Abstract
Cytochrome P450 2A6 (CYP2A6) is the primary human enzyme involved in nicotine metabolism. Our objective was to characterize two nonsynonymous single nucleotide polymorphisms (SNPs) in CYP2A6*24, 594G>C (Val110Leu) and 6458A>T (Asn438Tyr). We determined their haplotype, allele frequencies, effect on CYP2A6 activity in vivo, as well as their stability and ability to metabolize nicotine in vitro. CYP2A6*35 (6458A>T) occurred at a frequency of 2.5–2.9% among individuals of black African descent, 0.5–0.8% among Asians, and was not found in Caucasians. In addition, we identified two novel alleles, CYP2A6*36 (6458A>T and 6558T>C [Ile471Thr]) and CYP2A6*37 (6458A>T, 6558T>C, and 6600G>T [Arg485Leu]). In vivo, CYP2A6*35 was associated with lower CYP2A6 activity as measured by the 3HC/COT ratio. In vitro, CYP2A6.35 had decreased nicotine C-oxidation activity and thermal stability. In conclusion, we identified three novel CYP2A6 alleles (CYP2A6*35, *36, and *37); the higher allele frequency variant CYP2A6*35 was associated with lower CYP2A6 activity.
Keywords: Nicotine, Cotinine, CYP2A6, Smoking, Pharmacogenetics
INTRODUCTION
Cigarette’s addictive properties are largely due to the psychoactive effects of nicotine.1 Due to the relatively short half-life of nicotine, approximately 2 hours,2 dependent smokers smoke at regular intervals to maintain nicotine levels. Therefore, factors that influence the intake or removal (e.g. metabolism) of nicotine from the body can affect smoking behaviours such as the number of cigarettes smoked or the likelihood of cessation.3–6
In vivo, 70–80% of nicotine is metabolized to cotinine,2 and cytochrome P450 2A6 (CYP2A6) mediates approximately 90% of this reaction.7, 8 CYP2A6 also exclusively mediates cotinine’s (COT) hydroxylation to trans-3′-hydroxycotinine (3HC), making the metabolic ratio of 3HC/COT a specific and reliable marker of CYP2A6 activity.9–11 The 3HC/COT ratio correlates with the rate of nicotine clearance 10 and is stable over time, 12 thereby facilitating its use as a proxy measure for the rate of CYP2A6 mediated nicotine metabolism in population studies. A common feature of nicotine pharmacokinetic studies is the large interindividual variability in the rate of metabolism. 13–15 Sources of this large variability may include environmental (e.g. smoking), physiological (e.g. systemic diseases), and genetic (e.g. CYP2A6 polymorphisms) factors.16
Recently, we characterized the effect of a number of established and novel CYP2A6 alleles on CYP2A6 activity and nicotine disposition kinetics in a population of black African descent.15 Interestingly, most of the novel alleles were associated with lower in vivo CYP2A6 activity, accounting for some of the previously uncharacterized variability and slower nicotine metabolism in this population. Nonetheless, even after controlling for gender, smoking, and known CYP2A6 alleles, large variability in CYP2A6 activity still remains. One possible source of the remaining variation is the presence of additional genetic variants. Through sequencing, we found that CYP2A6*24 contains two nonsynonymous single nucleotide polymorphisms (SNPs): 594G>C (Val110Leu); and 6458A>T (Asn438Tyr).15 A question that remained is whether 594G>C or 6458A>T can occur on their own, and if so, what are their allele frequencies and functional impact on CYP2A6 activity.
In the current study we genotyped multiple ethnic populations for 594G>C (Val110Leu) and 6458A>T (Asn438Tyr), assessed their allele frequencies, and examined their impact on in vivo CYP2A6 activity using the 3HC/COT ratio. In addition, we created cDNA expressed variants CYP2A6.V110L (Val110Leu), CYP2A6.35 (Asn438Tyr), CYP2A6.24 (Val110Leu + Asn438Tyr) and assessed their thermal stability and ability to metabolize nicotine to cotinine in vitro using an E. coli heterologous expression system.
RESULTS
Allele frequency of CYP2A6*35 and identification of novel alleles CYP2A6*36 and CYP2A6*37
Since CYP2A6*24 is a haplotype containing 594G>C and 6458A>T,15 individuals positively genotyped for these two variants were considered to have CYP2A6*24, while individuals positively genotyped for 6458A>T alone formed the CYP2A6*35 group. We did not find any individuals with the 594G>C SNP alone in any of the ethnic groups. CYP2A6*24 occurred at an allele frequency of 0.7% among African American smokers (n = 1234 alleles), and was not found among Caucasians (n = 304 alleles), Japanese (n = 120 alleles), Chinese (n = 196 alleles), or Taiwanese (n = 334 alleles). Alleles here are inferred by assuming that each individual has 2 copies of chromosome 19 containing one allele on each chromosome. Previously we found CYP2A6*24 at a frequency of 1.3% among an African Canadian population (1.4% in smokers and 1.1% in nonsmokers) and characterized it as a low activity variant.15 CYP2A6*35 was found among individuals of African descent with a frequency of 2.9% and 2.5% in African American smokers and African Canadians (1.8% in smokers and 3.3% in nonsmokers), respectively. The allele was also found in Japanese (0.8%), Taiwanese (0.6%), and Chinese (0.5%) but not among Caucasians. The genotype frequencies of CYP2A6*24 and CYP2A6*35 did not significantly deviate from Hardy-Weinberg equilibrium in any population.
The variant CYP2A6*35 allele from one African Canadian, one African American, and one Taiwanese individual were fully sequenced. The variant allele CYP2A6*35A was identical among the African American and African Canadian individuals, while the allele CYP2A6*35B in the Taiwanese individual differed modestly (table 1). The allele frequencies of CYP2A6*35 presented herein include both CYP2A6*35A and CYP2A6*35B. In addition, we identified two novel alleles, CYP2A6*36 and CYP2A6*37. CYP2A6*36 contains 6458A>T in haplotype with 6558T>C present in CYP2A6*7, while CYP2A6*37 contains 6458A>T and two additional SNPs present in CYP2A6*10 (6558T>C and 6600G>T), table 1. CYP2A6*36 and CYP2A6*37 were only found among Taiwanese individuals, both at a frequency of 0.3%. These alleles were not included in estimating the allele frequency of CYP2A6*35.
