylation but not expression of IRS-1, a significant decrease of p66shc content and equal protein loading (actin band) throughout the gel. (E) Densitometric quantitation of p-IRS (Ser-307 + Ser-636–639) bands in Ins + glucose samples from three independent experiments; values are expressed as percentage of mock-silenced cells (siCtrl). (F) Overexpression of p66 in 3T3L1 increases serine phosphorylation of IRS-1. IRS phosphorylation (Ser-307 + Serine 636–639) and expression of p66shc and actin (loading control) were assessed by Western blotting with specific antisera. Representative of several experiments. (G) Immunoblot analyses revealing the presence of IRS-1 in anti p66shc immunoprecipitates. Phosphorylation of p66 on serine 36 in response to treatments [Ins, insulin 100 ng/mL; Per, pervanadate (sodium orthovanadate + H2O2, 1 mM)] is also displayed. All images are representative of at least two independent experiments. (H) Immunoblot analysis of total IRS-1 content in liver and visceral white fat homogenates, and in a PI3K–GST pull-down from visceral fat. Anti actin immunoblotting (Bottom) indicates equal protein content in the two WAT homogenates. (I) Immunoblot analysis comparing p66shc expression in the liver and white fat of p66WT mice. Actin was used as an internal protein loading control.
Unlike in the short-term studies on cultured adipose cells, IRS-1 levels were reduced (down to 40–50%) in white fat, but not in the liver of p66WT obese mice compared with p66KO littermates (Fig. 3H). This difference was mirrored by a reduced recovery of IRS-1 in a pull-down assay using a recombinant GST–PI3K fusion protein as a bait for the tyrosine-phosphorylated form of the protein. (Fig. 3H third panel).
Thus, this evidence collectively indicates that p66 participates in insulin desensitization of fat cells by excess nutrients and points to IRS-1 serine phosphorylation as the potential mechanism of this action. Phosphorylation eventually leads to IRS degradation in vivo, although this effect is not appreciable in short-term in vitro studies. Importantly, these effects seem to occur selectively in adipose tissue.
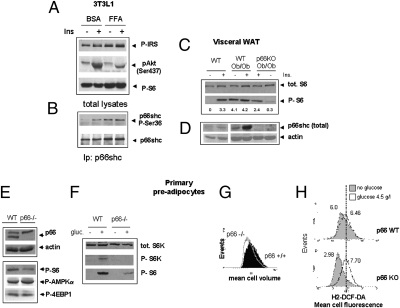
Among the protein kinases involved in the signal-inhibitory phosphorylation of IRS-1, the mTOR effector S6 kinase has attracted the most attention because S6K-deficient mice are resistant to genetic and diet-induced obesity and hypersensitive to insulin, although insulin deficient (26). Moreover, the mTOR inhibitor rapamycin prevents IRS phosphorylation on serine and degradation by prolonged exposure to insulin (23) and prevents adipocytic differentiation (27). We hypothesized that p66shc effects on obesity and insulin resistance may be mediated, at least in part, by the S6 kinase. In 3T3L1 cells exposed to excess fatty acids, S6 phosphorylation (Ser-285–286), an indirect correlate of S6 kinase activity, and phosphorylation of p66 on serine 36, an event reportedly crucial for most biological action of the protein (16), occurred concomitantly and in parallel with the establishment of insulin desensitization (Fig. 4 A and B). More importantly, constitutive activation of the S6 kinase-S6 cascade in the visceral white fat of p66WT obese mice (6) was nearly abolished in weight-matched obese animals lacking p66shc (Fig. 4 C and D). S6 phosphorylation, however, was responsive to insulin in this mouse strain, unlike in p66-proficient obese animals (Fig. 4C lanes 2, 4, and 5).
Fig. 4.
