Abstract
The Andes are the most species-rich global biodiversity hotspot. Most research and conservation attention in the Andes has focused on biomes such as rain forest, cloud forest, and páramo, where much plant species diversity is the hypothesized result of rapid speciation associated with the recent Andean orogeny. In contrast to these mesic biomes, we present evidence for a different, older diversification history in seasonally dry tropical forests (SDTF) occupying rain-shadowed inter-Andean valleys. High DNA sequence divergence in Cyathostegia mathewsii, a shrub endemic to inter-Andean SDTF, indicates isolation for at least 5 million years of populations separated by only ca. 600 km of high cordillera in Peru. In conjunction with fossil evidence indicating the presence of SDTF in the Andes in the late Miocene, our data suggest that the disjunct small valley pockets of inter-Andean SDTF have persisted over millions of years. These forests are rich in endemic species but massively impacted, and merit better representation in future plans for science and conservation in Andean countries.
Keywords: biogeography, dispersal limitation, Neotropics, phylogenetic niche conservatism, phylogeography
Geotemporal patterns of historical species assembly in the world's biodiversity hotspots and especially in species-rich tropical biomes are poorly understood. Many questions remain to be answered about how the biota of biodiversity hotspots were assembled and how comparable different hotspots are. To gain insights into these questions, we investigate plant species diversification in the tropical Andes, which contain ca. 40,000 plant species, ≈15% of the global total, in only 1% of the world's land area. Most of this diversity is found in the lowland and midelevation mesic forests of the Andean flanks. The “excess” of species in these forests largely explains why the Neotropics contain more plant species than both tropical Africa and Asia, and was hypothesized by Gentry (1, 2) to have been produced by recent, rapid species diversification coinciding with the accelerated orogeny of the Andes over the past 10 million years. Gentry's recent-species diversification model has received support from several dated phylogenies of plant clades with distributions centered in Andean mesic forests (e.g., refs. 3–5). In other Andean biomes such as the páramo, other instances of recent speciation have been documented, including one of the most rapid episodes of plant species diversification (6), although this is perhaps not surprising because these high-elevation habitats are geologically recent, dating from the Pliocene–Pleistocene. Considerable conservation attention has been rightly given to all these hyperdiverse Andean habitats that represent cradles of ongoing diversification (e.g., refs. 7 and 8). Here we present evidence for a very different and more ancient diversification history in Andean seasonally dry tropical forest (SDTF), a biome relatively neglected by scientists and conservationists in the Andes.
SDTF is found in scattered areas throughout the Neotropics from Mexico to Argentina that receive a 4–6-mo dry season too severe for rain forest species (9, 10). SDTF is deciduous or semideciduous in the dry season, often rich in cacti and other succulents, and with few grasses in the ground layer (Fig. 1). In the Andes, it occurs in valleys between ca. 500 m and ca. 2,500 m altitude, where a rain shadow effect creates highly seasonal rainfall (11). These valleys are separated physically by higher areas of the cordillera, which are clothed in more mesic vegetation: midelevation montane forest (MMF) from ca. 2,300 m to 3,400 m and high-altitude grasslands (HAG; páramo, puna, and jalca) above ca. 3,000 m. More occasionally, dry lowland valleys are separated ecologically by intervening lowland areas that receive much higher rainfall. Because of the fertile soils and pleasant dry climate, SDTF, including inter-Andean valleys, has been impacted by human activities over millennia (12), and it is the most threatened tropical forest biome (13).
Fig. 1.
Cactus-rich seasonally dry tropical forest in the Marañon valley, Peru.