Table 1.
CYP2A6*35(A and B), CYP2A6*36, and CYP2A6*37 alleles
| CYP2A6*35A | CYP2A6*35B | CYP2A6*36 | CYP2A6*37 |
|---|---|---|---|
| -1301A>C | -1301A>C | -1301A>C | -1301A>C |
| -1289G>A | -1289G>A | -1289G>A | -1289G>A |
| -745A>G | -745A>G | -745A>G | |
| -1013A>G | |||
| 22C>T (L8L, Exon 1) | 22C>T (L8L, Exon 1) | 22C>T (L8L, Exon 1) | |
| 720G>A, Intron 2 | |||
| 1137C>G, Intron 2 | |||
| 1620T>C, Intron 2 | 1620T>C, Intron 2 | 1620T>C, Intron 2 | 1620T>C, Intron 2 |
| 2483G>A, Intron 4 | |||
| 3225A>G, Intron 4 | |||
| 4084delA, Intron 5 | 4084delA, Intron 5 | 4084delA, Intron 5 | |
| 6218A>G, Intron 8 | |||
| 6282A>G, Intron 8 | |||
| 6293T>C, Intron 8 | |||
| 6354T>C, Intron 8 | 6354T>C, Intron 8 | ||
| 6458A>T (N438Y, Exon 9) | 6458A>T (N438Y, Exon 9) | 6458A>T (N438Y, Exon 9) | 6458A>T (N438Y, Exon 9) |
| 6558T>C (I471T, Exon 9) | 6558T>C (I471T, Exon 9) | ||
| 6600G>T (R485L, Exon 9) | |||
| 2A6*1B | 2A6*1B | 2A6*1B | 2A6*1B |
| 6782C>G | 6782C>G | 6782C>G | 6782C>G |
| 6835C>A | 6835C>A | 6835C>A | |
| 6936_6937insCACTT | |||
| 6961_6962insGAAAAG | |||
| 6989A>G | |||
| 6999T>C | 6999T>C | 6999T>C | |
| 7160A>G | 7160A>G | 7160A>G | 7160A>G |
Numbering of the nucleotides is in reference to the ATG start site of the CYP2A6 gene (reference genomic sequence NG_000008.7) in which A is numbered 1 and the base before A is numbered −1. Bolded nucleotides are synonymous SNPs. Bolded and italicized nucleotides are nonsynonymous SNPs. 2A6*1B refers to the 58 bp gene conversion in the 3′ untranslated region which was present in all alleles.
CYP2A6*35 is associated with lower CYP2A6 activity in vivo
In the African Canadian population, individuals with CYP2A6*1/*35 genotype had significantly lower mean 3HC/COT compared to those in the wildtype group (CYP2A6*1/*1), table 2. Four individuals were compound heterozygotes, one having CYP2A6*9/*35 and three having CYP2A6*17/*35. All four individuals had substantially lower 3HC/COT ratios compared to wildtype and heterozygote CYP2A6*1/*35 individuals (table 2).
Table 2.
CYP2A6*35 is associated with lower in vivo CYP2A6 activity.
| Population | Genotype | n | Mean adjusted 3HC/COT | S.D. | p-value | % of wildtype activity |
|---|---|---|---|---|---|---|
| African Canadian | *1/*1 | 151 | 1.20 | 0.63 | 100 | |
| *1/*35 | 10 | 0.84 | 0.50 | 0.02 | 70 | |
| *9/*35a | 1 | 0.47 | - | 39 | ||
| *17/*35a | 3 | 0.08 | 0.14 | 7 | ||
| African American | *1/*1 | 258 | 1.21 | 0.80 | 100 | |
| *1/*35 | 20 | 0.75 | 0.26 | 0.003b | 62 | |
| *35/*35a | 1 | 0.51 | - | 42 | ||
| *9/*35a | 2 | 0.78 | 0.51 | 62 | ||
| *17/*35a | 3 | 0.27 | 0.09 | 22 |
n=number of samples, S.D. = standard deviation.
the number of individuals with greater than one variant was too small for statistical analyses.
the p-value obtained does not include the homozygote CYP2A6*35 individual. The ratio was derived from plasma metabolite levels following oral nicotine intake or ad libitum smoking among the African Canadian and African American population, respectively. The effect of CYP2A6*35 remained significant in both populations when the wildtype group included individuals with at least one CYP2A6*1B allele (i.e. CYP2A6*1A/*1B and CYP2A6*1B/*1B). The 3H/COT ratios among individuals with CYP2A6*35 compared to individuals with only CYP2A6*1B were as follows: 0.84±0.50 vs. 1.25±0.66, p=0.03 among African Canadians and 0.75±0.26 vs. 1.38±0.75, p<0.001 among African Americans. The p-values are from the regression model as described in the methods section with the dependent variable being log 3HC/COT, controlling for gender and smoking. When the data was analyzed with a Mann-Whitney test where the dependent variable was 3HC/COT (i.e. not log normalized), and where the effects of gender and smoking were not considered, the effect of CYP2A6*35 remained significant in both populations (African Canadian p=0.016 and the African American p=0.001).
Among the African Americans, those with CYP2A6*1/*35 genotype also had significantly lower mean 3HC/COT compared to individuals in the wildtype group (CYP2A6*1/*1), table 2. In addition, an individual homozygous for CYP2A6*35 had lower 3HC/COT compared to the heterozygous individuals suggestive of a gene-dose effect. As seen in the African Canadian population, this population had compound heterozygote individuals (CYP2A6*9/*35, n = 2 and CYP2A6*17/*35, n = 3) with lower 3HC/COT values compared to wildtype.
In vitro enzymatic activities of the CYP2A6 variants
The apparent Km and Vmax values for nicotine C-oxidation by CYP2A6.1 and the variant constructs is presented in table 3. The Vmax value of CYP2A6.17 trended towards being lower compared to CYP2A6.1 (p=0.1), while the other variants (CYP2A6.V110L, CYP2A6.35, and CYP2A6.24) had similar values compared to CYP2A6.1. The apparent Km value of CYP2A6.17 was significantly higher than that of CYP2A6.1. The Km values of CYP2A6.V110L, and CYP2A6.35 trended to be higher compared to CYP2A6.1, while CYP2A6.24 trended to have a lower value. The resulting Vmax/Km (catalytic efficiency) values of CYP2A6.17, CYP2A6.V110L, and CYP2A6.35 were significantly lower compared to that of CYP2A6.1. In contrast, the Vmax/Km value of CYP2A6.24 was similar to that of CYP2A6.1.
