Abstract
The zinc finger transcription factor Miz1 is a negative regulator of TNFα-induced JNK activation and cell death through inhibition of TRAF2 K63-polyubiquitination in a transcription-independent manner. Upon TNFα stimulation, Miz1 undergoes K48-linked polyubiquitination and proteasomal degradation, thereby relieving its inhibition. However, the underling regulatory mechanism is not known. Here, we report that HECT-domain-containing Mule is the E3 ligase that catalyzes TNFα-induced Miz1 polyubiquitination. Mule is a Miz1-associated protein and catalyzes its K48-linked polyubiquitination. TNFα-induced polyubiquitination and degradation of Miz1 were inhibited by silencing of Mule and were promoted by ectopic expression of Mule. The interaction between Mule and Miz1 was promoted by TNFα independently of the pox virus and zinc finger domain of Miz1. Silencing of Mule stabilized Miz1, thereby suppressing TNFα-induced JNK activation and cell death. Thus, our study reveals a molecular mechanism by which Mule regulates TNFα-induced JNK activation and apoptosis by catalyzing the polyubiquitination of Miz1.
Keywords: Mule/ARF-BP1, TNFα signaling, Miz1 ubiquitination
The transcription factor Miz1 plays a critical role in regulation of cell cycle arrest, proliferation, differentiation, and apoptosis by activating or repressing transcription (1, 2). Miz1 binds to the core promoters of several genes, including p15Ink4b (3, 4), p21Cip1 (5–8), Mad4 (9), and Bcl-2 (10), and stimulates expression of these genes. However, Miz1 can also mediate transcriptional repression by interacting with other transcription factors, including Myc and Zbtb4. For instance, Miz1-mediated transcription of the p15Ink4b and p21Cip1 genes is repressed when Miz1 complexes with Myc, which displaces p300 from Miz1 (3, 8). Miz1 directly suppresses p21Cip1 transcription by binding to the POZ domain transcription factor Zbtb4, which recruits histone deacetylases (11). Miz1 contains multiple functional domains including a pox virus and zinc finger/bric a brac tramtrack broad complex (POZ/BTB) domain at its N terminus and 13 zinc fingers (1, 2). The POZ/BTB domain is essential for Miz1 to exert its transcriptional activation and repression functions (1, 5, 12). The activity of Miz1 is regulated by phosphorylation (13) and interactions with other cellular proteins, including Bcl-6 (10), the topoisomerase II binding protein TopBP1 (14), and Myc (3–9). Although Miz1 performs these transcriptional functions in the nucleus, Miz1 is also present in the cytoplasm, where its functions are incompletely understood (1).
Recent studies show that cytoplasmic Miz1 may function as a signal- and pathway-specific modulator or regulator (SMOR) to regulate the inflammatory cytokine tumor necrosis factor alpha (TNFα) signaling pathway (15). TNFα regulates a wide range of biological activities including inflammation, immune responses, apoptosis, and tumorigenesis through two cytoplasmic membrane receptors, TNF-R1 and TNF-R2 (16, 17). Engagement of TNF-R1 by TNFα induces the formation and recruitment of the receptor Complex 1 to TNF-R1. Complex 1 is composed of TNF-R1–associated death domain protein (TRADD), TNF-receptor–associated factor (TRAF) 2 and 5, receptor interacting protein 1 (RIP1), and two inhibitors of apoptosis cIAP1/cIAP2 (18–21). TRAF2/5 and RIP1 are involved in activation of multiple downstream effectors including JNK, also known as stress-activated protein kinase (SAPK), as well as p38 and the inhibitor of NF-κB kinase (IKK) complex (22–27). Prolonged JNK activation contributes to TNFα-induced apoptosis when NF-κB activation is impaired (28–32). Recently, we found that Miz1 selectively suppresses TNFα-induced JNK activation by interfering with TRAF2 K63-linked polyubiquitination (15). The suppression by Miz1 is signal and pathway specific, as it does not suppress JNK activation by other stimuli like UV, IL-1, and TPA, or TNFα-induced activation of other MAPKs and IKK (15). Interestingly, Miz1 undergoes proteosomal degradation upon TNFα stimulation, thereby relieving this inhibition. How Miz1 proteasomal degradation is regulated by TNFα is not known.
The HECT domain-containing ubiquitin ligase (E3) Mule (also known as Ureb1, LASU1, HUWE1, ARF-BP1, or HectH9) plays a critical role in proteasomal degradation of several proteins (33–37), and is involved in regulation of apoptosis (33). Mule has two Armadillo (ARM) repeat-like domains (ARLD1 spanning amino acids 104–374, and ARLD2 spanning amino acids 424–815), a ubiquitin-associated (UBA) domain (amino acids 1317–1355), a WWE (amino acids 1617–1678), and a well-conserved BH3 domain (amino acids 1972–1994) at its N terminus, and a HECT domain (amino acids 4016–4374) is at its C terminus (33, 38–40). Mule ubiquitinates Histone H2A (34), p53 (35), N-Myc (36), Mcl-1 (33) and Cdc6 (37) through K48-mediated linkage. Here, we report that Mule is the E3 ligase that interacts with and catalyzes K48-linked polyubiquitination of Miz1 in response to TNFα, and is required for TNFα-induced activation of JNK and cell death.
