Abstract
Our understanding of how mesodermal tissue is formed has been limited by the absence of specific and reliable markers of early mesoderm commitment. We report that mesoderm commitment from human embryonic stem cells (hESCs) is initiated by epithelial-to-mesenchymal transition (EMT) as shown by gene expression profiling and by reciprocal changes in expression of the cell surface proteins, EpCAM/CD326 and NCAM/CD56. Molecular and functional assays reveal that the earliest CD326−CD56+ cells, generated from hESCs in the presence of activin A, BMP4, VEGF, and FGF2, represent a multipotent mesoderm-committed progenitor population. CD326−CD56+ progenitors are unique in their ability to generate all mesodermal lineages including hematopoietic, endothelial, mesenchymal (bone, cartilage, fat, fibroblast), smooth muscle, and cardiomyocytes, while lacking the pluripotency of hESCs. CD326−CD56+ cells are the precursors of previously reported, more lineage-restricted mesodermal progenitors. These findings present a unique approach to study how germ layer specification is regulated and offer a promising target for tissue engineering.
Keywords: CD326, CD56, epithelial-to-mesenchymal transition, mesenchyme, hematopoiesis
The stages of early embryogenesis can be recapitulated during in vitro differentiation of human embryonic stem cells (hESCs) (1). Although hESCs are increasingly used in developmental biology and tissue engineering research, studies with hESCs are generally focused on the development of defined, fully differentiated tissues rather than the earliest events of germ-layer specification.
It is well established that hESCs have the potential to generate multiple mesodermal derivatives after several days in culture with specific morphogens (1). Cardiac and hematoendothelial progenitors can be identified during mouse and human ESC differentiation using the expression of the cell surface markers KDR (aka VEGFR2 or Flk-1) and CD34 (2–5). A primitive streak-like population generated from human hESCs was also isolated using simultaneous detection of the platelet-derived growth factor receptor alpha (PDGFR-α) protein and a Mixl-1–driven GFP reporter (6). The Mixl-1+PDGFR-α+ cells were shown to possess hematopoietic potential, whereas generation of cardiovascular and mesenchymal cell potential was not tested. As KDR expression was not described, it remains unclear whether PDGFR-α and KDR proteins are coexpressed on the emerging mesodermal progenitors or mark different subsets of mesoderm-committed cells. As yet, no population generated from hESC has been identified marking a stage in which full mesodermal potential still exists.
Epithelial-to-mesenchymal transition (EMT) occurs through a complex process that produces changes in tissue architecture, cell morphology, adhesion, and migratory capacity (7). In early embryogenesis, EMT plays a pivotal role during gastrulation and formation of the germ layers. In amniotes, mesoderm is the predominant germ layer formed via EMT (8). Although there are multiple genes associated with EMT progression, no reliable nonepithelial surface markers that identify cells undergoing this process have been reported to date. It was recently reported that the loss of E-cadherin expression during EMT is associated with up-regulation of neuronal cell adhesion molecule (NCAM)/CD56 in human epithelial breast carcinoma cells (9). Ablation of CD56 expression inhibited cell spreading and EMT, whereas forced expression of CD56 resulted in epithelial cell delamination and migration (9).
Similar to many other epithelial systems, hESCs usually establish apical–basal polarity in relation to the feeder layers on which they are cocultured (10, 11), and associate with each other through adhesion molecules such as E-cadherin and EpCAM (CD326) (11, 12), which have also been shown to be required for maintenance of pluripotency (13, 14). We hypothesized that the earliest stage of mesoderm commitment from pluripotent hESCs would be represented by a coordinated up-regulation of CD56 expression during the loss of epithelial cell adhesion.
Results
Identification of a CD56+ Subpopulation Generated from hESCs During Early Mesoendodermal Induction.