Table 3.
Kinetic parameters of the wildtype and variant CYP2A6 constructs for the metabolism of nicotine.
| Km (μM) | Vmax (pmol/min/pmol CYP2A6) | Vmax/Km (nl/min/pmol CYP2A6) | |
|---|---|---|---|
| CYP2A6.1 | 7.0±0.7 | 0.53±0.04 | 8.0±0.6 |
| CYP2A6.17 | 33.4±3.3* | 0.36±0.03 | 1.1±0.1* |
| CYP2A6.V110L | 11.3±1.4 | 0.52±0.06 | 4.7±0.3* |
| CYP2A6.35 | 9.9±0.8 | 0.49±0.05 | 5.0±0.2* |
| CYP2A6.24 | 5.4±1.8 | 0.53±0.09 | 10.5±1.9 |
Data are presented as mean ± SEM of 2–7 independent experiments.
p<0.05 when compared to CYP2A6.1.
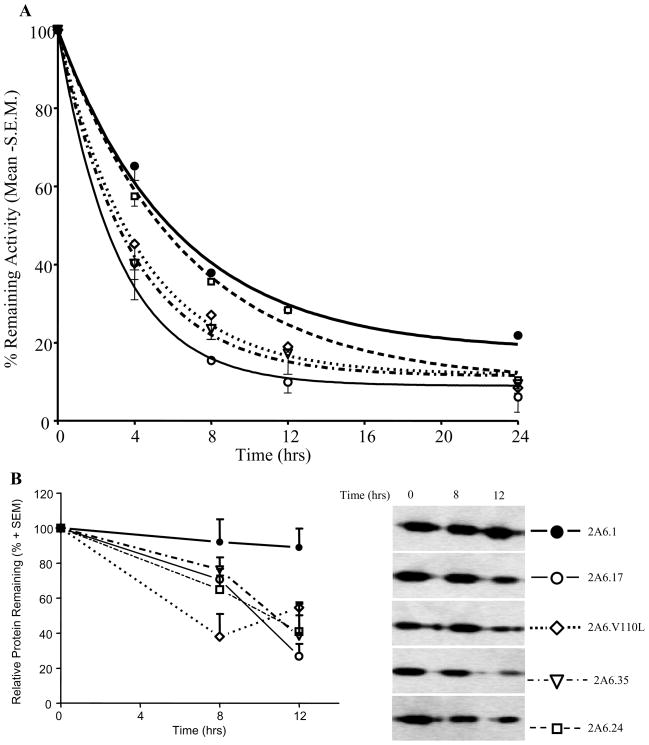
To determine whether the amino acid changes could affect protein stability we tested the thermal stability of the constructs by incubating them at 37°C and collecting aliquots at various times to measure nicotine C-oxidation activity and CYP2A6 protein levels. Generally, the activity of the variant constructs decreased to a greater extent compared to the wildtype construct (figure 1A). The time it took to decrease CYP2A6 activity by 50% (t1/2) was significantly shorter for CYP2A6.17 (t1/2 = 2.2 hrs), CYP2A6.V110L (t1/2 = 2.9 hrs), and CYP2A6.35 (t1/2 = 2.6 hrs) compared to CYP2A6.1 (t1/2 = 4.3 hrs, p<0.05), while the t1/2 of CYP2A6.24 was similar to CYP2A6.1 (4.6 vs. 4.3 hrs, respectively). With incubation time the CYP2A6.1 protein levels did not change, while the protein levels of CYP2A6.17, CYP2A6.V110L, CYP2A6.35, and CYP2A6.24 decreased (figure 1B). Of note, CYP2A6 protein expression following E. coli culture was lower among the variant constructs compared to wildtype, CYP2A6.1 > CYP2A6.V110L and CYP2A6.24 > CYP2A6.35 > CYP2A6.17 (0.44 > 0.38 > 0.20 > 0.10 pmol of CYP2A6/μg of membrane protein, respectively).
Figure 1. Thermal stability of the CYP2A6 wildtype and variant constructs at 37 °C.
(A) Activities are expressed as the percentage of remaining enzymatic activity for each construct with the activity at 0 hrs recorded as 100%. Aliquots (10 pmoles of CYP2A6 enzyme for each construct) were taken at 0, 4, 8, 12, and 24 hours to measure cotinine formation using 30 μM of nicotine. The activity of each construct at 0 hours is displayed as 100%, and over time, the activity was measured for each construct and expressed as a percent remaining. We used this approach to minimize potential impact of modestly different amounts of estimated CYP2A6 in the initial reaction. The lines through the points are stimulated using the plateau and rate of decay (k) values obtained for each CYP2A6 construct from the one phase decay equation as described in the methods. (B) Immunoreactive CYP2A6 protein at the indicated times of incubation. With time, the protein levels of the CYP2A6 variant constructs decreased while CYP2A6.1 protein levels remained the same. At 24 hrs, no CYP2A6 protein was detected with any of the CYP2A6 constructs (data not shown). 1.2 pmoles of CYP2A6 was loaded in each well.