Results
Mule Is Required for TNFα-Induced Miz1 Degradation.
Proteasomal degradation of proteins, in most cases, is the consequence of their K48-linked polyubiquitination (41, 42). We hypothesized that TNFα stimulates the interaction of Miz1 with an E3 ligase that catalyzes the polyubiquitination and degradation of Miz1. To test this hypothesis, HEK293 cells transfected with Xpress-tagged Miz1 were treated with or without TNFα in the presence of the proteasome inhibitor MG-132. The Xpress-Miz1 complex was immunoprecipitated using anti-Xpress antibody, separated by SDS/PAGE, and visualized by colloidal Coomassie blue staining. MS/MS tandem mass spectrometry analysis of the proteins coimmunoprecipitated with Xpress-Miz1 revealed that Mule, a HECT-domain–containing E3 ligase, associated with Miz1 (Fig. 1A).
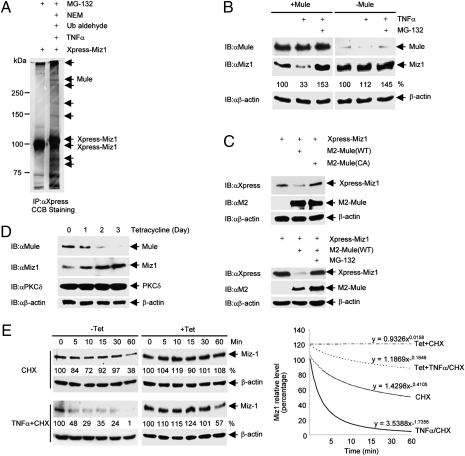
Fig. 1.
Mule regulates TNFα-induced Miz1 degradation. (A) HEK293 cells (1 × 107) were transfected with expression vector encoding Xpress-Miz1 (50 μg). After 36 h, cells were pretreated with MG-132 for 2 h before stimulation with TNFα (5ng/mL) for 10 min. Cells extracts were harvested in Co-IP buffer supplemented with NEM and ubiquitin aldehyde. Miz1 was immunoprecipitated by anti-Xpress antibody, and the immune complexes were resolved on a 7.5% gel and visualized by CCB staining. Bands for mass spectrometry analysis to obtain tryptic peptides for the E3 ligase Mule were indicated by arrows. (B) WT MEFs transfected with control siRNA or siRNA for Mule (100 nM each) were treated with TNFα (5 ng/mL, 15 min) in the absence or presence of MG-132. Expression levels of Mule, Miz1 and β-actin were determined by immunoblotting. Miz1 protein levels were quantitated and normalized to β-actin levels, and the untreated controls were calculated as 100%. Similar results were obtained from at least three independent experiments. (C) HEK293 cells (1 × 105) were transfected with Xpress-Miz1, along with M2-Mule (WT or the C4341A mutant) or empty vector (2 μg each), in the absence (Upper) or presence (Lower) of MG-132. Expression levels of Xpress-Miz1 and M2-Mule were determined. (D) U2OS stable cells harboring tet-inducible shMule (U2OS/shMule) were treated with tetracycline (2 μg/mL) for the indicated times. Expression levels of Mule, Miz1, PKCδ, and β-actin were determined. (E) U2OS/shMule stable cells were treated with or without tetracycline for 3 d followed by the addition of CHX (3 μg/mL) with or without TNFα (10 ng/mL) for the indicated times. Expression levels of Miz1 and β-actin were determined (Left). Miz1 protein levels were quantitated and normalized to β-actin levels, as shown in power trendlines (Right). Slope constants were indicated. Similar results were obtained from at least three independent experiments.
To test whether Mule is involved in TNFα-induced Miz1 degradation, expression of Mule was silenced with specific siRNA (Fig. 1B). Immunoblotting analysis with anti-Miz1 antibody revealed that TNFα induced degradation of Miz1, which was blocked by MG-132, consistent with our previous finding (15). Interestingly, knockdown of Mule inhibited TNFα-induced Miz1 degradation (Fig. 1B). These results suggest that Mule targeted TNFα-induced degradation of Miz1 in a proteasome-dependent manner. Conversely, ectopic expression of WT Mule, but not its E3 ligase defective (C4341A) mutant (33), induced degradation of cotransfected Miz1. This degradation was also blocked by MG-132 (Fig. 1C). These results demonstrate that the E3 ligase activity of Mule is required for Miz1 degradation. However, the treatment with TNFα did not completely eliminate Miz1, consistent with our previous finding (15). It is possible that partial degradation of Miz1 may provide a fine control on the strength of JNK1 activation in response to TNFα.
In U2OS stable cells harboring tetracycline-inducible shMule (33), tetracycline treatment decreased expression of Mule in a time-dependent manner. After treatment with tetracycline for 3 d, Mule expression was almost undetectable (Fig. 1D). Under these conditions, the expression level of Miz1 increased, whereas PKCδ and β-actin expression levels were constant (Fig. 1D). These results demonstrate that Mule also regulates basal Miz1 degradation.