Involvement of p66shc in nutrient signaling through mTOR/S6K. (A and B) Immunoblot analysis of signaling events relevant to insulin responses in 3T3L1 cells exposed to excess FFA (oleic + linoleic + palmitic acid) for 16 h. Phosphorylation of Akt, p66shc, S6 (serine 235–236), and IRS-1 (Ser-636–639 and 307) were monitored. Image representative of at least two independent experiments. (C and D) Immunodetection of the S6 kinase substrate S6 (phospho-Ser and total), of p66shc and of actin as a loading control in WAT homogenates from mice of the indicated genotypes. Samples are from single animals. Mice in lanes 2, 4, and 5 were stimulated with 10 U/kg insulin. The blot is representative of two/three independent experiments. Numbers indicate band intensities (OD values, background subtracted) for phospho-S6. (E) Immunoblot analysis showing expression of p66shc in freshly isolated (1 g/L glucose, 20% FCS) adipocyte precursor cells. The p66shc band, missing in p66KO-derived cells, is indicated. Reduced phosphorylation of S6, but unchanged phosphorylation of 4EBP1 and AMPK α in p66KO cells is also shown. (F) Reactivation of the S6 kinase–S6 pathway in nutrient-starved preadipocytes in response to glucose (60 min). (G) Flow cytometry analysis of cell volume in exponentially growing P66-deficient and -proficient cells (passage 1–3). (H) Generation of reactive oxygen species in glucose-refed adipocyte precursors of different p66 genotype. Serum and glucose-starved cells were refed with 2 g/L glucose for 2 h (unshaded curve) or left untreated (shaded curve). Fluorescence of cells loaded with the redox sensitive dye DCF-DA was analyzed by flow cytometry. Numbers are mean cell fluorescence intensities. The vertical line indicates overlapping peak positions for stimulated cell populations. Data representative of two independent experiments.
Thus, nutrient signaling derangement associated with obesity is attenuated in the absence of p66shc in vivo. Additionally, in freshly isolate adipocyte precursor cells (1 g/L glucose, 20% FCS) from p66KO obese mice, the basal phosphorylation of S6 on serines 285–286, but not that of another major mTOR substrate, 4EBP1, and of the upstream mTOR regulator AMPK, was consistently lower than in p66shc-proficient cells (Fig. 4E). Furthermore, when these mesenchymal precursors were glucose starved for 5 h and subsequently restimulated with 25 mM glucose, the presence of p66shc allowed a stronger phosphorylation of the S6 kinase and its substrate S6 (Fig. 4F). Finally, glucose-induced increase in cell size, another readout of the mTOR–S6K cascade, was blunted in p66KO cells (Fig. 4G). Of note, p66KO cells are permeant to glucose, because they displayed a prompt metabolic response to refeeding as indicated by a raise in the generation of reactive oxygen species (Fig. 4H). In a complementary set of experiments, overexpression of p66 in 3T3L1 cells by retroviral transduction led to a marked and selective increase of S6 kinase response to insulin (Fig. S3B) and IGF1 (Fig. S3C) with no or marginal effects on activation of ERK 1/2 (Fig. S3 B and C). In this cell line, P66 robust overexpression (over twofold, Fig. S3A) did not elicit significant oxidative stress in mitochondria, as determined by the oxidation profile of a mitochondrially targeted redox-sensitive fluorescent protein (rxYFP) (Fig. S4A); however, microspectrofluorometric analysis of cellular autofluorescence (Fig. S4D) indicated that p66 slightly but significantly reduced cellular accumulation of NAD(P)H in cells exposed to glucose, in keeping with a role for this protein in accelerating NAD(P)H oxidation in mitochondria (28). Moreover, overexpression of p66shc and p66shcqq, a mutant p66shc reportedly devoid of prooxidant activity (15), activated S6K to a comparable extent in serum-stimulated 3T3L1 cells, while having no effect on the redox-sensitive stress kinase p38 MAPK (Fig. S4B). Finally, S6K activation by p66shc was not inhibited by cell treatment with high elevated concentrations of oxidant scavengers N-acetyl cysteine and catalase (Fig. S4C).