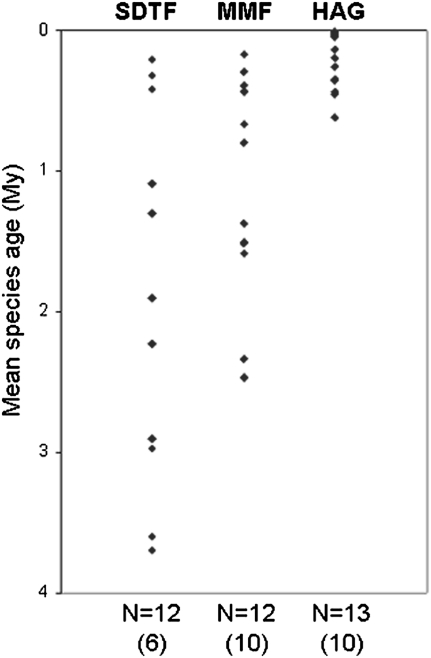
Remarkable late Miocene (8–12 Ma) fossils from southern Ecuador include multiple plant taxa characteristic of SDTF that are highly similar to or even potentially conspecific with extant Andean SDTF species (e.g., in the genera Tipuana, Loxopterygium, and Ruprechtia) (14, 15). These fossils imply that Andean SDTF existed in the Ecuadorean Andes before the final, rapid phase of the Andean orogeny during the late Miocene, ca. 10 Ma (16, 17). The age of Andean SDTF is further suggested by Miocene–Pliocene ages of plant lineages endemic to the inter-Andean valleys in Ecuador, Peru, and Bolivia (18). Using the limited number of published, dated phylogenetic trees for Andean plants (Table S1), a comparative examination of ages of species diversification in lineages endemic to HAG, MMF, and SDTF indicates that SDTF contains the oldest species and lineages with significantly longer times for speciation, in stark contrast to the HAG biome, which shows significantly younger mean species ages and lineages with higher rates of species diversification (Fig. 2; two-tailed t test, P = 0.001; Fig. S1). We believe that the differences between SDTF and MMF would be more marked if dated phylogenies were available for the species-rich genera that are characteristic of MMF (e.g., Calceolaria, Ceratostema, and Bomarea) in which mean species ages might be younger. In contrast, the Andean SDTFs harbor few species-rich clades (19), and we suggest our sample to be representative.
Fig. 2.
Ages of species diversification in lineages endemic to the tropical Andes. Crown node ages are divided by species number to give a mean species age per lineage. Lineages were classified into three major Andean biomes: SDTF, MMF, and HAG. Number of data points (N) refers to the number of lineages identified from published dated phylogenies, and the number in parentheses refers to the number of phylogenies these data points were derived from.
These old SDTF species may have been affected by the final phase of the Andean orogeny. Mountain-building could have either separated into smaller pockets a once more-widespread SDTF formation, or caused rain shadow effects that created suitable conditions for drought-adapted SDTF species in some valleys. The relative antiquity of Andean SDTF, its long-term separation in different valleys, and lack of dispersal between them may also explain the startlingly high plant species endemism in some of these valleys (e.g., 38% of plant species in the SDTF of the Marañon valley, Peru) (19).
Here we use phylogenetic dating to examine whether lineages of Cyathostegia mathewsii (Benth.) Schery (Leguminosae-Swartzieae) found in separate inter-Andean valleys (Fig. 3) are old enough for their biogeography to have been influenced by the final phase of the Andean uplift. C. mathewsii is a shrub or small tree that grows at 500–2,300 m in present-day Andean SDTF (20, 21). It is not known in the fossil record. We examine whether its widespread distribution reflects recent dispersal between valleys or is a more ancient pattern of dispersal limitation caused by the Andean orogeny, using time-calibrated molecular phylogenetic analyses based on plastid matK and nuclear ribosomal internal transcribed spacer (ITS) sequences generated from individuals of C. mathewsii sampled from throughout the species’ range. We deliberately biased our phylogenetic dating analyses to younger dates to test most rigorously the hypothesis that the distribution of C. mathewsii reflects dispersal limitation caused in part by the Andean orogeny.
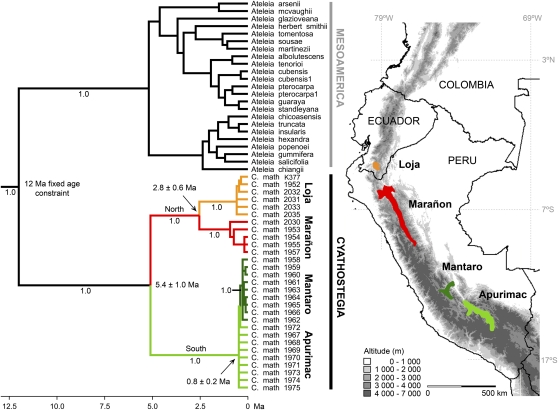
Fig. 3.
Chronogram for Cyathostegia derived from penalized likelihood rate smoothing of a 50% majority rule Bayesian likelihood tree estimated from the ITS sequence data. Clade labels (Loja, Marañon, Mantaro, Apurimac) indicate the dry inter-Andean valleys where accessions were collected, the locations of which are shown on the map. Numbers (1.0) below branches indicate posterior probabilities.