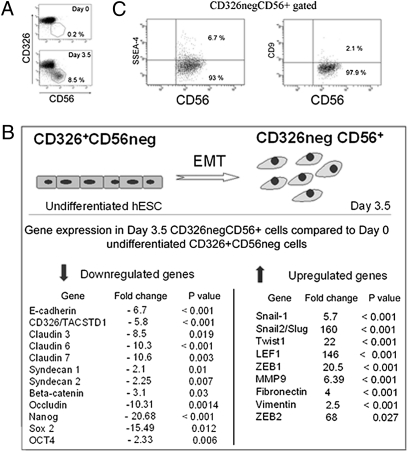
To explore the process of mesodermal commitment in the context of EMT, we used candidate cell surface markers to identify populations generated from hESCs during the earliest stages of differentiation. The epithelial marker CD326 was uniformly expressed at high levels in undifferentiated cells from three different human embryonic stem cell lines (H9, H1, and HES3), whereas CD56 was not expressed in undifferentiated hESCs (Fig. 1A). By day 3.5 of differentiation in mesoendoderm induction conditions, a population marked by loss of CD326 expression and acquisition of CD56 (CD326−CD56+) was clearly detectable (Fig. 1A). Emerging CD56+ cells were organized in clusters (Fig. S1A), and after FACS isolation attached to Matrigel-coated plates and exhibited polygonal morphology (Fig. S1B).
Fig. 1.
Generation of day 3.5 CD326−CD56+ cells from hESCs occurs during the process of EMT. (A) Flow cytometry analysis of CD326 and CD56 expression in undifferentiated hESCs (H9) at day 0, and cells generated from hESCs after 3.5 d in A-BVF induction conditions. (B) Microarray analysis demonstrates changes in gene expression typical of EMT during the generation of the day 3.5 CD32−CD56+ population from undifferentiated hESCs. (C) FACS analysis of day 3.5 CD326−CD56+ cells showing down-regulation of expression of SSEA4 and CD9, two cell surface markers associated with pluripotent phenotype.
The efficiency and timing of the generation of CD326−CD56+ cells from hESCs was tested using various mesoendoderm-inducing signals (Fig. S1 C and D). A small population (∼3%) of CD326−CD56+ cells was spontaneously produced without the addition of any exogenous morphogens. The most efficient production of CD326−CD56+ cells was accomplished with the combination of BMP4, VEGF, and bFGF and a transient (1 d) exposure to activin A (“A-BVF conditions”) (n = 3, P < 0.001) (Fig. S1 C and E). Activin/nodal signaling was critical for the generation of CD326−CD56+ cells; the presence of the ALK 4/5/7 inhibitor SB-431542 significantly reduced the number of CD326−CD56+ cells generated from hESCs (Fig. S1F, P < 0.001). The optimal A-BVF conditions (Fig. S1C) were used for all subsequent experiments.
hESCs Undergo EMT During the Generation of CD326−CD56+ Cells.
Microarray analysis confirmed that the population of cells that down-regulate CD326 and acquire CD56 cell surface expression at day 3.5 of culture had undergone the process of EMT. Significant up-regulation of the key transcriptional regulators of EMT, Snail-1, Snail-2, and Twist coincided with down-regulation of E-cadherin and CD326 and tight junction-related genes such as claudins, syndecans, and occludins (Fig. 1B). A significant reorganization of cytoskeletal proteins was documented on the basis of vimentin up-regulation (Fig. 1B). Up-regulation of fibronectin expression was also seen (Fig. 1B), consistent with the role this extracellular matrix protein plays in facilitating cell migration. More global analysis of the microarray data showed a total of 1,623 genes were differentially expressed fivefold or more between the CD326−CD56+ population and hESCs, (signal difference ≥ 100, P < 0.05), of which 851 genes were up-regulated and 772 down-regulated (please see normalized data at the publicly open database NCBI GEO, record GSE21668 at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21668). Significantly enriched biofunctional groups of genes differentially expressed between the hESCs and CD326−CD56+ cells are presented in Fig. S2A. The quality control analysis of the microarray data are shown in Fig. S2 B–D.
Gene Expression and Functional Assays Demonstrate That the Day 3.5 CD326−CD56+ Population Has Lost Pluripotency.
Flow cytometry demonstrated that CD9 and SSEA-4, cell surface markers often used to identify undifferentiated cells, were down-regulated in the CD326−CD56+ population (Fig. 1C). Microarray analysis of CD326−CD56+ cells also demonstrated significant down-regulation of Nanog (20-fold, P < 0.001), Sox-2 (15-fold, P = 0.012) and Oct-4/Pou5f1 (2-fold, P = 0.006) (Fig. 1B), three key transcriptional factors associated with the pluripotency of hESCs. Microarray data were additionally validated with real-time PCR (Fig. S3A).