Thus, although p66shc elicits the formation of oxygen species and increases cell oxidative stress in several pathophysiological settings, data displayed in Fig. 4H and Fig. S4 suggest that redox properties of p66shc unlikely account for the observed stimulatory effects on the mTOR–S6 kinase cascade in response to nutrients and insulin. Alternatively, p66shc may operate as an adaptor molecule by participating in the assembly of supramolecular complexes relevant to the propagation of insulin signaling.
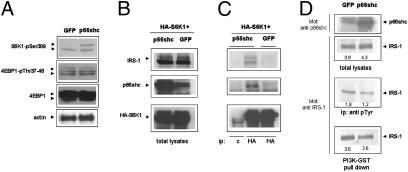
Prompted by evidence of physical association between p66shc and IRS-1 in 3T3 L1 preadipocytes (Fig. 3G), we investigated the possibility that p66 may promote the interaction of IRS-1 with S6 kinase. To facilitate biochemical analysis of protein interactions, p66shc and an HA-tagged form of the human S6 kinase 1 were overexpressed in HEK293T cells, a cell line responsive to insulin and largely used for the investigation of metabolic signaling cascades (29). Importantly, also in this cell line overexpression of p66 is accompanied by robust phosphorylation of S6K on Thr-389, whereas another major mTOR substrate, 4EBP1, appears to be unaffected (Fig. 5A). We readily detected both IRS-1 and p66shc in anti-HA-S6K immunocomplexes precipitated from insulin-stimulated 293-T cells (Fig. 5C). Of note, the amount of S6K-bound IRS was greatly increased by the concomitant expression of p66shc (Fig. 5C Upper).Transient overexpression of the adaptor, however, did not impact on the total input IRS-1 (Fig. 5B), indicating that the efficiency of the IRS–S6K interaction, rather than IRS stability or expression, were being modified by p66shc in these experimental conditions. In parallel with in vivo findings displayed in Fig. 3H, IRS-1 tyrosine phosphorylation and IRS recovery in a GST–PI3K pulldown assay were reduced by p66shc in this cell line (Fig. 5D), confirming that S6K recruitment to IRS-1 negatively impacts on downstream signal propagation through the PI3 kinase cascade. Of note, interaction between endogenous p66shc, S6K, and IRS-1, expressed at physiological levels, could also be demonstrated in 3T3L1 cells (Fig. S2).
Fig. 5.
Analysis of the p66-IRS-S6K interaction in HEK-293T cells. (A) Phosphorylation of endogenus S6K, but not of the other major mTOR substrate 4EBP1, in 293-T cells overexpressing p66shc. (B Left). Immunoblot analysis of total protein lysates confirming equal levels of IRS-1 and S6K1 and overexpression of p66shc in cells cotransfected with an HA-tagged form of the S6K1 kinase together with p66shc or GFP as a transfection control. (C) Coimmunoprecipitation assay; S6K was immunoprecipitated with an anti HA antibody, and the presence of IRS1 (Top) and p66shc (Middle) in the immunocomplexes was verified by specific immunoblotting. Interactions displayed are specific as confirmed by the complete absence of bands in the mock immunoprecipitation (lane marked c). (D) p66 overexpression has no effect on the IRS-1 cellular level in these conditions, but reduces IRS-1 precipitation with an antiserum specific for phosphotyrosine, and its interaction with a GST–PI3K fusion protein, consistent with reduced IRS-1-PI3K signaling. Numbers are band intensities (volumes) in arbitrary units.
Conclusion
Taken together, the above evidence provides a mechanism for two major observed effects of p66 (i) increased inhibitory phosphorylation of IRS-1 and (ii) enhanced activity of S6K in the context of nutrient replenishment. These effects conceivably account, at least in part, for improved glucose tolerance and insulin action in the context of obesity and for reduced adipose cell differentiation and triglyceride accumulation by excess nutrients in p66-deficient mice (model in Fig. S5).