To determine the rate of Mule-mediated Miz1 degradation in response to TNFα, U2OS stable cells harboring tetracycline-inducible shMule were treated with or without tetracycline for 3 d, followed by treatment with the protein synthesis inhibitor cycloheximide (CHX), or TNFα plus CHX. Analysis of the slope constants of the power trendlines representing Miz1 protein levels revealed that TNFα significantly increased the rate of Mule-mediated degradation of Miz1 by 3.8-fold ([SlopeTNFα/CHX – SlopeTet+TNFα/CHX]/[SlopeCHX – SlopeTet+CHX]) (Fig. 1E). Taken together, these data demonstrate that Mule promotes Miz1 degradation.
Mule Ubiquitinates Miz1 in Vitro.
To determine whether Mule catalyzes K48-linked polyubiquitination of Miz1, we performed in vitro ubiquitination assays. HEK293 cells were transfected with expression vectors encoding M2-Mule (WT or C4341A mutant) or empty vector. M2-Mule was immunoprecipitated and used as the source of E3 ligase in an in vitro ubiquitination assay with GST-Xpress-Miz1 as substrate. High-molecular-weight bands of Miz1 were appeared when incubated with WT but not the C4341 mutant of Mule in the presence of recombinant E1/E2 and ATP (Fig. 2A, Left). These high-molecular-weight bands were also recognized by anti-HA antibody, indicating that they were polyubiquitin conjugates of Miz1 (Fig. 2A, Right). Thus, Mule ubiquitinates Miz1 in vitro.
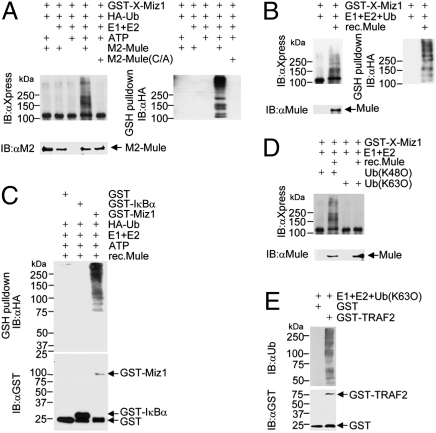
Fig. 2.
Mule ubiquitinates Miz1 in vitro. (A) GST-Xpress-Miz1 was incubated with HA-ubiquitin in the presence or absence of immunoprecipitated Mule (WT or C4341A mutant) in an in vitro ubiquitination assay in the presence or absence of ATP, and E1/E2, as indicated. Ubiquitination of GST-Xpress-Miz1 was analyzed by immunoblotting with anti-Xpress antibody (Left) or anti-HA antibody (Right). (B) In vitro ubiquitination assays with GST-Miz1 and recombinant Mule. (C) Ubiquitination of GST, GST-IκBα(1-54), or GST-Miz1 by recombinant Mule in an in vitro ubiquitination assay. (D) In vitro ubiquitination assays with GST-Miz1 in the presence of ubiquitin K48O or K63O. (E) Ubiquitination catalyzed by GST-TRAF2 in an in vitro ubiquitinaiton assay in the presence of ubiquitin K63O. The polyubiquitin chain formation was analyzed by immunoblotting with antiubiquitin antibody.
To exclude the possibility that GST-Miz1 was ubiquitinated by other E3 ligases coimmunoprecipitated with Mule, we tested whether recombinant Mule (33) could ubiquitinate GST-Miz1 in the in vitro ubiquitination assay. High-molecular-weight bands of Miz1 were detected when incubated with recombinant Mule (Fig. 2B, Left). As expected, these high-molecular-weight Miz1 bands were recognized by anti-HA antibody, indicating that they were polyubiquitin conjugates of GST-Miz1 (Fig. 2B, Right). This demonstrates that Mule can directly ubiquitinate Miz1 in vitro. The ubiquitination of Miz1 by the recombinant Mule was specific, as GST or GST-IκBα (1–54) were not ubiquitinated under the same conditions (Fig. 2C). The bands below the molecular weight of GST-Miz1 (∼110 kDa) are likely to be the products of partial degradation of polyubiquitinated GST-Miz1.
Mule belongs to the HECT-domain–containing E3 ligase family, which is known to catalyze only one type of polyubiquitination linkages (43), K48-linked polyubiquitination of substrates including p53, Mcl-1, N-Myc, and Cdc6 (33, 35–37). To determine whether Mule catalyzes K48-linked polyubiquitination of Miz1, we performed the in vitro ubiquitination assay in the presence of Ub (K48O), in which all lysines in ubiquitin were mutated to arginines except K48, or Ub (K63O). As expected, Mule ubiquitinated GST-Miz1 in the presence of Ub (K48O) but not Ub (K63O) (Fig. 2D). Under the same conditions, purified GST-TRAF2 was able to catalyze polyubiquitination by forming K63-linked polyubiquitin chains in the presence of Ub (K63O) (Fig. 2E), as reported previously (15, 44). Taken together, these data suggest that Mule is a K48-E3 ligase for Miz1.