DISCUSSION
The focus of the present study was two SNPs found in CYP2A6, 594G>C and 6458A>T. Here, we did not find individuals with 594G>C alone, suggesting that 594G>C is in obligate haplotype with 6458A>T forming CYP2A6*24. Conversely, individuals with only 6458A>T were found. The allele containing 6458A>T was sequenced and assigned the name CYP2A6*35 by the CYP-allele nomenclature committee (http://www.cypalleles.ki.se/). CYP2A6*35 was predominantly found among individuals of African descent and to a lesser extent among individuals of Asian descent. The CYP2A6*35 allele sequence differed slightly between African and Asian descent distinguishing it as CYP2A6*35A and CYP2A6*35B, respectively. We also discovered two novel alleles among Taiwanese individuals where the 6458A>T SNP was in haplotype with 6558T>C in CYP2A6*7 or 6558T>C + 6600G>T in CYP2A6*10, forming CYP2A6*36 and CYP2A6*37, respectively. We could not test the functional in vivo consequence of CYP2A6*36 and CYP2A6*37, but since both of these rare alleles contain the amino acid substitution (Ile471Thr) found in CYP2A6*7 and CYP2A6*10 it is likely they will result in a loss of function. The substitution Ile471Thr has been associated with lower CYP2A6 activity and stability in vitro (E. coli system) 17 in addition to lower in vivo CYP2A6 activity in multiple studies.18–21
Our in vivo data strongly suggests that CYP2A6*35 is associated with lower CYP2A6 activity. In both African populations, individuals heterozygous for CYP2A6*35 had ~70% of the activity of the wildtype CYP2A6 group, a reduction in activity that is similar to well characterized decreased function alleles CYP2A6*9, CYP2A6*12, and CYP2A6*17.14, 15, 22 Several case-control studies have associated CYP2A6 genetic variants that result in lower CYP2A6 activity with a lower likelihood of smoking;5, 23 however, negative finding have also been reported.24, 25 Among the African Canadians, a trend was observed in which individuals with CYP2A6*35 were less likely to be current adult smokers (OR = 0.51; 95% C.I. = 0.17–1.60, p = 0.19). The allele frequency of CYP2A6*35 was 3.3% (n = 9/269 alleles) in smokers compared to 1.8% (n = 5/275 alleles) in nonsmokers. The magnitude of the effect was the same when analysis was restricted to individuals with only CYP2A6*1/*1 and CYP2A6*1/*35 (OR = 0.39, 95% C.I. 0.01–1.60, p = 0.19). All the African American individuals were participants in a smoking cessation trial (i.e. smokers) limiting our ability to further test this hypothesis.
Our in vitro enzymatic function data (table 3) suggests that the variants CYP2A6.V110L (Val110Leu) and CYP2A6.35 (Asn438Tyr) result in lower nicotine C-oxidation activity compared to the wildtype. This was driven by the higher apparent Km values observed for CYP2A6.V110L and CYP2A6.35. In addition, CYP2A6.V110L and CYP2A6.35 had reduced thermal stability compared to the wildtype. The most dramatic reduction in stability was seen following 4 hrs of incubation, in which the constructs CYP2A6.17, CYP2A6.V110L, and CYP2A6.35 lost more than 50% of their initial activity, while the wildtype (CYP2A6.1) lost only about 30% of its initial activity. It is interesting to note that the amino acid substitutions Val110Leu and Asn438Tyr are not highly conserved among the CYP2A family. For instance, the human CYP2A13 and the mouse CYP2A5, both of which metabolize nicotine efficiently, have leucine (Leu) at position 110 and tyrosine (Tyr) at position 438. In addition, the physical and chemical properties of valine (Val) and leucine (Leu) and asparginine (Asn) and tyrosine (Tyr) are similar to each other (e.g. Val and Leu are similarly aliphatic, whereas Asp and Tyr are similarly polar). It is possible though that the positions of the following amino acid changes are important. Valine 110 resides within the substrate recognition site 1 (SRS1, residues 101–120)26 and is in very close proximity to the active site of CYP2A6.27, 28 Thus, this substitution (Val110Leu) might alter the active site cavity.28 On the other hand, asparginine 438 resides on the surface o CYP2A6 and is situated next to the conserved heme-binding cysteine 439.26, 29 Thus, it is possible that the amino acid substitution Asn438Tyr might affect the binding of the heme. In our study the reduction in protein levels did not fully complement the reduction in CYP activity suggesting that the heat treatment might have also facilitated the loss of heme. We were unable to test this (due to the low sensitivity of our assay); however it would be valuable in the future to determine the stability of the holoenzyme by obtaining the CO-difference spectrum for the constructs.
It was surprising for us to find that individually Val110Leu and Asn438Tyr reduced CYP2A6 activity and stability, while in CYP2A6.24 the occurrence of both amino acid changes appeared to have stabilized the enzyme. This is in contrast to our previously published study15, in which individuals with CYP2A6*24 were associated with lower CYP2A6 activity measured by the 3HC/COT ratio in vivo. There are several possible reasons for the discordance between the in vivo and in vitro data for CYP2A6*24. It is possible that the coding SNPs might be affecting protein degradation (discussed below). Alternatively, the coding SNPs might be in linkage disequilibrium with polymorphisms in the gene promoter or within introns that affect transcription and mRNA splicing, and as a result might influence function in vivo. For example, CYP2A6*35A and CYP2A6*24 contain -1013A>G, which is thought to disrupt a putative enhancer region30, while CYP2A6*35B contains -745A>G that is thought to disrupt a CCAAT box to which the regulatory element NF-Y binds.31 Both of these promoter SNPs have been shown to result in decreased expression in a reporter construct system.30, 31.