In our opinion, our findings are not in conflict nor overlap with data generated by Berniakovitch and colleagues, indicating a ROS-dependent positive role of p66shc in normal insulin signaling (17). In fact, those authors have not investigated the role of p66shc in the context of insulin resistance, and it is conceivable that p66shc promotes insulin responses in physiological contexts by its redox activity, while favoring, as an adaptor, molecular interactions relevant to IR desensitization under nutrient and insulin overload. Interestingly, the same authors reported, in keeping with our results in obese animals, that p66KO mice fed a high-fat diet have better glucose control in spite of lower plasma insulin levels, compared with WT mice (17).
Molecular aspects of the signaling complex involving IRS-1, S6 kinase, and p66shc certainly need to be further elucidated; in particular, it is not clear whether these interactions are direct or involve other yet unidentified molecular partners and whether they also operate in physiological nutrient signaling or exclusively in the context of obesity-associated hyperinsulinemia and dysmetabolism.
Notwithstanding these limitations, we propose that the above outlined circuitry represents a critical component of the nutrient signaling derangement underlying metabolic syndrome and insulin resistance, with crucial implications for the understanding of the intricate connection between diabetes and aging.
Materials and Methods
Mice.
The c129/sv mice harboring a p66shc homozygous null mutation, previously described (13), were kindly provided by P. G. Pelicci and M. Giorgio (European Institute of Oncology, Milan, Italy). C57BL/6 LepOb/Ob mice were obtained from the Jackson Laboratories through Charles River Laboratories (Calco, Italy). P66KO/LepOb/Ob double-mutant mice were derived by initially crossing p66KO males with LepOb/+ females. [(p66−/+; LepOb/+)] offspring were further breeded to obtain the desired genotypes. Only littermates were compared in the experimental groups, to minimize potential confounding effects deriving from the mixed (c129 × C57Bl6) genetic background of the animals.
Mouse genotyping was performed by allele-specific PCR using the following primer sets, as previously described (13, 30):
p66shc WT Fw 5′-CCT CCC CAG GTC ATC TGT TAT CAC-3′;
p66shc MUT Fw 5′-GGG TGG AGA GGC TTT TTG CTT C-3′;
p66shc Rv 5′-CTC GTG TGG GCT TAT TGA CAA AG-3′;
Lep WT Fw 5′-TGA CCT GGA GAA TCT CC-3′;
Lep MUT Fw 5′-TGA CCT GGA GAA TCT CT-3′; and
Lep Rv 5′-CAT CCA GGC TCT CTG GC-3′.
Metabolic studies.
All studies involving animals were previously approved by the institutional ethical committee and the Italian Ministry of Health. P66 (WT)-LepOb/Ob and p66KO-LepOb/Ob mice were fed ad libitum a standard chow and monitored weekly for weight gain and food consumption, starting from weaning. Survival curves were calculated by the Kaplan-Meyer method, using WINSTAT software (Microsoft).
Glucose tolerance test.
Groups of 4- to 5-male 15- to 20-wk-old mice were starved for 16 h (overnight) and basal values of blood glucose measured in a drop of blood from tail vein, using a microglucometer (Accu-check, Aviva, Roche Diagnostics). Glucose was given intraperitoneally at 1 g/kg body weight and glycemia again measured after 30, 60, and 120 min. Data from two or three independent experiments were pooled for analysis.
Insulin tolerance test.
Groups of mice as above were starved for 16 h (overnight) and basal glycemia measured. Insulin (Humulin, Eli-Lilly, 100 U/mL) was given intraperitoneally at 2 U/kg body weight, and blood samples collected from the tail vein at 30, 60, and 120 min for blood glucose determination. A few mice displaying seizures by hypoglycemia were immediately given glucose and excluded from the study. Data were collected and expressed as percentage of basal (i.e., unstimulated) glycemia to minimize mouse-to-mouse variability. Data from independent experiments conducted in identical conditions were pooled and analyzed together.