C. mathewsii was previously considered endemic to the Loja depression of southern Ecuador and the Marañon valley of northern Peru, but fieldwork carried out as part of this study has extended the known range of the species to also include the Apurimac and Mantaro valleys considerably further south in Peru (Fig. 3). The Mantaro and Apurimac areas are adjacent, separated only by ca. 200 km of lowland mesic vegetation. They are separated from the Marañon area by ca. 600 km of high Andean cordillera that reaches altitudes of 4,000–5,000 m. The Marañon valley is also separated from the Loja depression by high cordillera, although in this area some passes reach only ca. 3,000 m. The hypothesis that the biogeography of C. mathewsii has been molded by the Andean orogeny predicts that the depth of genetic divergence should reflect the height and date of mountain barriers separating the SDTF areas. We test this prediction that suggests recent dispersal will have been most likely across the lowland mesic forest separating the Apurimac and Mantaro areas, but will have been unlikely across the high Andean cordilleras that separate Loja from Marañon, and these areas from Mantaro/Apurimac.
Results and Discussion
The ITS phylogenetic tree (Fig. 3) has a topology consistent with long isolation of populations of C. mathewsii in all inter-Andean valleys except the Apurimac and Mantaro. Northern (Loja/Marañon) and southern (Apurimac/Mantaro) populations are reciprocally monophyletic, a signature that there has been sufficient time for lineage sorting (22–24). Within the northern group, Loja and Marañon populations are also reciprocally monophyletic, indicating another ancient split. In contrast, the Mantaro accessions are nested within a clade that also includes the Apurimac accessions, rendering the latter paraphyletic, a signature of recency—either of recent colonization from the Apurimac to the Mantaro or recent vicariance of a wider population in this area. The matK-trnK phylogenetic tree (Figs. S2 and S3) corroborates the ITS results, showing strongly supported monophyly for each of the Loja, Marañon, and Apurimac/Mantaro populations. Such population level resolution is remarkable for the relatively slowing evolving matK-trnK locus that is often used to resolve relationships among genera.
The fossil-calibrated matK rates analysis of all Leguminosae estimated the mean age of the crown node (most recent common ancestor) of C. mathewsii as 9.4 Ma (SD 2.3), and of the stem node (its divergence from Ateleia) as 16.6 Ma (SD 1.8) (Table 1). Sampling of matK was sufficient to estimate the age the Apurimac–Mantaro crown at 1.4 Ma (SD 0.7). These age estimates are derived from rates of substitution (Table 1) that are at least three times faster than those reported by Lavin et al. (ref. 25; nodes 14–15), thus suggesting that they are biased young.
Table 1.
Penalized likelihood age and rate estimates derived from analysis of matK sequences of all Leguminosae and ITS data
| Clade | Mean age (matK) | SD (matK) | Mean age (ITS) | SD (ITS) |
| Cyathostegia stem | 16.6 | 2.3 | 12 (fixed) | — |
| Cyathostegia crown | 9.4 | 1.8 | 5.4 | 1.0 |
| Loja crown | 1.8 | 1.0 | 0.8 | 0.3 |
| Mantaro–Apurimac crown | 1.4 | 0.7 | 0.8 | 0.2 |
| Mantaro crown | — | — | 0.4 | 0.1 |
| Clade | Mean age (matK) | SD (matK) | Mean rate (ITS) | SD (ITS) |
| Cyathostegia stem | 0.0018 | 0.0002 | — | — |
| Cyathostegia crown | 0.0019 | 0.0002 | 0.0118 | 0.0017 |
| Loja crown | 0.0019 | 0.0002 | 0.0112 | 0.0018 |
| Mantaro–Apurimac crown | 0.0019 | 0.0002 | 0.0122 | 0.0018 |
| Mantaro crown | — | — | 0.0128 | 0.0017 |
Age is reported as million years ago (Ma) and rate as substitution per site per Ma. See Fig. 3 for clade designations.