The protein levels of CYP2A6.1 did not appear to decrease during the first 12 hrs of incubation, while its activity did. This is likely due to the fact that immunoblotting not only detected functional holoenzyme, but also apoenzyme and misfolded CYP2A6. However, it is interesting to note that unlike CYP2A6.1, the CYP2A6 protein levels of the variant constructs all decreased with incubation time suggesting that these constructs are more susceptible to degradation. It was also noted that the CYP2A6 variant constructs had lower CYP2A6 protein expression (pmoles of CYP2A6/μg of membrane protein) compared to the wildtype construct following culturing of the E. coli. There is a growing body of evidence for nonsynonymous coding SNPs influencing enzymatic function by decreasing the amount of protein through accelerated degradation.32–35 Much of this evidence comes from cytoplasmic proteins such as thiopurine S-methyltransferase (TPMT),32 and has also been shown for cytochromes P450. For example, Bandiera et al. demonstrated that CYP1B1.4 is expressed at lower levels compared to CYP1B1.1 in COS-1 cells largely due to its increased rate of degradation.35
The present study suggests that CYP2A6*35 is associated with lower in vivo CYP2A6 activity, likely due to its lower nicotine C-oxidation activity, stability, and greater susceptibility to protein degradation. Because of its lower stability, CYP2A6*35 is likely to result in lower in vivo activities toward all CYP2A6 substrates; however this remains to be tested. In conclusion, we have identified several novel alleles (CYP2A6*35, CYP2A6*36, and CYP2A6*37), one of which (CYP2A6*35) is found at a relatively high frequency among individuals of African descent and is associated with lower CYP2A6 activity in vivo. Identifying and characterizing novel CYP2A6 variants among different ethnic groups will help refine the relationship between CYP2A6 genotype and nicotine metabolism phenotype, thus increasing the utility of genotyping in epidemiological and clinical treatment studies.15, 36, 37
MATERIALS and METHODS
CYP2A6 6458A>T genotype assay
A two-step PCR assay was developed to detect the 6458A>T SNP based on the reference genomic sequence NG_000008.7. The first PCR reaction amplified a gene specific region from intron 6 to the 3′-flanking region utilizing the primers 2A6in6F1 and 2A6R0 as described previously.38 In the second PCR reaction, an allele specific region (1327 bp) from exon 9 to the 3′-flanking region of CYP2A6 was amplified utilizing one of two forward primers 2A6in8ex9F6458W or 2A6in8ex9F6458V in combination with one reverse primer 2A6R0 (table 4). The 25 μl reaction consisted of: 1 X PCR buffer (10 mM Tris pH 8.8, 50 mM KCl), 0.1 mM of each dNTP, 1.25 mM MgCl2, 0.15 μM of each primer, 0.25 U of Taq polymerase (MBI Fermentas, Burlington, Canada), 0.8 μl of undiluted 1st amplification PCR product, and H2O. Initial denaturation at 95°C for 1 min was followed by 20 cycles, each consisting of, denaturation at 95°C for 15 sec, annealing at 55°C for 20 sec, and extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min. 594G>C was detected as described previously.15 The PCR products were analyzed by electrophoresis on a 1.2 % agarose gel (OnBio, Richmond Hill, Canada) stained with ethidium bromide. The assay was developed using multiple cosmids containing genomic clones of CYP2A6 as a positive control and CYP2A7 and CYP2A13 as negative controls (gratefully provided by Dr. Linda Ashworth, Human Genome Centre, Liverpool).39
Table 4.
Primers used.
| Primer | Sequence | Location |
|---|---|---|
| 2A65Pr1Fa | 5′-ACC TAG ACT TAA TCT TCC CGT ATA C-3′ | 5′-flanking region |
| 2A6in8ex9F6458W | 5′-TCC TCA GGA AAG CGG A-3′ | Intron8/Exon9 |
| 2A6in8ex9F6458V | 5′-TCC TCA GGA AAG CGG T-3′ | Intron8/Exon9 |
| 2A6R0b | 5′-AGG TCA TCT AGA TTT TCT CCT ACA-3′ | 3′-flanking region |
| 2A6_594G>C | 5′-GCC ACC TTC GAC TGG CTC TTC AAA GGC TAT G-3′ | Exon 2 |
| 2A6_594G>C Anti | 5′-CAT AGC CTT TGA AGA GCC AGT CGA AGG TGG C-3′ | Exon 2 |
| 2A6_6458A>T | 5′-TTC CAT CGG AAA GCG GTA CTG TTT CGG AGA AGG-3′ | Exon 8/9 |
| 2A6_6458A>T Anti | 5′-CCT TCT CCG AAA CAG TAC CGC TTT CCG ATG GAA-3′ | Exon 8/9 |
Pitarque et. al. 2001.
Mwenifumbo et. al 2008.
CYP2A6 sequencing
To confirm our genotyping assay and determine the haplotype of the allele, we amplified (Long PCR) and sequenced a 9.2 kb fragment containing the CYP2A6 gene from 3 individuals heterozygous for 6458A>T (one African Canadian, one African American, and one Taiwanese). The 25 μl PCR reaction utilized the primers 2A65Pr1F and 2A6R0 (table 4) and was performed following the manufacturer’s instructions (long PCR enzyme mix, MBI Fermentas, Burlington, Canada) with the following minor modifications. The annealing temperature was 57°C and the reaction contained 75 ng of DNA, 0.3 mM of each dNTP, and 0.25 μM of each primer. The PCR product was subcloned and sequenced as described previously.15 In addition, the CYP2A6 gene (9.2 kb) of two individuals was amplified. One was genotyped as having 6458A>T as well as 6558T>C present in CYP2A6*7, while the other was genotyped as having 6458A>T and 6558T>C + 6600G>T present in CYP2A6*10. Following CYP2A6 gene (9.2 kb) amplification the products were cloned and then genotyped or sequenced to determine whether 6458A>T is in haplotype with the SNPs in CYP2A6*7 and CYP2A6*10.
CYP2A6 genotyping and phenotyping
Individuals from different ethnic backgrounds, African Canadian (n=279), African American (n=617), Japanese (n=60), Taiwanese (n=167), Chinese (n=98), and Caucasian (n=152) were genotyped for the 6458A>T variant. In addition, the African Americans and African Canadians were genotyped for other CYP2A6 variants including CYP2A6*1B, *2, *4, *9, *12, *14, *17, *20, *21, *23, *24, *25, *26, *27, and *28), while the Taiwanese, Chinese, Japanese, and Caucasians were genotyped for CYP2A6*1B, *2, *4, *7, *8, *9, *10, and *12. The demographics of the African Canadian, African American, Japanese, Chinese, Taiwanese, and Caucasian populations have been previously described.5, 40, 41
Phenotypic measures were available for the African Canadian and African American populations. The plasma 3HC/COT ratio collected at 270 min following oral nicotine (4 mg capsule) was used as a proxy measure for CYP2A6 activity among the African Canadians,41 while the plasma 3HC/COT ratio collected from ad libitum smoking was used in the African American population.10 In both populations CYP2A6 genotype has been shown to be significantly associated with the ratio (3HC/COT), supporting its use as a measure of CYP2A6 activity.15, 42 All studies were approved by the University of Toronto ethics board.