To test whether the E3 ligase activity of Mule is stimulated by TNFα, we isolated M2-Mule from control or TNFα-treated HEK293 cells. In an in vitro ubiquitination assay, the ubiquitination of purified His-tagged Miz1 by Mule was enhanced when Mule was isolated from TNFα-treated cells (compare lane 2 with lane 1, Fig. S1). Similar results were obtained with two other known Mule substrates, GST-Mcl-1 and GST-p53 (33, 35) (Fig. S2). Thus, TNFα appears to modestly stimulate the ability of Mule to catalyze the ubiquitination of Miz1, Mcl-1, or p53. However, the underlying mechanism remains to be determined. Interestingly, the ubiquitination of His-tagged Miz1 proteins by Mule was further enhanced (compare lane 3 with lane 2, Fig. S1) when Miz1 was also purified from TNFα-treated cells, under which conditions Miz1 would have been subjected to TNFα-induced posttranslational modification. The enhancement was abolished when Miz1 was preincubated with λ-phosphatase (compare lane 5 with lane 4, Fig. S1). Taken together, these results suggest that TNFα may induce Miz1 ubiquitination through both stimulation of Mule E3 ligase activity and induction of Miz1 phosphorylation.
Mule Regulates Miz1 K48-Linked Polyubiquitination and Degradation in Vivo.
To determine whether Mule regulates Miz1 K48-linked polyubiquitination in vivo, HEK293 cells were transfected with expression vectors encoding Xpress-Miz1 along with HA-Ub (K48O), M2-Mule (WT or the C4341A mutant), or empty vector, followed by the treatment with TNFα in the presence of MG-132. K48-linked polyubiquitination of Xpress-Miz1 was augmented when Miz1 was cotransfected with Mule under nonstimulation conditions. This ubiquitination was further significantly enhanced by TNFα stimulation (Fig. 3A). In contrast, the mutant Mule (C4341A) failed to augment Miz1 ubiquitination (Fig. 3A). These results demonstrate that Mule can promote Miz1 ubiquitination in vivo.
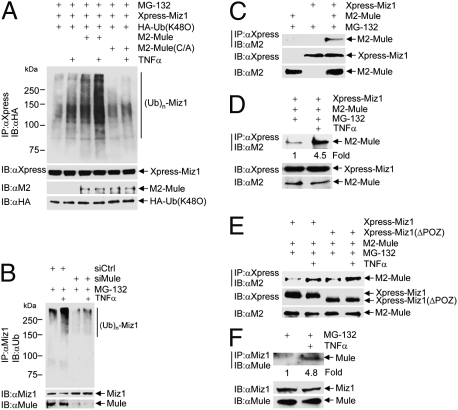
Fig. 3.
Mule ubiquitinates Miz1 in vivo. (A) HEK293 cells were transfected with expression vector encoding Xpress-Miz1, along with HA-Ub (K48O) and/or M2-Mule (WT or the C4341A mutant, 5 μg each), as indicated. Cells were treated with TNFα (5 ng/mL, 15 min) in the presence of MG-132. Ubiquitination of immunoprecipitated Xpress-Miz1 and expression levels of Xpress-Miz1, HA-Ub (K48O), and M2-Mule were determined. (B) HEK293 cells were transfected with control siRNA or siRNA for Mule (100 nM each), followed by treatment with or without TNFα (5 ng/mL, 15 min) in the presence of MG-132. Ubiquitination of immunoprecipitated Miz1 was analyzed by immnoblotting with antiubiquitin antibody. (C–E) HEK293 cells were transfected with Xpress-Miz1 WT (C and D) or the POZ deletion mutant (ΔPOZ) (E), with or without M2-Mule. Cells were treated with or without TNFα (5 ng/mL, 10 min) in the presence of MG-132. The interaction between Xpress-Miz1 and M2-Mule was determined by immnoprecipitation with anti-Xpress antibody, followed by immunoblotting with anti-M2 antibody. (F) HEK293 cells were treated with or without TNFα (5 ng/mL, 10 min) in the presence of MG-132. Interaction between endogenous Mule and Miz1 was determined by immunoprecipitation with anti-Miz1 antibody, followed by immunoblotting with anti-Mule antibody.
To determine whether Mule is necessary for TNFα-induced Miz1 ubiquitination in vivo, HEK293 cells were transfected with control siRNA or siRNA specific for Mule, followed by the treatment with TNFα. Miz1 ubiquitination was significantly augmented by TNFα in cells transfected with control siRNA, but was not in cells transfected with siMule (Fig. 3B). Taken together, these results demonstrate that the E3 ligase Mule is required for TNFα-induced Miz1 K48-linked polyubiquitination in vivo.
TNFα Promotes Interaction Between Mule and Miz1.