The ITS rates analysis yielded an age estimate for the Cyathostegia crown node, which represents the divergence of southern (Mantaro/Apurimac) from northern populations (Marañon/Ecuador) of 5.4 Ma. The Ecuadorean populations diverged from the Marañon populations 2.8 Ma, and the Apurimac–Mantaro crown is 0.8 million years old. That these ages are likely to have been biased young is indicated by the elevated substitution rate of ca. 1.1–1.2 × 10−8 substitutions per site per year (ssy). This rate exceeds those reported for herbs by Kay et al. (26), who reviewed ITS substitution rates in 28 angiosperm groups. Herbs would be expected to have a faster substitution rate than a woody plant such as Cyathostegia because of their faster generation time (26, 27). To approximate rates of ITS substitution estimated for woody plants including tropical legumes (ca. 2.0–4.0 × 10−9 ssy; ref. 26), the Cyathostegia stem age would need to be set at 35 million years, and the estimated age of the Cyathostegia crown would then be 18 million years.
Recent studies suggest that the Andean orogeny consisted of long phases of elevational stability, separated by rapid (1–4 million years) changes of 1,500 m or more (16). The last such rapid change is thought to have started in the central Andes ca. 10 Ma (16, 17), and the dates calculated for divergences in Cyathostegia are consistent with this late Miocene–Pliocene period of rapid uplift resulting in the isolation of separate populations of C. mathewsii. Mountain chains greater than 2,500 m would present a dispersal barrier for C. mathewsii based on its current ecology because it does not grow at altitudes greater than ca. 2,300 m (21). Multiple proxies indicate that the Central Andean Plateau (currently ca. 4,000 m) and its surrounding cordilleras (up to 6,000 m) were only at approximately half their current elevation 10 Ma (16), before this phase of rapid uplift commenced. As predicted, the depth of genetic divergence between populations of C. mathewsii reflects the height of the altitudinal barrier to dispersal. The valleys separated by high mountains (Loja, Marañon, and Apurimac/Mantaro) contain well-supported monophyletic groups of populations subtended by long branches (Fig. 3 and Figs. S2, and S3). In the ITS tree, the deepest divergence is found between the southern (Apurimac/Mantaro) and northern (Marañon/Ecuador) populations because these are separated by mountains of 4,000–5,000 m altitude. If these mountains were half their current elevation 10 Ma, then they already would have presented a barrier to C. mathewsii soon after the late Miocene onset of mountain-building, which is corroborated by our dates. The Mantaro and Apurimac populations, separated by only ca. 200 km of lowland rain forest vegetation and no mountains, are shown by our data to have been connected much more recently, perhaps during drier climates of glacial periods of the Pleistocene, which would have favored spread of SDTF at the expense of rain forest (9). A Pleistocene scenario is consistent with the age of migration from the Apurimac to the Mantaro, which is estimated with ITS data as between 0.8 Ma (Apurimac–Mantaro crown) and 0.4 Ma (Mantaro crown; Fig. 3 and Table 1). These young ages are in stark contrast to the older dates for the Cyathostegia crown of 5.4 Ma (ITS) and 9.4 Ma (matK).
Although age estimation based on branch lengths on phylogenetic trees is approximate, even our biased minimum age estimates based on ITS suggest that northern and southern populations of C. mathewsii have been isolated for at least 5 million years. The old monophyletic clades and coalescence within small inter-Andean valleys suggests long persistence of small, isolated, endemic SDTF populations. The inter-Andean SDTF system has been remarkably persistent, fragmented, and dispersal-limited for long geological periods. Field observations, coupled with inventory data (e.g., ref. 28), demonstrate that C. mathewsii and many other species endemic to inter-Andean valley SDTF are locally abundant within individual valleys. This local abundance may reflect ecological drift to higher abundances, as predicted in a system that is little perturbed by immigration (29, 30). The restriction of historical immigration by Andean mountain barriers may be reinforced in SDTF by phylogenetic niche conservatism (e.g., ref. 31) because the SDTF biome may only be open to immigration by lineages that already have drought adaptations to survive a seasonal environment (32, 33), adaptations which are lacking in species from adjacent Amazonian rain forest and Andean cloud forest biomes. Furthermore, the SDTF ecosystem, unlike tropical savannas, is not prone to fire disturbance in the absence of humans, so saturation of the woody plant community (sensu ref. 29) may further restrict immigration. The ecology of SDTF may therefore accentuate the historically high degree of dispersal limitation and exert influence on phylogenetic geographic structure, which may be more marked in this biome than in rain forests (where no drought adaptations are required) and tropical savannas (which are disturbed by fire).