Evidence that the pluripotency of undifferentiated hESCs was lost during generation of the CD326−CD56+ population was further verified using an in vivo teratoma formation assay. No tumor formation was observed 2 mo after injection of day 3.5 CD326−CD56+ sorted cells, whereas undifferentiated hESCs produced teratomas in all tested animals (n = 6 animals in each arm, in two independent experiments) (Fig. S3 B and C).
The Day 3.5 CD326−CD56+ Population Generates Cells Expressing KDR, PDGFR-α, and CD34.
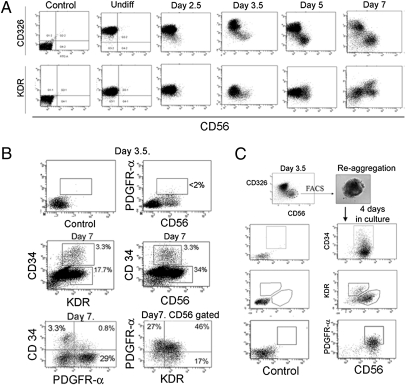
The onset of expression of known mesodermal markers relative to the generation of CD326−CD56+ cells was next examined. KDR, PDGFR-α, and CD34 are three markers previously reported to identify hematoendothelial or cardiac cells during early hESC differentiation (3, 4, 6). Flow cytometric analysis indicated that undifferentiated cells of all three tested hESC lines uniformly expressed low levels of KDR (Fig. 2A), but had no detectable PDGFR-α or CD34 expression. At day 3.5 after mesoderm induction, KDR expression was down-regulated (Fig. 2A) and protein expression levels of PDGFR-α (Fig. 2B) and CD34 remained undetectable on the CD326−CD56+ population. Other markers of more differentiated mesodermal derivatives, e.g., CD45, CD31, CD117, VE-cadherin, CD105, and CD73 were also not detectable on the day 3.5 CD326−CD56+ population. After 4 d of mesoderm induction (day 5 cultures), KDR (Fig. 2A), CD34, and PDGFR-α began to be detected on small subsets within the CD326−CD56+ population. By day 7, several different subpopulations could be distinguished on the basis of KDR, PDGFR-α, and CD34 expression. A total of 52.1 ± 6.3% of the CD326−CD56+ cells (∼20% of total cells) could be readily identified as KDRhi (Fig. 2A). Within the KDRhi fraction, 15–20% of cells (∼3–4% of total cells) coexpressed CD34 by day 7 (Fig. 2B). A large subset of CD326−CD56+ cells (47.9 ± 7.4%) showed no up-regulation of either CD34 or KDR expression throughout all 7 d of culture, but did express PDGFR-α at high levels (Fig. 2B). Another subset of PDGFR-α+ cells expressed KDR at high levels but did not express CD34. A subset of CD34+ cells expressed low but detectable levels of PDGFR-α, consistent with previous reports (6). Thus, although the percentage of CD326−CD56+ cells peaked at day 14 of induction culture, from day 5 onward they represented a highly heterogeneous pool of cells. It should be noted that the differentiation conditions used did not involve generation of embryoid bodies (EB). Thus differentiation kinetics of CD56+ cells may be different from those observed when differentiation is induced by EB formation.
Fig. 2.
Time course analysis of expression of the early mesodermal markers KDR, PDGFR-α, and CD34 in the CD326−CD56+ population. (A) Time course of CD326 and CD56 expression in A-BVF induction conditions (Top). At day 3.5 of culture, CD326−CD56+ cells were detectable as a distinct population demonstrating lower levels of KDR (Bottom). The control panel shows undifferentiated hESCs that have not been stained with antibody. (B) PDGFR-α and CD56 expression on ungated day 3.5 cells generated in A-BVF (Top). CD34, KDR, and CD56 expression on ungated day 7 cells (Middle row and Lower Left panel). PDGFR-α and KDR expression on CD56+ gated cells at day 7 (Lower Right). (C) Subpopulations marked by increased KDR, CD34, and PDGFR-α expression were generated from the CD326−CD56+ population. Shown are FACS analyses of H9 hESC-derived cultures, representing one of three experiments conducted on different passages of hESCs (using also HES3 and H1 lines). Controls are unstained samples.