The above observations prompted us to examine whether TNFα regulates the interaction between Mule and Miz1, thereby inducing Miz1 K48-linked polyubiquitination and proteasomal degradation. To test this hypothesis, HEK293 cells were transfected with expression vectors encoding M2-Mule, Xpress-Miz1, or empty vector, followed by treatment with TNFα in the presence of MG-132. Immunoblotting analysis of the immnoprecipitated Xpress-Miz1 complex with anti-M2 antibody revealed that M2-Mule coprecipitated with Xpress-Miz1 (Fig. 3C). This suggests that ectopically expressed Mule and Miz1 interact with each other in resting cells. The interaction was significantly enhanced by TNFα (Fig. 3D) and was independent of the POZ domain of Miz1 (Fig. 3E). Unlike ectopically expressed Miz1 and Mule, the interaction between endogenous Mule and Miz1 was undetectable in nonstimulated cells (Fig. 3F). However, endogenous Mule and Miz1 interacted with each other after TNFα stimulation, as demonstrated by coimmunoprecipitation of Mule with Miz1 (Fig. 3F). It is possible that in the cotransfection experiments, the ectopic expression of Miz1 and Mule was much higher than endogenous levels, allowing the detection of weak basal interactions in nonstimulated cells. These results demonstrate that TNFα induces K48-linked polyubiquitination of Miz1, at least in part, by promoting the interaction of Mule with Miz1.
To test whether TNFα regulates the subcellular localization of Miz1 and Mule, control or TNFα-treated whole cell extracts were fractionated into cytoplasmic and nuclear fractions. Immunoblotting analysis of Miz1 and Mule revealed that Miz1 was present in both the cytoplasm and nucleus, whereas Mule was present only in the cytoplasm, consistent with the previous finding (45). Treatment with TNFα did not change the cytoplasmic/nuclear distribution of Miz1 and Mule (Fig. S3).
Mule Regulates TNFα-Induced JNK Activation by Targeting Miz1 for Degradation.
Miz1 is a SMOR that selectively inhibits TNFα-induced JNK activation (15). Upon TNFα stimulation, Miz1 itself undergoes K48-linked polyubiquitination and subsequent degradation by the proteasome (15) (Fig. 3A). If Mule is the K48-E3 ligase that is responsible for TNFα-induced K48-linked polyubiquitination of Miz1, then Mule should affect TNFα-induced JNK activation. To test this hypothesis, WT mouse embryonic fibroblasts (MEFs) were transfected with control siRNA or siRNA specific for Mule, followed by the treatment with TNFα. Immunoblotting analysis with anti-phospho-JNK antibody revealed that knockdown of Mule significantly inhibited TNFα-induced JNK activation (Fig. 4A, Left). Similar results were obtained when U2OS stable cells harboring the tet-inducible shMule were treated with tetracycline (Fig. 4B). To exclude the possibility that Mule might regulate TNFα-induced JNK activation via a yet-to-be identified protein other than Miz1, Miz1−/− MEFs were transfected with control siRNA or siRNA for Mule. Immunoblotting analysis of TNFα-induced JNK phosphorylation revealed that knockdown of Mule was no longer able to inhibit TNFα-induced JNK activation in Miz1 null cells (Fig. 4A, Right). This result demonstrates that Mule regulates TNFα-induced JNK activation by stimulating the ubiquitination and degradation of Miz1.
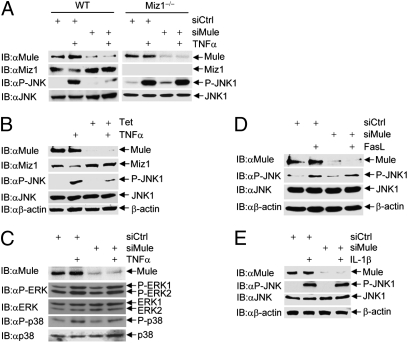
Fig. 4.
Silencing of Mule specifically inhibits TNFα-induced JNK activation. (A and C) WT fibroblasts (A, Left, and C) and Miz1 null MEFs (A, Right) were transfected with control siRNA or siRNA for Mule (100 nM each) and treated with or without TNFα (5 ng/mL, 15 min). Phosphorylation of JNK1 (A), p38 and ERK (C), and expression levels of Mule, JNK1, p38, and ERK were determined. (B) U2OS/shMule stable cells were treated with tetracycline (2 μg/mL) for 3 d and then treated with or without TNFα (5 ng/mL, 15 min). Expression levels of Mule, Miz1, JNK1, and β-actin, as well as phosphorylation of JNK1, were determined. (D and E) HEK293 cells transfected with control siRNA or siRNA for Mule (100 nM each) were treated with or without FasL (50 ng/mL, 60 min) (D), or IL-1β (5 ng/mL, 15 min) (E), as indicated. Phosphorylation of JNK1 and expression levels of JNK1, Mule, and β-actin were determined.
Mule appears to specifically regulate TNFα-induced JNK activation. We found that knockdown of Mule had no detectable effects on TNFα-induced activation of ERK or p38 (Fig. 4C), FasL-induced JNK activation (Fig. 4D), or IL-1β–induced JNK activation (Fig. 4E). These results are consistent with our previous observations that Miz1 selectively inhibits TNFα-induced JNK activation (15).