The deep DNA sequence divergences in C. mathewsii are consistent with a view of SDTF as a biome that has been remarkably persistent in isolated valleys in the Andes since the late Miocene. This view is further corroborated by the large number of endemic species found in some of these valleys, which we suggest have accumulated by a gradual process of allopatric speciation in evolutionarily persistent small patches of SDTF.
The Andean biodiversity hotspot has been highlighted as an evolutionary cradle of recent plant speciation (1, 2, 4–6), but within it we suggest that different biomes have contrasting diversification histories and that the history of this hotspot is complex. The Andean SDTF biome is distinct from neighboring biomes, and on the basis of the presence of late Miocene lineages such as those of C. mathewsii in separate dry valleys, it represents a museum of relatively ancient Andean biodiversity and a possible model system for studying the effect of vicariant or dispersal limitation processes on plant populations. Much attention has been given to the conservation of the Andean biodiversity hotspot (e.g., by Conservation International; http://www.biodiversityhotspots.org/xp/hotspots/andes/Pages/conservation.aspx;), but the evolutionary distinctiveness of the SDTF biome within the Andes has been neglected. Some of the most species-rich SDTF areas, such as the Marañon valley in Peru, are, to date, biologically very poorly known and entirely unprotected. We hope that further understanding of the distinct diversification history of SDTF may add impetus to its urgent conservation. Given scenarios of future warmer, drier climates in some areas of the tropical Andes (34), plants such as C. mathewsii, which have adapted to drought and have survived upheavals of Andean uplift and the vicissitudes of Pleistocene climate fluctuations over million-year time scales, may represent important resources for the future.
Methods
Datasets and Sampling.
We sequenced 29 accessions of C. mathewsii (Table S2), collected from diverse locations in Mantaro (9 accessions), Apurimac (9 accessions), Marañon (5 accessions), and Loja (6 accessions) for the ITS region, including the 5.8S subunit, according to methods of Schrire et al. (35). To assess the monophyly of C. mathewsii and to ensure that age estimation was not biased by inadequate sampling of related taxa (cf. ref. 36) we sampled 23 ITS sequences representing 20 of 31 species of Ateleia, which is sister to Cyathostegia in phylogenetic studies involving chloroplast and nuclear DNA sequences and morphology (37–39). Analyses were rooted using ITS sequences representing three species of Trischidium, estimated as sister to Ateleia and Cyathostegia (37, 38). With respect to inferring species phylogenies, the utility of ITS sequences can be compromised by the presence of paralogs or pseudogenes within individuals (e.g., ref. 40). In Cyathostegia, Ateleia, and Trischidium, PCR products were sequenced directly in both the forward and reverse direction, and these contiged cleanly. The 5.8S region revealed no evidence of indels and divergent sequences, and therefore no evidence of pseudogenes and other paralogs, intra- and interindividual, was detected in Cyathostegia, Ateleia, or Trischidium.
In the absence of a fossil record for Cyathostegia or its sister genera Ateleia and Trischidium that could provide direct age constraints for a dating analysis using ITS, we used age estimates derived from a molecular rates analysis of matK sequences representing the entire Leguminosae (25). The original matK sequence data set of 335 sequences and 1,674 aligned sites (25) was expanded to 510 sequences by 1,713 aligned sites and included five additional sequences of Ateleia and Cyathostegia, contributing to a total of six accessions of each genus (Table S2). These accessions were chosen to sample the full phylogenetic and geographic diversity of Cyathostegia as determined by the analysis of ITS sequences here and of Ateleia as outlined in the molecular and morphological phylogeny of Ireland et al. (39). PCR and sequencing protocols were according to Wojciechowski et al. (41).
As a means corroborating the nuclear ITS phylogeny with an independent plastid dataset, we generated sequences for the matK gene and its flanking trnK introns for 15 accessions of C. mathewsii (4 Loja, 3 Marañon, 4 Apurimac, 4 Mantaro) and six Ateleia outgroup species. PCR and sequencing protocols were according to Wojciechowski et al. (41).
Details of taxon names, voucher samples, and GenBank accession numbers are presented in Table S2.
Phylogenetic Analyses and Dating.