Plasma insulin concentration was determined by an Ultrasensitive (Mouse) Elisa kit (Alpco immunoassays). Blood samples were collected from the tail vein with an heparizined micropipette tip. After centrifugation, 5 μL of plasma were used for the assay.
For determination of body composition, lightly anesthesized obese mice were subjected to single frequency bioelectrical impedance analysis (BIA) (21) (SI Materials and Methods).
For biochemical analysis of tissue response to insulin, mice were given 10 U/kg insulin i.p. and killed 10 min after insulin injection. Tissues (liver, muscle, and visceral fat pads) were quickly removed and immediately frozen in liquid nitrogen.
Isolation of primary adipocytes and of WAT stromal cells.
Mature adipocytes and their precursors were isolated from visceral fat pads of obese mice by standard procedure. Briefly, pads were rinsed in cold PBS and blood vessels carefully removed. Fat tissue was minced with scissors and resuspended in complete DMEM medium containing 2.5 mg/mL Collagenase IV (Sigma-Aldrich, cat. no. C-5138). After 60 min incubation at 37 °C with occasional agitation, tissue suspensions were passed through nylon filters (100 μm diameter, BD Biosciences) and centrifuged at 500 × g for 5 min. Floating mature adipocytes were rinsed twice in PBS and kept for further processing. Stromal cell pellets containing adipocyte precursors were resuspended for 2 min in hypotonic buffer to lyse red blood cells, washed once with PBS, and plated in complete DMEM containing 20% vol/vol FCS.
Confocal analysis of cell oxidation and intracellular NAD(P)H.
3T3L1 cells were seeded in 35-mm glass bottom dishes (Ibidi, Integrated Biodiagnostic) and transduced with a retroviral construct encoding a mitochondrially targeted variant of the redox-sensitive Yellow Fluorescent Protein (mt-rxYFP). After 48 h, cells were serum starved for 2 h and fluorescent cells analyzed with a DM-IRE2 (Leica) confocal microscope as described in detail elsewhere (31). Average R values were calculated over several regions of interest (ROIs) each comprising single cells or small cell clusters.
Intracellular NAD(P)H was measured by two-photon confocal analysis of cell green autofluorescence upon excitation at 366 nm, as described by Patterson et al. (32).
Statistics.
Experimental groups of animals were compared by Student's t-test, two-way ANOVA, or two-way ANOVA with repeated measures for one factor, where appropriate, using a web-based computation tool (http://faculty.vassar.edu/lowry/VassarStats.html) or Microsoft Excel software. Survival curves of p66WT and p66KO obese mice were compared by the Kaplan-Meyer method and differences evaluated by the log-rank (Cox-Mantel) test.
Differences were considered significant for a probability of the null hypothesis (P < 0.05).
Supplementary Material
Acknowledgments
We thank Prof. P. G. Pelicci and Dr. Marco Giorgio (European Institute of Oncology, Milan, Italy) for providing the p66shc KO mouse strain and the pBabe-p66shc expression construct, Drs. Fabio Martelli and Valeria Di Stefano (Molecular Cardiology Laboratory, Istituto di Ricovero e Cura a Carattere Scientifico-Policlinico San Donato, Milan, Italy) for the kind gift of the p66shcqq mutant, Drs. G. Nolan (University of Stanford, Palo Alto, CA), and Katherine Siminovitch (Mount Sinai Hospital, Toronto) for reagents, members of the laboratory for suggestions and experimental contributions, Egidio Stigliano for valuable technical support, and Drs. Renata Colavitti and Barbara Bedogni (Stanford University, Palo Alto, CA) for critically reading the manuscript. This work was supported by the European Association for the Study of Diabetes, European Foundation for the Study of Diabetes/Glaxo Smith Kline Programme for the Study of Metabolic Toxicity in Diabetes (G.P.) and the Italian Ministry of University and Research and Catholic University (MIUR ex 60% linea D1 to G.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008647107/-/DCSupplemental.