Construction and expression of CYP2A6 constructs in E. coli
A bicistronic construct containing the full length cDNA of CYP2A6 followed by the human NADPH-cytochrome P450 reductase (hNPR) inserted into a pCW expression vector was generously provided by Dr. E. Gillam.43, 44 Two amino acid substitutions V110L and N438Y were introduced either alone or in combination to create CYP2A6.V110L, CYP2A6.35 (Asn438Tyr), and CYP2A6.24 (Val110Leu + Asn438Tyr), respectively. The commercial kits QuickChange II XL Site-Directed Mutagenesis and QuickChange Multi Site-Directed Mutagenesis were used to introduce the single and multiple amino acid substitutions respectively according to manufacturer’s directions (Stratagene, La Jolla, California, USA). Primers used to introduce Val110Leu (CYP2A6.V110L) were 2A6_594G>C and 2A6_594G>C Anti (table 4). Asn438Tyr (CYP2A6.35) was introduced using the primers 2A6_6458A>T and 2A6_6458A>T Anti (table 4). The combination of 2A6_594G>C Anti and 2A6_6458A>T Anti were used to construct CYP2A6.24 (Val110Leu+Asn438Tyr). Introduction of the correct changes to the variant constructs were confirmed by sequencing. In addition, a construct with 802 bp deleted from the CYP2A6 cDNA (CYP2A6.NEG) was used as a negative control, while a CYP2A6.17 construct served as a positive control.44 All five variant constructs (CYP2A6.NEG, CYP2A6.17, CYP2A6.V110L, CYP2A6.35, and CYP2A6.24) in addition to the wildtype CYP2A6 (CYP2A6.1) were expressed in E. coli and membrane fractions were prepared as described previously.44 Membrane protein content was determined by the Bradford protein assay (Bio-Rad Labs, Mississauga, Canada) and the amount of CYP2A6 protein was determined by immunoblotting with a monoclonal CYP2A6 antibody (BD Biosciences, Mississauga, ON) as described previously.44
In vitro nicotine assay
Nicotine C-oxidation activity was assessed as previously described with minor modifications.44 The reaction mixture (final volume of 0.5 ml) contained E. coli membrane preparations (10 pmol CYP2A6), 20 nmol/l of expressed cytochrome b5 (Invitrogen; Burlington, Canada), 50 mmol/l Tris-HCL buffer (pH 7.4), 1 mmol/l NADPH (Sigma-Aldrich; Oakville, Canada), nicotine substrate (ranging from 2–100 μM), and 1.0 mg protein/ml human liver cytosol (added in excess so that CYP-mediated oxidation would be rate limiting). The reaction was initiated by the addition of 1 mmol/l NADPH following a 2 min preincubation at 37°C. After 15 min incubation, the reaction was stopped by the addition of 100 μM of Na2CO3. Cotinine formation was determined by high-pressure liquid chromatography as described previously.45 Cotinine formation increased linearly with CYP2A6 protein concentrations (5–70 pmoles) and time (5–30 min) at 100 and 500 μM of nicotine (data not shown). The minimal amounts of cotinine detected in the reaction with CYP2A6.NEG were used as baseline correction to control for any contamination of the substrate or cotinine formation by the E. coli system. The kinetic parameters (Vmax and Km) were estimated from the high affinity site using the Michaelis-Menten equation in the computer program GraphPad Prism (GraphPad Software Inc., version 5.0 for windows)
In vitro thermal stability assay
To test the thermal stability of the different CYP2A6 constructs, E. coli membrane preparations were incubated at 37°C for 24 hrs. The experiment was carried out twice independently. On one day, single aliquots were collected, while on the other day duplicate aliquots were collected. Data was consistent within and between days. Aliquots were taken at 0, 4, 8, 12, and 24 hrs to measure nicotine C-oxidation activity. Activity assays were carried out with 30 μM of nicotine as described above. In addition, aliquots were taken at 0, 8, 12, and 24 hrs to measure CYP2A6 protein levels by immunoblotting.
Statistical analyses
Hardy-Weinberg equilibrium was tested using Chi-square, or Fisher’s exact test if five or fewer individuals were in one genotype group. Since gender and smoking status significantly influence the 3HC/COT ratio in the African Canadian population 46 and other populations,47–49 we controlled for gender and smoking. The African American population were all smokers, thus we only controlled for gender. Throughout the manuscript the 3HC/COT ratio is presented as the mean adjusted value. The adjustment for the impact of gender and smoking status was made as previously described.15 Briefly, the metabolic ratio for each individual was adjusted by dividing the 3HC/COT value with the overall mean from their respective group. For example, the 3HC/COT of each female nonsmoker was divided by the overall mean of their respective group (i.e. female nonsmokers). The mean adjusted ratios were not used in any statistical analyses. Instead we used the raw 3H/COT values and controlled for the effects of gender and smoking using a regression model. We adopted a forward stepwise regression strategy in which gender and/or smoking status were in the first block and CYP2A6 genotype in the second block. The reference for CYP2A6 genotype in the regression analyses were those with wildtype genotype (CYP2A6*1/*1) and the dependent variable was log 3HC/COT. The wildtype genotype group (CYP2A6*1/*1) included individuals with undetected CYP2A6 variants and the wild-type variant CYP2A6*1B. The CYP2A6*1B allele was included in the wildtype group (CYP2A6*1/*1) for two main reasons. Firstly, the SNPs in CYP2A6*35 are found on the CYP2A6*1B background, meaning they are in linkage with the 58bp gene conversion found in CYP2A6*1B (table 1). Secondly, CYP2A6*1B is a wildtype allele usually included in the wildtype reference group in the literature15, 18, along with other variants of the CYP2A6*1 allele (CYP2A6*1B to*1K, listed at http://www.cypalleles.ki.se/cyp2a6.htm). In order to compare the impact CYP2A6*35 to other variants in the literature we have included the individuals with CYP2A6*1B in the wildtype (CYP2A6*1/*1) group. The 3HC/COT values were log normalized as the 3HC/COT values were not normally distributed.
In vitro kinetic parameters (Vmax, Km, Vmax/Km) of the variant CYP2A6 constructs were compared to the wildtype CYP2A6 construct using one-way analysis of variance followed by the Bonferroni post hoc test. The half-life (t1/2) for the thermally induced loss of CYP2A6 activity was determined according to the one phase decay equation (GraphPad Prism Version 5). The t1/2 values reported herein are a measure of the thermal activity of the enzyme as previously determined 17, 50 and not the t1/2 of the CYP2A6 protein. Time courses of multiple experiments performed were analyzed in concert to obtain the value of the rate of decay k; which was subsequently used to determine the half life (t1/2 = ln2/k). By fixing the value of k obtained for the wildtype (CYP2A6.1), the changes in the absolute sum of squares of the variant constructs were compared to their baseline fits to determine the level of significance. All analyses were performed on the collected data, while the data were normalized as the percentage remaining activity for presentation. All p-values are one tailed with p<0.05 considered statistically significant. Statistical analyses were performed using SPSS (SPSS Inc., version 15.0 for Windows) and GraphPad Prism (GraphPad Software Inc., version 5.0 for windows).