To confirm that the various KDR-, PDGFR-α–, and CD34-expressing populations that appeared by day 7 were generated from the CD326−CD56+ population, CD326−CD56+ cells were isolated by flow cytometry at day 3.5 of A-BVF induction and reaggregated for further differentiation (Fig. 2C). After an additional 4 d of culture in the presence of BMP4, VEGF, and bFGF, the aggregates were dissociated and analyzed by FACS. Subpopulations marked by increased KDR, PDGFR-α, and CD34 expression were generated from the day 3.5 CD326−CD56+ population (Fig. 2C). Thus the day 3.5 CD326−CD56+ population is a precursor of other reported mesodermal phenotypes previously identified with the surface markers KDR, PDGFR-α, and CD34. It should be noted that although large numbers of CD326−CD56+ cells continued to be generated for at least 14 d of culture (Fig. S1C), immunophenotypically they were otherwise dissimilar to day 3.5 cells as more than 90% of day 14 CD326−CD56+ cells had high expression of PDGFR-α and coexpressed mesenchymal stem cell markers CD73 and CD105.
CD326−CD56+ Cells Generated by Day 3.5 of Culture Have Committed to the Mesodermal Germ Layer.
As the CD326−CD56+ population precedes and generates cells expressing the mesodermal markers KDR, PDGFR-α, and CD34, we next explored the mesoderm specificity of the CD326−CD56+ immunophenotype. Microarray analysis of gene expression in day 3.5 CD326−CD56+ cells, indicated consistent and dramatic up-regulation of genes essential for the earliest stages of mesoderm specification, including T, Tbx-6, Snail-1, and Mesp-1 and -2 (Table 1).
Table 1.
Gene expression analysis demonstrates mesoderm commitment in CD326−CD56+ cells
| Mesoderm |
Ectoderm |
Endoderm |
||||||
| Gene | Fold change | P value | Gene | Fold change | P value | Gene | Fold change | P value |
| T | 14.41 | 0.001 | PAX6 | 1.35 | NS | AFP | 0.68 | NS |
| MIXL1 | 32.18 | 0.026 | NES | 1.27 | NS | FOXA2 | 1.64 | NS |
| SNAI1 | 5.76 | 0.000 | PAX7 | 1.05 | NS | ALB | 1.17 | NS |
| SNAI2 | 160.1 | 0.000 | SOX1 | 0.53 | NS | SOX17 | 11.61 | 0.011 |
| HLX | 40.24 | 0.003 | MSI1 | 1.33 | NS | GATA4 | 2.91 | NS |
| EOMES | 49.63 | 0.001 | NKX2-2 | 0.87 | NS | FOXA1 | 1.62 | NS |
| MESP1 | 74.19 | 0.017 | NKX6-1 | 1.90 | NS | PDX1 | 0.52 | NS |
| MESP2 | 16.25 | 0.001 | FOXP2 | 0.59 | NS | TMPRSS4 | 0.30 | NS |
| TBX6 | 3.41 | 0.045 | FOXD3 | 0.17 | NS | CLIC6 | 0.60 | NS |
| MEST | 2.44 | 0.005 | SOX10 | 1.55 | NS | RIPK4 | 0.95 | NS |
| NKX2-5 | 35.67 | 0.040 | ZIC1 | 1.02 | NS | CLDN8 | 0.66 | NS |
| KDR | 2.09 | 0.008 | TUBB3 | 1.02 | NS | ST14 | 0.07 | 0.007 |
Microarray analysis of undifferentiated hESCs (H9) vs. CD326−CD56+ cells isolated from A-BVF induction conditions at day 3.5. Fold change represents the ratio of transcript levels in CD326−CD56+ cells/day 0 hESCs. Values shown are average fold changes of separate cell isolations from three independent experiments for each cell type. P < 0.05 was considered statistically significant.
In contrast, no pattern of either ectodermal- or endodermal-specific differentiation was found in gene expression analyses of the CD326−CD56+ cells (Table 1 and Fig. S4).
Using differentiation systems previously shown to be efficient for ectodermal or endodermal specification (15, 16), the nonmesodermal potential of the CD326−CD56+ population was next examined. Neither endoderm- (Fig. S4A) nor ectoderm- (Fig. S4B) associated genes were up-regulated in cultures initiated by day 3.5 CD326−CD56+ cells. Similarly, cells expressing endodermal proteins (FOXA-2, α-Feto-Protein [AFP]), and ectodermal proteins (Pax-6 and Sox-1) were not detectable in cultures initiated by day 3.5 CD326−CD56+ cells, whereas robust generation of both endodermal and neuroectodermal cells was achieved in the same conditions from undifferentiated hESCs (Fig. S4C). Thus, consistent with the gene expression profile, the CD326−CD56+ phenotype produced by day 3.5 in A-BVF conditions, identifies an early stage of hESC differentiation, specifically committed to the mesoderm germ layer.