Sequences were aligned manually using Se-Al (42). ModelTest 3.7 (43) was used to select the best-fitting substitution model. Markov chain Monte Carlo (MCMC) analyses were run using MrBayes (44) for 5,000,000 generations with four simultaneous MCMC chains, sampling one tree every 10,000 generations such that 500 trees were sampled. The initial trees discarded as burnin were identified using the sump command in MrBayes, which identifies the generation at which separate runs merge and successive samples are not autocorrelated. Maximum parsimony analyses were according to Ireland et al. (39).
In the matK dating analysis using the entire Leguminosae, the same 10 fossil constraints used by Lavin et al. (25) were used, and the root was fixed at 60 Ma, which represents the minimal age of the legume family (25). Fixing an older root age would cause all age estimates more distal in the tree to be older, however slightly (25). As such, age estimates reported in this study from matK are biased young. To calibrate the timing of divergences within Cyathostegia using the ITS phylogeny, we used the minimum age estimate gained from this matK analysis (12 Ma) for the divergence of Ateleia and Cyathostegia as a calibration point (Fig. 3 and Table 1). Because we wished to test the hypothesis that divergences in Cyathostegia are old enough to be explained by the Andean orogeny, our strategy was, therefore, to bias toward estimating younger ages. A second dating analysis for matK was carried out to investigate temporal congruence between markers using all 15 available sequences for C. mathewsii and the same 12 Ma root calibration (Fig. S3 and Table S3).
Because substitution rates in the matK and ITS phylogenies were identified as not rate constant with a likelihood ratio test, molecular dating analyses used the software package r8s (45) and the data-driven penalized likelihood rate-smoothing method and “pruning” the accessions of Trischidium from the trees. Input trees were derived from sampling 100 unautocorrelated trees at likelihood stationarity during a Bayesian analysis. The SD for age and rate estimates were derived from 100 Bayesian trees, thereby incorporating into the variance uncertainty in both topology and branch lengths.
Ages of Andean Plant Species Diversifications.
Published dated molecular phylogenies of endemic Andean plant lineages were identified using the Institute for Scientific Information Web of Science database. Lineages largely found in the flanking lowland (below ca. 2,300 m) mesic rain forest areas on the eastern Amazonian flanks and in the Chóco biogeographic region of Pacific coastal Ecuador and Colombia were not included. A total of 22 studies were identified that included 38 Andean lineages (Table S1). Each lineage was assigned to one of the three major biomes found in the Andean cordilleras according to Cuatrecasas (46): SDTF ca. 0–2,500 m elevation; MMF (e.g., cloud forests) ca. 2,300–3,400 m; and HAG (including Páramo, Puna, and Jalca) ca. 3,000–4,800 m. To categorize biomes for each Andean lineage, the elevational range of each species within a lineage was recorded from Brako and Zarucchi (47) and Jørgensen et al. (48). Elevational ranges of species were then used to plot a frequency distribution of numbers of species present in 500-m elevational bands, and the elevational range of maximum species diversity was used to categorize each lineage into a biome (Table S1). To account for species diversity within Andean lineages, crown node age estimates were divided by the number of species, to derive mean species ages for each Andean clade. We also calculated time for speciation—the inverse of net species diversification rate—for each lineage, using both linear and logarithmic tree models to account for variation in tree topology (49).
Supplementary Material
Acknowledgments
We thank Carlos Reynel, Aniceto Daza, and Alex Monro for help collecting plant material; the Peruvian and Ecuadorean authorities for permission to collect plant material; Susana Magallón for help with analyses and Alex Antonelli for data on Hedyosmum and Campanulaceae. R.T.P. was supported by Leverhulme Trust Study Abroad Fellowship RF/2/2006/0142 for a 6-mo study visit to M.L.’s laboratory, during which many of the ideas in this article were developed.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HM347452–HM347494).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001317107/-/DCSupplemental.