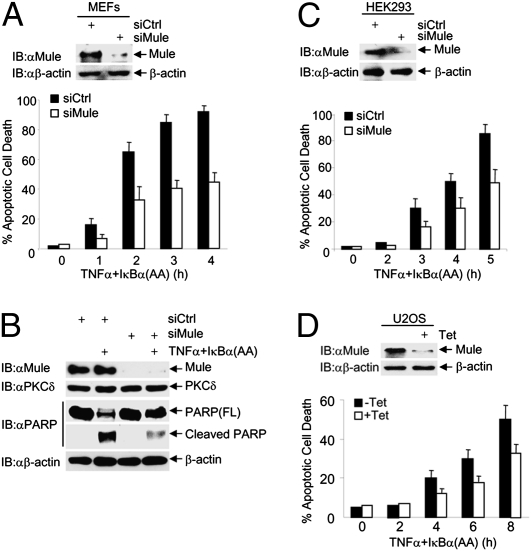
Mule Regulates TNFα-Induced Cell Death.
JNK is essential for TNFα to induce cell death when NF-κB activation is impaired (28–30, 46). To determine whether Mule regulates TNFα-induced cell death, WT MEFs were transfected with control siRNA or siRNA for Mule, followed by the infection with an adenoviral vector encoding the IκBα(AA) mutant, which is the “super-repressor” of NF-κB (47), or LacZ control. Immunoblotting analysis revealed that Mule was significantly silenced (Fig. 5A, Upper). Under these conditions, knockdown of Mule significantly inhibited TNFα-induced apoptosis of WT MEFs, as analyzed by Hoechst staining (Fig. 5A, Lower) and PARP cleavage (Fig. 5B). The inhibition was partial, as the residual JNK activity may still be able to promote a certain degree of cell death. Similar results were obtained in HEK293 cells (Fig. 5C) and U2OS cells (Fig. 5D).
Fig. 5.
Depletion of Mule inhibits TNFα-induced cell death. (A–C) WT fibroblasts (A and B), or HEK293cells (C) were transfected with control siRNA or siRNA for Mule (100 nM). Cells were treated with or without TNFα (5 ng/mL for MEFs and 20 ng/mL for HEK293 cells) plus adenovirus IκBα(AA) (200 MOI) for the indicated times. Apoptotic cell death was determined by Hoechst staining (A and C) or immunoblotting with anti-PARP antibody (B). (D) U2OS/shMule stable cells were treated with or without tetracycline (2 μg/mL) for 3 d and then treated with TNFα (20 ng/mL) plus adenovirus IκBα(AA) (200 MOI) for the indicated times. Apoptotic cell death was determined. Results are presented as mean ± SE and represent three independent experiments.
Discussion
The zinc finger transcription factor Miz1 is known to regulate gene expression and repression (1, 5, 12). Recently it has been reported that Miz1 can also function as a novel SMOR to inhibit TNFα-induced JNK1 activation by suppressing TRAF2 K63-linked polyubiquitination (15). Upon TNFα stimulation, Miz1 itself undergoes proteasomal degradation (15). However, the underlying mechanism has remained elusive. Here we report that the HECT-domain–containing E3 ligase Mule acts as the E3 ligase that ubiquitinates Miz1 and triggers its proteasomal degradation, thereby contributing to TNFα-induced JNK activation and cell death. This conclusion is based on the following evidence.
First, silencing of Mule prevented TNFα-induced Miz1 degradation, whereas ectopic expression of Mule facilitated the degradation of cotransfected Miz1 (Fig. 1 B–E). Second, recombinant and immunoprecipitated Mule ubiquitinated Miz1 in vitro (Fig. 2). Third, silencing of Mule inhibited TNFα-induced K48-linked polyubiquitination of Miz1 in vivo, whereas ectopic expression of WT Mule but not its E3 ligase defective (C4341A) mutant enhanced the K48-linked polyubiquitination of cotransfected Miz1 (Fig. 3). Finally, silencing of Mule specifically inhibited TNFα-induced JNK activation and cell death (Figs. 4 and 5).
Is Mule a K48-specific E3 ligase for Miz1? Our data show that Mule catalyzes K48- but not K63-mediated linkage of polyubiquitin chains on Miz1 in vitro and in vivo (Figs. 2 and 3). In addition, Mule is required for Miz1 degradation in resting cells or in response to TNFα stimulation (Fig. 1). Thus, Mule is a K48-specific E3 for Miz1. This is consistent with the general notion that HECT-domain–containing E3 ligases form only homogeneous ubiquitin chains, i.e., either K48- or K63-linked ubiquitin chains on their substrates (43). Indeed, Mule is the E3 ligase that ubiquitinates histone H2A (34), p53 (35), N-Myc (36), Mcl-1 (33) and Cdc6 (37) through K48-mediated linkage. The only exception to this trend is that a truncated Mule with the deletion of its first 2,470 amino acids (ΔN-Mule) ubiquitinates c-Myc through K63-mediated linkage (12). This suggests that the N-terminal truncation alters the substrate specificity of Mule, as the N-terminal domains of HECT-domain–containing E3 ligases often confer the substrate recognition (48).