A subset of cells that retained the original CD326+CD56− phenotype showed progressive down-regulation of the marker SSEA4 beginning at day 3.5. The pluripotency genes Nanog, OCT-4, and Sox-2 were significantly down-regulated by day 3.5 (Fig. S5A). By day 14 of A-BFV culture, the endodermal genes HNF4-α, AFP, GATA-4, and GATA-6 were up-regulated in cells that remained CD326+ (Fig. S5C). In the same conditions, CD326+ cells showed either no change or down-regulation of ectoderm- and mesoderm-associated genes (Fig. S5 B and D). These data suggest that in the A-BVF conditions, the persistence of CD326 most likely defines primitive and visceral endoderm.
The CD326−CD56+ Population Can Generate All Mesodermal Lineages.
Having established that the process of EMT defines the loss of pluripotency and mesodermal commitment, an exhaustive analysis was conducted to test the full mesodermal potential of the day 3.5 CD326−CD56+ population. The TGF-β inhibitor SB-431542 has been shown in recent studies to enhance the generation of endothelial cells from hESCs (17). In the current study we found that TGF-β inhibition also enhanced generation of hematopoietic cells (fivefold, compared with control) from the day 3.5 CD326−CD56+ population (Fig. S6). SB-914542 was thus added in combination with cytokines and OP9 stroma coculture to optimize the assay of hematoendothelial differentiation potential (SI Materials and Methods).
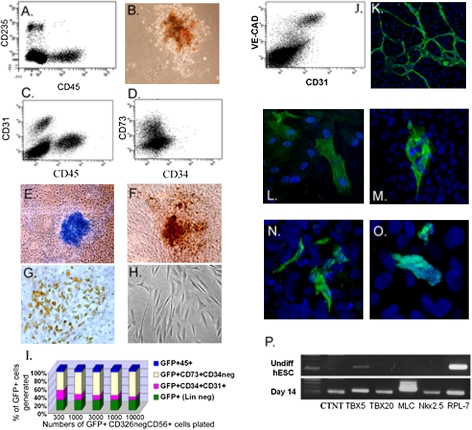
The day 3.5 CD326−CD56+ population generated robust cultures of hematopoietic and endothelial cells on the basis of both morphology and expression of hematopoietic cell surface markers (CD45 and CD235) or endothelial markers (CD31 and VE-cadherin) in stromal coculture and semisolid assays (Fig. 3 A–C, J and K and Fig. S6 B–E). The CD45+ hematopoietic cells coexpressed dim levels of surface CD31, whereas CD45 expression was largely absent on CD34+ cells (Fig. S6 B, C, and E).
Fig. 3.
The CD326−CD56+ population can generate all mesodermal lineages. CD326−CD56+ cells were isolated at day 3.5 from hESC cultures in A-BVF conditions and placed in specific differentiation conditions to test mesoderm lineage potential. (A–C) Hematopoietic and endothelial potential as shown by: (A) CD45+ and CD235+ cells generated after 2 wk of culture on OP9 stroma. (B) Erythroid colony generated in semisolid culture. (C) Generation of CD31+CD45− endothelial cells on OP9 stroma. (D) Generation of CD73+CD34− mesenchymal cells from day 3.5 CD326−CD56+ cells after an additional 7-d culture in Matrigel. (E–H) Mesenchymal potential as shown by histochemical staining of (E) cartilage (Alcian blue), (F) bone (von Kossa staining), and (G) adipose (Oil Red O) tissues. (H) Fibroblastic cells generated in day 7 culture. (I) Proportion of GFP+ cells, produced after 14 d from day 3.5 GFP+CD326−CD56+ cells, which represent hematopoietic, endothelial, or mesenchymal lineages based on flow cytometry. Data were compiled from limiting dilution analysis as described in Fig. S8. (J and K) Endothelial potential was confirmed, shown on FACS plot (J) by generation of CD31+VE-cadherin+ endothelial cells after 2 wk in culture. (K) The formation of branching capillaries in Matrigel-coated chamber slides, expressing CD31 (green). (L) Smooth muscle potential shown as cells expressing α-SMA (green). (M–P) Cardiac potential of CD326−CD56+ cells. (M) Myosin heavy chain, (N) CTNT, and (O) α-actinin protein (all as green) expressing cells at day 14 of culture. TOPRO3-staining (blue) showed cell nuclei. (P) Expression of genes involved in cardiomyocyte specification demonstrated by semiquantitative RT-PCR at day 14 of culture. RPL-7 is a housekeeping gene. Shown are data from H9, HES3, and H1 lines. Magnification 200× in B–H and 400× in K, N, and O).