References
- 1.Gentry AH. Neotropical floristic diversity: Phytogeographical connections between Central and South America. Pleistocene climatic fluctuations or an accident of Andean orogeny? Ann Missouri Bot Gard. 1982;69:557–593. [Google Scholar]
- 2.Gentry AH. In: Biological Relationships Between Africa and South America. Goldblatt P, editor. New Haven, CT: Yale University Press; 1993. pp. 500–547. [Google Scholar]
- 3.Kay KM, Reeves PA, Olmstead RG, Schemske DW. Rapid speciation and the evolution of hummingbird pollination in neotropical Costus subgenus Costus (Costaceae): Evidence from nrDNA ITS and ETS sequences. Am J Bot. 2005;92:1899–1910. doi: 10.3732/ajb.92.11.1899. [DOI] [PubMed] [Google Scholar]
- 4.Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Recent and rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- 5.Särkinen TE, et al. Recent oceanic long-distance dispersal and divergence in the amphi-Atlantic rain forest genus Renealmia L.f. (Zingiberaceae) Mol Phylogenet Evol. 2007;44:968–980. doi: 10.1016/j.ympev.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Hughes C, Eastwood R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc Natl Acad Sci USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks TM, et al. Habitat loss and extinction in the hotspots of biodiversity. Conserv Biol. 2002;16:909–923. [Google Scholar]
- 8.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 9.Pennington RT, Prado DA, Pendry C. Neotropical seasonally dry forests and Pleistocene vegetation changes. J Biogeogr. 2000;27:261–273. [Google Scholar]
- 10.Pennington RT, Ratter JA, Lewis GP. In: Neotropical Savannas and Seasonally Dry Forests: Plant Diversity, Biogeography and Conservation. Pennington RT, Lewis GP, Ratter JA, editors. Boca Raton, FL: CRC Press; 2006. pp. 1–29. [Google Scholar]
- 11.Killeen TJ, Douglas M, Consiglio T, Jørgensen PM, Mejia J. Dry spots and wet spots in the Andean hotspot. J Biogeogr. 2008;34:1357–1373. [Google Scholar]
- 12.Piperno DR, Dillehay TD. Starch grains on human teeth reveal early broad crop diet in northern Peru. Proc Natl Acad Sci USA. 2008;105:19622–19627. doi: 10.1073/pnas.0808752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson DM, et al. The Global 200: A Representation Approach to Conserving the Earth's Distinctive Ecoregions. Washington, DC: Conservation Science Program, World Wildlife Fund-US; 2000. [Google Scholar]
- 14.Burnham RJ. A new species of winged fruit from the Miocene of Ecuador: Tipuana ecuatoriana (Leguminosae) Am J Bot. 1995;82:1599–1607. [Google Scholar]
- 15.Burnham RJ, Carranco N. Miocene winged fruits of Loxopterygium (Anacardiaceae) from the Ecuadorean Andes. Am J Bot. 2004;91:1767–1773. doi: 10.3732/ajb.91.11.1767. [DOI] [PubMed] [Google Scholar]
- 16.Garzione CN, et al. Rise of the Andes. Science. 2008;320:1304–1307. doi: 10.1126/science.1148615. [DOI] [PubMed] [Google Scholar]
- 17.Gregory-Wodzicki KM. Uplift history of the Central and Northern Andes: A review. Geol Soc Am Bull. 2000;112:1091–1105. [Google Scholar]
- 18.Pennington RT, et al. Historical climate change and speciation: neotropical seasonally dry forest plants show patterns of both tertiary and quaternary diversification. Philos Trans R Soc Lond B. 2004;359:515–537. doi: 10.1098/rstb.2003.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linares-Palomino R. In: Neotropical Savannas and Seasonally Dry Forests: Plant Diversity, Biogeography and Conservation. Pennington RT, Lewis GP, Ratter JA, editors. Boca Raton, FL: CRC Press; 2006. pp. 257–279. [Google Scholar]
- 20.Ireland H. PhD thesis. United Kingdom: University of Reading; 2001. The taxonomy and systematics of Ateleia and Cyathostegia (Leguminosae-Swartzieae) [Google Scholar]
- 21.Linares-Palomino R Annotated checklist of the woody plants in Peruvian seasonally dry forests. 2005. Available at: http://rbg-web2.rbge.org.uk/dryforest/database.htm.