Acknowledgments
This study was supported by the Centre for Addiction and Mental Health, Canadian Institutes for Health Research (CIHR) MOP86471, and CA91912 (JSA). N.K. receives funding from CIHR-Strategic Training Program in Tobacco use in Special Population (TUSP) and Ontario Graduate Scholarship program (OGS). R.F.T. holds a Canada Research Chair in Pharmacogenetics. We thank Bin Zhao, Ewa B. Hoffmann, Qian Zhou, Jill C. Mwenifumbo, Man Ki Ho, and Evan Dorey for their technical assistance, Rabindra Shivnaraine for his valuable input in data analyses, and Sandy Faheim for her careful review of the manuscript. We also thank Dr. Elizabeth Gillam for her generous gift of the CYP2A6 construct and Dr. Linda Ashworth for generously providing us with cosmid DNA clones 19296, 19019, and 27292 that contain CYP2A6, CYP2A7, and CYP2A13. Dr. Rachel F. Tyndale and Dr. Edward M. Sellers hold shares in Nicogen Research Inc., a company that is focused on novel smoking cessation treatment approaches. None of the data contained in this manuscript alters or improves any commercial aspect of Nicogen and no Nicogen funds were used in this work.
References
- 1.Benowitz NL. Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction. Clin Pharmacol Ther. 2008;83(4):531–541. doi: 10.1038/clpt.2008.3. [DOI] [PubMed] [Google Scholar]
- 2.Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56(5):483–493. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 3.Minematsu N, Nakamura H, Furuuchi M, Nakajima T, Takahashi S, Tateno H, et al. Limitation of cigarette consumption by CYP2A6*4, *7 and *9 polymorphisms. Eur Respir J. 2006;27(2):289–292. doi: 10.1183/09031936.06.00056305. [DOI] [PubMed] [Google Scholar]
- 4.Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79(6):600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14(9):615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther. 2008;84(3):320–325. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24(11):1212–1217. [PubMed] [Google Scholar]
- 8.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;282(3):1608–1614. [PubMed] [Google Scholar]
- 9.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277(2):1010–1015. [PubMed] [Google Scholar]
- 10.Dempsey D, Tutka P, Jacob P, 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Yamanaka H, Nakajima M, Nishimura K, Yoshida R, Fukami T, Katoh M, et al. Metabolic profile of nicotine in subjects whose CYP2A6 gene is deleted. Eur J Pharm Sci. 2004;22(5):419–425. doi: 10.1016/j.ejps.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Lea RA, Dickson S, Benowitz NL. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J Anal Toxicol. 2006;30(6):386–389. doi: 10.1093/jat/30.6.386. [DOI] [PubMed] [Google Scholar]
- 13.Benowitz NL, Jacob P, 3rd, Jones RT, Rosenberg J. Interindividual variability in the metabolism and cardiovascular effects of nicotine in man. J Pharmacol Exp Ther. 1982;221(2):368–372. [PubMed] [Google Scholar]
- 14.Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80(5):457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Mwenifumbo JC, Al Koudsi N, Ho MK, Zhou Q, Hoffmann EB, Sellers EM, et al. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat. 2008;29(5):679–688. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 16.Mwenifumbo JC, Tyndale RF. Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 2007;8(10):1385–1402. doi: 10.2217/14622416.8.10.1385. [DOI] [PubMed] [Google Scholar]
- 17.Ariyoshi N, Sawamura Y, Kamataki T. A novel single nucleotide polymorphism altering stability and activity of CYP2a6. Biochem Biophys Res Commun. 2001;281(3):810–814. doi: 10.1006/bbrc.2001.4422. [DOI] [PubMed] [Google Scholar]
- 18.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, et al. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80(3):282–297. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Peamkrasatam S, Sriwatanakul K, Kiyotani K, Fujieda M, Yamazaki H, Kamataki T, et al. In vivo evaluation of coumarin and nicotine as probe drugs to predict the metabolic capacity of CYP2A6 due to genetic polymorphism in Thais. Drug Metab Pharmacokinet. 2006;21(6):475–484. doi: 10.2133/dmpk.21.475. [DOI] [PubMed] [Google Scholar]
- 20.Xu C, Rao YS, Xu B, Hoffmann E, Jones J, Sellers EM, et al. An in vivo pilot study characterizing the new CYP2A6*7, *8, and *10 alleles. Biochem Biophys Res Commun. 2002;290(1):318–324. doi: 10.1006/bbrc.2001.6209. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida R, Nakajima M, Watanabe Y, Kwon JT, Yokoi T. Genetic polymorphisms in human CYP2A6 gene causing impaired nicotine metabolism. Br J Clin Pharmacol. 2002;54(5):511–517. doi: 10.1046/j.1365-2125.2002.01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, Tyndale RF. Association of CYP2A6 genotype and phenotype with smoking behaviors and treatment outcomes in African-American light smokers. Clin Pharmacol Ther. 2008 doi: 10.1038/clpt.2009.19. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwahashi K, Waga C, Takimoto T. Whole deletion of CYP2A6 gene (CYP2A6AST;4C) and smoking behavior. Neuropsychobiology. 2004;49(2):101–104. doi: 10.1159/000076418. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Amemo K, Ameno S, Iwahashi K, Kinoshita H, Kubota T, et al. Lack of association between smoking and CYP2A6 gene polymorphisms in A Japanese population. Nihon Arukoru Yakubutsu Igakkai Zasshi. 2001;36(5):486–490. [PubMed] [Google Scholar]
- 25.Tan W, Chen GF, Xing DY, Song CY, Kadlubar FF, Lin DX. Frequency of CYP2A6 gene deletion and its relation to risk of lung and esophageal cancer in the Chinese population. Int J Cancer. 2001;95(2):96–101. doi: 10.1002/1097-0215(20010320)95:2<96::aid-ijc1017>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Lewis DF, Dickins M, Lake BG, Eddershaw PJ, Tarbit MH, Goldfarb PS. Molecular modelling of the human cytochrome P450 isoform CYP2A6 and investigations of CYP2A substrate selectivity. Toxicology. 1999;133(1):1–33. doi: 10.1016/s0300-483x(98)00149-8. [DOI] [PubMed] [Google Scholar]