Endothelial differentiation was confirmed with LDL-Dil-Ac uptake studies and immunohistochemical staining for e-NOS and von Willebrand factor.
When cultured either on Matrigel-coated plates (Fig. 3D) or cocultivated on OP9 stroma, CD326−CD56+ cells could also efficiently generate cells that expressed the mesenchymal marker CD73 and did not express CD34. In contrast to the endothelial cells, CD73+ cells expressed high levels of PDGFR-α as well as the mesenchymal markers PDGFR-β, CD146, CD10, CD105, and CD166, but had little or no KDR expression (Fig. S7). To test further whether the CD73+CD34− population represented mesenchymal progenitors, we isolated the CD73+CD34− cells from Matrigel or OP9 cultures and replated them into osteogenic, chondrogenic, and adipogenic culture conditions. After an additional 3 wk of culture, generation of bone, cartilage, and adipocytes was demonstrated, confirming the presence of a separate mesenchymal branch of differentiation from the CD326−CD56+ population (Fig. 3 E–H).
To assess smooth muscle and cardiac potential, the day 3.5 CD326−CD56+ cells were isolated by FACS and reaggregated in large cell clumps and then transferred to gelatin in cardiovascular differentiation conditions. After 10–14 d of culture, smooth muscle actin (α-SMA) (Fig. 3L) and H-caldesmon expressing smooth muscle cells were generated. These cultures also contained endothelial cells that expressed CD31 and VE-cadherin (Fig. 3 J and K). Cardiomyocyte differentiation was demonstrated by myosin heavy chain (Fig. 3M), CTNT (Fig. 3N), and α-actinin expression (Fig. 3O). Based on CTNT expression, the percentage of cardiomyocytes in these cultures was ≈5–10%. Further confirmation of cardiac differentiation was shown by detection of message for the transcriptional factors TBX5, TBX20, and Nkx2.5, as well as transcripts for muscle-associated proteins myosin (MLC2A) and CTNT (Fig. 3P). Beating colonies of cardiomyocytes were generated from CD326−CD56+ cells by 11–14 d of culture (Movie S1).
The generation of endothelial and blood lineages was respectively ∼30- and 50-fold more efficient from cultures initiated with CD326−CD56+ cells than the reciprocal CD326+CD56− population (Fig. S8A), confirming that mesodermal potential is highly enriched in the CD326−CD56+ population.
For a more quantitative assessment of hematopoietic, endothelial, and mesenchymal cell potential, day 3.5 CD326−CD56+ cells were generated using an hESC subclone, which stably expresses GFP. GFP+ day 3.5 CD326−CD56+ cells were seeded onto OP9 cells in serial dilution after reaggregation with GFP−CD326−CD56+ cells from the H9 parent line. High differentiation efficiency was simultaneously demonstrated for all tested mesodermal lineages (Fig. S8B). Of note, as maximal numbers of hematopoietic and mesenchymal cells are generated in different culture conditions to those optimal for the endothelial lineage, simultaneous culture of all three lineages does not represent the maximal efficiency for each. In addition, cardiomyocyte differentiation conditions are sufficiently different to prevent simultaneous readout of this lineage using this assay. Nonetheless, after 14 d in the conditions tested, more than 80% of all GFP+ cells generated from CD326−CD56+ cells represented either mesenchymal, hematopoietic, or endothelial cells (Fig. 3I).
Discussion
We report the identification and characterization of a unique population of human embryonic mesodermal progenitors (hEMP), which arise from hESCs through the process of EMT. These events can be tracked through the combined loss of the epithelial adhesion marker EpCAM/CD326 and up-regulation of NCAM/CD56.