Miz1 is a SMOR that selectively inhibits TNFα-induced JNK activation (15) and thereby has to be eliminated to allow TNFα to activate JNK. Our results show that Mule is required for TNFα-induced K48-linked polyubiquitination and proteasomal degradation of Miz1 (Figs. 1–3), and that depletion of Mule suppresses TNFα-induced JNK activation (Fig. 4B). The effect of Mule on TNFα-induced JNK activation depends on Miz1, as knock-down of Mule has no detectable effects on TNFα-induced JNK activation in Miz1 null MEFs (Fig. 4A). Thus, ubiquitination of Miz1 by Mule is a key step in TNFα-induced JNK activation, and may provide another potential target for selectively intervening with TNFα signaling. Future studies are needed to test this hypothesis in a physiological or pathological setting.
The mechanism by which TNFα induces Miz1 K48-linked polyubiquitination by Mule has yet to be determined, but appears to involve, at least in part, enhancing the E3 ligase activity of Mule. The initiation of the ubiquitination process depends on posttranslational modifications that either activate the E3 ligases or increase the accessibility of the substrates (41, 42). In addition, the recognition between E3 ligases and their substrates also depends on the association of the substrates with ancillary proteins such as molecular chaperones (42). An emerging theme regarding the regulation of the HECT E3 ligases is that phosphorylation of the HECT E3 ligases may increase their activity, probably by relieving the inhibitory intramolecular interactions (49, 50). Our data show that TNFα-treated Mule was more active for ubiquitinating Miz1 in vitro (Fig. S1), suggesting the E3 ligase activity of Mule may be stimulated by TNFα. Future studies are needed to investigate the underlying mechanism.
In vivo, ubiquitination of Miz1 by Mule in response to TNFα may be regulated by multiple mechanisms. Our results show that TNFα induces the interaction between Mule and Miz1 (Fig. 3 C–E). It is possible that Mule is posttranslationally modified, which leads to its activation and interaction with its substrate. Another possibility is that Miz1 may be posttranslationally modified (most likely phosphorylated, as shown in Fig. S1), thereby increasing its accessibility to Mule. Because Mule and Miz1 both have conserved or putative BH3 protein–protein interaction domains, respectively, they may interact with each other through these BH3 domains.
Our results demonstrate that the regulation of TNFα-induced JNK1 activation by Mule is mediated by its catalysis of Miz1 K48-linked polyubiquitination (Fig. 4). However, Mule is also known to contribute to degradation of the antiapoptotic Bcl-2 family protein Mcl-1 (33). Thus, Mule regulates TNFα-induced cell death by at least two mechanisms. First, K48-linked polyubiquitination of Miz1 by Mule triggers Miz1 proteasomal degradation, thereby releasing Miz1-mediated inhibition on TNFα-induced JNK activation and apoptosis (28, 51). Second, K48-linked polyubiquitination of Mcl-1 by Mule leads to proteasomal degradation of Mcl-1, thereby contributing to TNFα-induced apoptosis (33). Thus, it appears that Mule is a key regulator in TNFα-induced cell death. Future studies will explore how Mule is regulated by TNFα.
Methods
Reagents, Plasmids, and siRNA.
Antibodies against phospho-JNK, p38, phospho-p38, ERK, and phosphor-ERK were from Cell Signaling. Antibody against JNK was from Pharmingen. Antibody against Miz1 was as described previously (15). Antibodies against ubiquitin, Tubulin, LaminA/C, Xpress, PARP, PKCδ, and HA were from Santa Cruz. Antibody against IKKβ was from Upstate. Antibody against Mule was from ProSci. All siRNAs were purchased from Dharmacon. Glutathione (GSH)-agarose beads, tetracycline, N-ethylmaleimide (NEM), ubiquitin aldehyde, purified E1, imidazole, cycloheximide (CHX), MG-132, Hoechst 33258, and antibodies against M2 and β-actin were from Sigma. Fas ligand, IL-1β, and murine TNFα were from R&D Systems. TRAIL was from Biomol. Recombinant E2 (UbcH5), HA-ubiquitin and ubiquitin (K63O or K48O) were from Boston Biochem. Ni-NTA Magnetic Agarose beads was from QIAGEN. λ-phosphatase was from New England Biolabs. GST-TRAF2, GST-Miz1, Xpress-Miz1, HA-ubiquitin (WT, K48O, K63O), M2-Mule (WT and the C4341A mutant) and recombinant Mule were as described previously (15, 33). GST-Mcl-1 and GST-p53 were generated by PCR and subcloned into pGEX-KG vector. All constructs were verified by DNA sequencing. The sequence of siMule is GAGCAGAAGTGGAGAGGAT dTdT.
Mass Spectrometry.
Protein in-gel digestion and nano-HPLC electrospray ion trap mass spectrometry (MS/MS) were performed as previously described (15). The acquired MS/MS spectra were searched against NCBI-nr protein sequence database. One tryptic peptide from the E3 ligase Mule was identified, (R)SSLLTEKLLRLLSLISIALPENK(V).
Other materials and methods are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Alexander Schilling for the mass spectrometry analysis and Anning Lin (University of Chicago) and Xiaodong Wang (University of Texas Southwestern Medical Center at Dallas) for valuable reagents that made this work possible. We also thank Frank Aguilar for helpful discussions and Samuel Jang for excellent technical support. This work was partially supported by National Institutes of Health Grant GM081603 (to J.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913690107/-/DCSupplemental.