- 22.Cunningham C, Collins T. In: Molecular Ecology and Evolution: Approaches and Applications. Schierwater B, Streit B, Wagner GP, DeSalle R, editors. Basel: Birkhäuser; 1994. pp. 405–433. [Google Scholar]
- 23.Cunningham C, Collins T. In: Molecular Approaches to Ecology and Evolution. DeSalle R, Schierwater B, editors. Basel: Birkhäuser; 1998. pp. 297–321. [Google Scholar]
- 24.Barraclough TG. Evolving entities: Towards a unified framework for understanding diversity at the species and higher levels. Philos Trans R Soc Lond B. 2010;365:1801–1813. doi: 10.1098/rstb.2009.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- 26.Kay KM, Whittall JB, Hodges SA. A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: An approximate molecular clock with life history effects. BMC Evol Biol. 2006;6:36. doi: 10.1186/1471-2148-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- 28.Bridgewater S, Pennington RT, Reynel C, Daza A, Pennington TD. A preliminary floristic and phytogeographic analysis of the woody flora of seasonally dry forests in northern Peru. Candollea. 2003;58:129–148. [Google Scholar]
- 29.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton: Princeton University Press; 2001. [Google Scholar]
- 30.Latimer AM, Silander JA, Jr, Cowling RM. Neutral ecological theory reveals isolation and rapid speciation in a biodiversity hot spot. Science. 2005;309:1722–1725. doi: 10.1126/science.1115576. [DOI] [PubMed] [Google Scholar]
- 31.Donoghue MJ. Colloquium paper: A phylogenetic perspective on the distribution of plant diversity. Proc Natl Acad Sci USA. 2008;105(Suppl 1):11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrire BD, Lavin M, Lewis GP. Global distribution patterns of the Leguminosae: Insights from recent phylogenies. Biol Skrift. 2005;55:375–422. [Google Scholar]
- 33.Pennington RT, Lavin M, Oliveira-Filho AT. Woody plant diversity, evolution and ecology in the tropics: Perspectives from seasonally dry tropical forests. Annu Rev Ecol Evol Syst. 2009;40:437–457. [Google Scholar]
- 34.Urrutia R, Vuille M. Climate change projections for the tropical Andes using a regional climate model: Temperature and precipitation simulations for the end of the 21st century. J Geophys Res. 2009 10.1029/2008JD011021. [Google Scholar]
- 35.Schrire B, Lavin M, Barker NP, Forest F. Phylogeny of the tribe Indigofereae (Leguminosae-Papilionoideae): Geographically structured more in succulent-rich and temperate settings than in grass-rich environments. Am J Bot. 2009;96:816–852. doi: 10.3732/ajb.0800185. [DOI] [PubMed] [Google Scholar]
- 36.Linder HP, Hardy CR, Rutschmann F. Taxon sampling effects in molecular clock dating: An example from the African Restionaceae. Mol Phylogenet Evol. 2005;35:569–582. doi: 10.1016/j.ympev.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Ireland H, Pennington RT, Preston J. In: Advances in Legume Systematics, Part 9. Herendeen PS, Bruneau A, editors. Kew, United Kingdom: Royal Botanic Gardens; 2000. pp. 217–231. [Google Scholar]
- 38.Torke BM, Schaal BA. Molecular phylogenetics of the species-rich neotropical genus Swartzia (Leguminosae-Papilionoideae) and related genera of the swartzioid clade. Am J Bot. 2008;95:215–228. doi: 10.3732/ajb.95.2.215. [DOI] [PubMed] [Google Scholar]
- 39.Ireland HE, et al. Biogeographic, ecological, and morphological structure of an Ateleia (Swartzieae-Leguminosae) phylogenetic analysis derived from combined molecular and morphological/chemical data. Bot J Linn Soc. 2010;162:39–53. [Google Scholar]
- 40.Bailey CD, Carr TG, Harris SA, Hughes CE. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol Phylogenet Evol. 2003;29:435–455. doi: 10.1016/j.ympev.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analyses of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- 42.Rambaut A. Se-Al, ver. 2.0a11, sequence alignment editor 1996. Available at: http://evolve.zoo.ox.ac.uk/evolve/software.html.
- 43.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 44.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 46.Cuatrecasas J. In: Tropical Botany. Larsen K, Holm-Nielsen LB, editors. London: Academic Press; 1979. pp. 397–410. [Google Scholar]
- 47.Brako L, Zarucchi JL. Catalogue of the Flowering Plants and Gymnosperms of Peru. St. Louis, MO: Missouri Botanical Garden; 1993. [Google Scholar]
- 48.Jørgensen PM, Neill DA, León-Yánez S. Catalogue of the Vascular Plants of Ecuador. St. Louis, MO: Missouri Botanical Garden; 1999. [Google Scholar]
- 49.McCune AR. In: Molecular Evolution and Adaptive Radiation. Givnish TJ, Sytsma KJ, editors. Cambridge, UK: Cambridge University Press; 1997. pp. 585–610. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.