Several transcription factors that orchestrate EMT during epiblast-to-mesoderm transition in vivo in the primitive streak during gastrulation (18, 19), are also involved in the initial stages of mesodermal differentiation of hESCs. The gene expression profile of CD326−CD56+ cells showed profound loss of the epithelial genes encoding tight junctions and adherin junction proteins responsible for belt-like structures at the lateral interface of epithelial cells. The down-regulation of epithelial genes coincided with a dramatic up-regulation of Snail-1, Snail-2, Twist, Eomes, Mesp-1 and -2, and other transcriptional regulators of EMT.
It has recently been found that the loss of E-cadherin during EMT is associated with up-regulation of expression of CD56 in human epithelial breast carcinoma cells (9). The finding that CD56 is also up-regulated during mesodermal differentiation of hESCs suggests that CD56 is likely to be fundamental to the reorganization of cell assembly rather than specific to mesodermal commitment per se. Expression of CD56 has also been documented in neuroectodermal progenitors derived from hESCs (20). Thus, the significance of surface markers is context specific and may differ dramatically in different culture conditions.
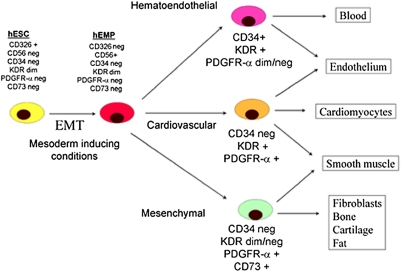
The generation from hESCs of several progenitor populations with limited mesodermal potential has been previously described using combinations of markers of hematoendothelial (CD34 and PDGFR-α), cardiovascular (KDR+ CD117− cells), and mesenchymal (CD73) differentiation (3, 4, 6, 21). We now demonstrate that day 3.5 CD326−CD56+ cells represent the earliest multipotent mesodermal progenitors reported to date, being generated before the appearance of the previously described populations and capable of giving rise to all of the above progenitor cell phenotypes and related mesodermal lineages. On the basis of these data, a schema can be proposed of the lineage relationships between more restricted mesoderm progenitors that are generated later in culture (Fig. 4).
Fig. 4.
Proposed model of cell surface marker expression during mesodermal specification from hESCs. The initial stage of mesoderm commitment is marked by the process of EMT during which the CD326−CD56+ population is generated. Subsequent commitment to mesoderm populations with more restricted potential is identified by day 7 of induction cultures by differential expression of the surface markers KDR, PDGFR-α, CD34, and CD73. The phenotype of precursors to the day 7 populations shown is yet to be delineated.
A mesoendodermal stage of mouse ES differentiation has been previously proposed (22). Up-regulation of Sox-17 seen in our analyses of CD326−CD56+ cells may suggest that this population is derived from a stage that possesses both mesodermal and endodermal potential. In addition to its role in endoderm differentiation, Sox-17 has been shown to be essential for mesoderm specification (23). Sox-17 has also been recently shown to play an important role in the maintenance of fetal and neonatal, but not adult hematopoietic stem cells (24). Thus, the finding of up-regulation of Sox-17, but not other endodermal genes in the CD326−CD56+ population, (Table 1), and the lack of functional evidence of endodermal potential (Fig. S4) is entirely consistent with the assignment of mesoderm commitment to this population.
The separation of hEMP from hESCs and other germ layers at an early stage of differentiation allows their manipulation in more defined culture conditions than those present within the bulk colonies of differentiating hESCs or embryoid bodies. The isolation of hEMP may also allow generation of functional tissue units or niches, composed of, for example, hematopoietic cells, endothelium, and supportive mesenchymal stroma, recapitulating the microenvironmental interactions present during normal embryogenesis.
In conclusion, this study has demonstrated the existence of a primitive population of hEMP, generated from hESCs by the process of epithelial-to-mesenchymal transition. The CD326−CD56+ cells emerge before more lineage-restricted mesodermal populations, when full mesodermal potential still exists, thereby providing a unique opportunity for understanding the mechanisms regulating mesoderm specification in humans. Isolation and manipulation of this population in controlled and defined culture conditions may also significantly improve existing protocols for tissue engineering.