- 27.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12(9):822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 28.Johnson EF, Stout CD. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338(1):331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- 29.Juvonen RO, Iwasaki M, Negishi M. Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and site-directed mutagenesis. J Biol Chem. 1991;266(25):16431–16435. [PubMed] [Google Scholar]
- 30.Pitarque M, von Richter O, Rodriguez-Antona C, Wang J, Oscarson M, Ingelman-Sundberg M. A nicotine C-oxidase gene (CYP2A6) polymorphism important for promoter activity. Hum Mutat. 2004;23(3):258–266. doi: 10.1002/humu.20002. [DOI] [PubMed] [Google Scholar]
- 31.von Richter O, Pitarque M, Rodriguez-Antona C, Testa A, Mantovani R, Oscarson M, et al. Polymorphic NF-Y dependent regulation of human nicotine C-oxidase (CYP2A6) Pharmacogenetics. 2004;14(6):369–379. doi: 10.1097/00008571-200406000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Salavaggione OE, Wang L, Wiepert M, Yee VC, Weinshilboum RM. Thiopurine S-methyltransferase pharmacogenetics: variant allele functional and comparative genomics. Pharmacogenet Genomics. 2005;15(11):801–815. doi: 10.1097/01.fpc.0000174788.69991.6b. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics. 2003;13(9):555–564. doi: 10.1097/01.fpc.0000054124.14659.99. [DOI] [PubMed] [Google Scholar]
- 34.Weinshilboum R, Wang L. Pharmacogenetics: inherited variation in amino acid sequence and altered protein quantity. Clin Pharmacol Ther. 2004;75(4):253–258. doi: 10.1016/j.clpt.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Bandiera S, Weidlich S, Harth V, Broede P, Ko Y, Friedberg T. Proteasomal degradation of human CYP1B1: effect of the Asn453Ser polymorphism on the post-translational regulation of CYP1B1 expression. Mol Pharmacol. 2005;67(2):435–443. doi: 10.1124/mol.104.006056. [DOI] [PubMed] [Google Scholar]
- 36.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11 (4):400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 37.Fujieda M, Yamazaki H, Saito T, Kiyotani K, Gyamfi MA, Sakurai M, et al. Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis. 2004;25(12):2451–2458. doi: 10.1093/carcin/bgh258. [DOI] [PubMed] [Google Scholar]
- 38.Mwenifumbo JC, Lessov-Schlaggar CN, Zhou Q, Krasnow RE, Swan GE, Benowitz NL, et al. Identification of novel CYP2A6*1B variants: the CYP2A6*1B allele is associated with faster in vivo nicotine metabolism. Clin Pharmacol Ther. 2008;83(1):115–121. doi: 10.1038/sj.clpt.6100246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman SM, Fernandez-Salguero P, Gonzalez FJ, Mohrenweiser HW. Organization and evolution of the cytochrome P450 CYP2A-2B-2F subfamily gene cluster on human chromosome 19. J Mol Evol. 1995;41(6):894–900. doi: 10.1007/BF00173169. [DOI] [PubMed] [Google Scholar]
- 40.Ahluwalia JS, Okuyemi K, Nollen N, Choi WS, Kaur H, Pulvers K, et al. The effects of nicotine gum and counseling among African American light smokers: a 2 × 2 factorial design. Addiction. 2006;101(6):883–891. doi: 10.1111/j.1360-0443.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 41.Mwenifumbo JC, Sellers EM, Tyndale RF. Nicotine metabolism and CYP2A6 activity in a population of black African descent: impact of gender and light smoking. Drug Alcohol Depend. 2007;89(1):24–33. doi: 10.1016/j.drugalcdep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Ho MK, Mwenifumbo JC, Zhou Q, Hoffmann EB, Okuyemi K, Ahluwalia JS, Benowitz NL, Tyndale RF. CYP2A6 activity and its association with baseline smoking behaviors and treatment outcomes in a clinical trial of African-American light smokers. Society for Research on Nicotine and Tobacco (SRNT 14th Annual Meeting); Portland, Oregon. 2008. p. 87. [Google Scholar]
- 43.Gillam EM, Aguinaldo AM, Notley LM, Kim D, Mundkowski RG, Volkov AA, et al. Formation of indigo by recombinant mammalian cytochrome P450. Biochem Biophys Res Commun. 1999;265(2):469–472. doi: 10.1006/bbrc.1999.1702. [DOI] [PubMed] [Google Scholar]
- 44.Ho MK, Mwenifumbo JC, Zhao B, Gillam EM, Tyndale RF. A novel CYP2A6 allele, CYP2A6*23, impairs enzyme function in vitro and in vivo and decreases smoking in a population of Black-African descent. Pharmacogenet Genomics. 2008;18(1):67–75. doi: 10.1097/FPC.0b013e3282f3606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siu EC, Wildenauer DB, Tyndale RF. Nicotine self-administration in mice is associated with rates of nicotine inactivation by CYP2A5. Psychopharmacology (Berl) 2006;184(3–4):401–408. doi: 10.1007/s00213-006-0306-6. [DOI] [PubMed] [Google Scholar]
- 46.Mwenifumbo JC, Sellers EMRFT. The association of demographic characteristics with CYP2A6 mediated nicotine metabolism in a population of black African descent. Drug and Alcohol Dependence. 2006 doi: 10.1016/j.drugalcdep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Johnstone E, Benowitz N, Cargill A, Jacob R, Hinks L, Day I, et al. Determinants of the rate of nicotine metabolism and effects on smoking behavior. Clin Pharmacol Ther. 2006;80(4):319–330. doi: 10.1016/j.clpt.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, et al. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80(3):282–297. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 49.Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther. 2006;79 (5):480–488. doi: 10.1016/j.clpt.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura K, Ariyoshi N, Yokoi T, Ohgiya S, Chida M, Nagashima K, et al. CYP2D6.10 present in human liver microsomes shows low catalytic activity and thermal stability. Biochem Biophys Res Commun. 2002;293(3):969–973. doi: 10.1016/S0006-291X(02)00328-5. [DOI] [PubMed] [Google Scholar]