References
- 1.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 2.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozcan U, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 4.Picard F, Guarente L. Molecular links between aging and adipose tissue. Int J Obes (Lond) 2005;29(Suppl 1):S36–S39. doi: 10.1038/sj.ijo.0802912. [DOI] [PubMed] [Google Scholar]
- 5.Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 7.Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- 8.Vellai T, et al. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 9.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapahi P, Zid B. TOR pathway: Linking nutrient sensing to life span. Sci Aging Knowledge Environ. 2004;2004:PE34. doi: 10.1126/sageke.2004.36.pe34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blüher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 13.Migliaccio E, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 14.Trinei M, et al. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 15.Giorgio M, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Pinton P, et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315:659–663. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- 17.Berniakovich I, et al. p66Shc-generated oxidative signal promotes fat accumulation. J Biol Chem. 2008;283:34283–34293. doi: 10.1074/jbc.M804362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87:99–109. doi: 10.1016/j.biochi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Napoli C, et al. Deletion of the p66Shc longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc Natl Acad Sci USA. 2003;100:2112–2116. doi: 10.1073/pnas.0336359100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westman S. Development of the obese-hyperglycaemic syndrome in mice. Diabetologia. 1968;4:141–149. doi: 10.1007/BF01219435. [DOI] [PubMed] [Google Scholar]
- 21.Rutter K, Hennoste L, Ward LC, Cornish BH, Thomas BJ. Bioelectrical impedance analysis for the estimation of body composition in rats. Lab Anim. 1998;32:65–71. doi: 10.1258/002367798780559356. [DOI] [PubMed] [Google Scholar]
- 22.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 23.Haruta T, et al. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol Endocrinol. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- 24.Smith U. Impaired (‘diabetic’) insulin signaling and action occur in fat cells long before glucose intolerance—is insulin resistance initiated in the adipose tissue? Int J Obes Relat Metab Disord. 2002;26:897–904. doi: 10.1038/sj.ijo.0802028. [DOI] [PubMed] [Google Scholar]
- 25.Giorgetti S, Pelicci PG, Pelicci G, Van Obberghen E. Involvement of Src-homology/collagen (SHC) proteins in signaling through the insulin receptor and the insulin-like-growth-factor-I-receptor. Eur J Biochem. 1994;223:195–202. doi: 10.1111/j.1432-1033.1994.tb18983.x. [DOI] [PubMed] [Google Scholar]
- 26.Pende M, et al. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature. 2000;408:994–997. doi: 10.1038/35050135. [DOI] [PubMed] [Google Scholar]
- 27.Yeh WC, Bierer BE, McKnight SL. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci USA. 1995;92:11086–11090. doi: 10.1073/pnas.92.24.11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemoto S, et al. The mammalian longevity-associated gene product p66shc regulates mitochondrial metabolism. J Biol Chem. 2006;281:10555–10560. doi: 10.1074/jbc.M511626200. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 30.Namae M, et al. New method for genotyping the mouse Lep(ob) mutation, using a polymerase chain reaction assay. Lab Anim Sci. 1998;48:103–104. [PubMed] [Google Scholar]
- 31.Maulucci G, et al. High-resolution imaging of redox signaling in live cells through an oxidation-sensitive yellow fluorescent protein. Sci Signal. 2008;1:pl3. doi: 10.1126/scisignal.143pl3. [DOI] [PubMed] [Google Scholar]
- 32.Patterson GH, Knobel SM, Arkhammar P, Thastrup O, Piston DW. Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc Natl Acad Sci USA. 2000;97:5203–5207. doi: 10.1073/pnas.090098797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.