References
- 1.Peukert K, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell VJ, Treisman R. The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 3.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 4.Seoane J, et al. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 5.Herold S, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 6.Seoane J, Le HV, Massagué J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 7.van de Wetering M, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 8.Wu S, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 9.Kime L, Wright SC. Mad4 is regulated by a transcriptional repressor complex that contains Miz-1 and c-Myc. Biochem J. 2003;370:291–298. doi: 10.1042/BJ20021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito M, et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2009;106:11294–11299. doi: 10.1073/pnas.0903854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber A, et al. Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J. 2008;27:1563–1574. doi: 10.1038/emboj.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Wanzel M, et al. Akt and 14-3-3eta regulate Miz1 to control cell-cycle arrest after DNA damage. Nat Cell Biol. 2005;7:30–41. doi: 10.1038/ncb1202. [DOI] [PubMed] [Google Scholar]
- 14.Herold S, et al. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J. 2008;27:2851–2861. doi: 10.1038/emboj.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Zhao Y, Eilers M, Lin A. Miz1 is a signal- and pathway-specific modulator or regulator (SMOR) that suppresses TNF-alpha-induced JNK1 activation. Proc Natl Acad Sci USA. 2009;106:18279–18284. doi: 10.1073/pnas.0906328106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tartaglia LA, Goeddel DV. Two TNF receptors. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 17.Tracey KJ, Cerami A. Tumor necrosis factor, other cytokines and disease. Annu Rev Cell Biol. 1993;9:317–343. doi: 10.1146/annurev.cb.09.110193.001533. [DOI] [PubMed] [Google Scholar]
- 18.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 19.Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 20.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 21.Muppidi JR, Tschopp J, Siegel RM. Life and death decisions: Secondary complexes and lipid rafts in TNF receptor family signal transduction. Immunity. 2004;21:461–465. doi: 10.1016/j.immuni.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 23.Kyriakis JM, et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, et al. Differential activation of ERK, JNK/SAPK and P38/CSBP/RK map kinase family members during the cellular response to arsenite. Free Radic Biol Med. 1996;21:771–781. doi: 10.1016/0891-5849(96)00176-1. [DOI] [PubMed] [Google Scholar]
- 25.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 26.Yeh KW, Chen JC, Lin MI, Chen YM, Lin CY. Functional activity of sporamin from sweet potato (Ipomoea batatas Lam.): A tuber storage protein with trypsin inhibitory activity. Plant Mol Biol. 1997;33:565–570. doi: 10.1023/a:1005764702510. [DOI] [PubMed] [Google Scholar]
- 27.Tada K, et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J Biol Chem. 2001;276:36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Lin A. Wiring the cell signaling circuitry by the NF-kappa B and JNK1 crosstalk and its applications in human diseases. Oncogene. 2007;26:3267–3278. doi: 10.1038/sj.onc.1210417. [DOI] [PubMed] [Google Scholar]
- 29.Tang G, et al. Inhibition of JNK activation through NF-kappaB target genes. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 30.Tang F, et al. The absence of NF-kappaB-mediated inhibition of c-Jun N-terminal kinase activation contributes to tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2002;22:8571–8579. doi: 10.1128/MCB.22.24.8571-8579.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda S, et al. IKKbeta is required for prevention of apoptosis mediated by cell-bound but not by circulating TNFalpha. Immunity. 2003;19:725–737. doi: 10.1016/s1074-7613(03)00301-7. [DOI] [PubMed] [Google Scholar]
- 32.De Smaele E, et al. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 33.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol Cell Biol. 2005;25:2819–2831. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 36.Zhao X, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall JR, et al. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell. 2007;18:3340–3350. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aravind L. The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann K, Bucher P. The UBA domain: A sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 40.Wilkinson CR, et al. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 41.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 42.Ciechanover A, Iwai K. The ubiquitin system: From basic mechanisms to the patient bed. IUBMB Life. 2004;56:193–201. doi: 10.1080/1521654042000223616. [DOI] [PubMed] [Google Scholar]
- 43.Kim HT, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 44.Xia ZP, Chen ZJ. TRAF2: A double-edged sword? Sci STKE. 2005;2005:pe7. doi: 10.1126/stke.2722005pe7. [DOI] [PubMed] [Google Scholar]
- 45.Parsons JL, et al. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 2009;28:3207–3215. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamata H, et al. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Minemoto Y, Lin A. c-Jun N-terminal protein kinase 1 (JNK1), but not JNK2, is essential for tumor necrosis factor alpha-induced c-Jun kinase activation and apoptosis. Mol Cell Biol. 2004;24:10844–10856. doi: 10.1128/MCB.24.24.10844-10856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cyr DM, Höhfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 49.Gao M, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 50.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 51.Chang L, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 52.Adhikary S, et al. Miz1 is required for early embryonic development during gastrulation. Mol Cell Biol. 2003;23:7648–7657. doi: 10.1128/MCB.23.21.7648-7657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, et al. NF-kappaB is required for UV-induced JNK activation via induction of PKCdelta. Mol Cell. 2006;21:467–480. doi: 10.1016/j.molcel.2005.12.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.