Materials and Methods
hESC lines H1, H9, and HES3 obtained from WiCell were maintained on irradiated primary mouse embryonic fibroblasts (MEFs) in medium recommended by WiCell. hESCs were routinely characterized and found to have a normal karyotype and expression of pluripotency markers SSEA4, CD9, OCT4, and alkaline phosphatase. To induce mesoderm differentiation, colonies of H1, H9, or HES3 were cut into uniform-sized pieces using the StemProEZPassage tool (Invitrogen), transferred into 6-well plates precoated for 1 h with Matrigel (growth factor reduced, no phenol red; BD Biosciences), and cultured initially in TESR medium (Stem Cell Technologies) until 50–60% confluent (typically 2 d). To induce differentiation, TESR medium was replaced with basal induction medium Stemline II (Sigma-Aldrich). Basal induction medium was supplemented with optimal concentrations of human VEGF, bFGF, BMP4, and activin A (all at 10 ng/mL; R&D Systems, see SI Materials and Methods for details) (A-BVF conditions). After mesoderm induction, CD326−CD56+ cells were typically isolated by fluorescence activated cell sorting (FACS) at day 3.5 (unless otherwise described) and further differentiated into mesodermal lineages in hematoendothelial, cardiac, or mesenchymal stem cell conditions (SI Materials and Methods). Differentiation potential was assessed with microarray analysis, PCR (Table S1), FACS, morphology, and immunohistochemistry (SI Materials and Methods).
Supplementary Material
Acknowledgments
We acknowledge the University of California Los Angeles Clinical Microarray Core and Broad Stem Cell Research Center FACS Core, Mr. Ben Van Handel, and Mr. Tan Ke Duong for their excellent technical assistance and Dr. Don Kohn for his thoughtful contributions to this manuscript. This research was made possible by California Institute for Regenerative Medicine Grant RC1-00108-1 and support from a University of California Los Angeles Broad Stem Cell Research Center award.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002077107/-/DCSupplemental.
References
- 1.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy M, D'Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 5.Kataoka H, et al. Expressions of PDGF receptor alpha, c-Kit and Flk1 genes clustering in mouse chromosome 5 define distinct subsets of nascent mesodermal cells. Dev Growth Differ. 1997;39:729–740. doi: 10.1046/j.1440-169x.1997.t01-5-00009.x. [DOI] [PubMed] [Google Scholar]
- 6.Davis RP, et al. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- 7.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 8.Tam PP, Behringer RR. Mouse gastrulation: The formation of a mammalian body plan. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- 9.Lehembre F, et al. NCAM-induced focal adhesion assembly: A functional switch upon loss of E-cadherin. EMBO J. 2008;27:2603–2615. doi: 10.1038/emboj.2008.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krtolica A, et al. Disruption of apical-basal polarity of human embryonic stem cells enhances hematoendothelial differentiation. Stem Cells. 2007;25:2215–2223. doi: 10.1634/stemcells.2007-0230. [DOI] [PubMed] [Google Scholar]
- 11.Ullmann U, et al. GSK-3-specific inhibitor-supplemented hESC medium prevents the epithelial-mesenchymal transition process and the up-regulation of matrix metalloproteinases in hESCs cultured in feeder-free conditions. Mol Hum Reprod. 2008;14:169–179. doi: 10.1093/molehr/gan001. [DOI] [PubMed] [Google Scholar]
- 12.Ng VY, Ang SN, Chan JX, Choo AB. Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells. 2010;28:29–35. doi: 10.1002/stem.221. [DOI] [PubMed] [Google Scholar]
- 13.González BDS, Denzel S, Mack B, Conrad M, Gires O. EpCAM is involved in maintenance of the murine embryonic stem cell phenotype. Stem Cells. 2009;27:1782–1791. doi: 10.1002/stem.97. [DOI] [PubMed] [Google Scholar]
- 14.Lu TY, et al. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J Biol Chem. 2010;285:8719–8732. doi: 10.1074/jbc.M109.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasonkin IO, Koliatsos VE. Nonhuman sialic acid Neu5Gc is very low in human embryonic stem cell-derived neural precursors differentiated with B27/N2 and noggin: Implications for transplantation. Exp Neurol. 2006;201:525–529. doi: 10.1016/j.expneurol.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Borowiak M, et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James D, et al. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFbeta inhibition is Id1 dependent. Nat Biotechnol. 2010;28:161–166. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 19.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundberg M, et al. CD marker expression profiles of human embryonic stem cells and their neural derivatives, determined using flow-cytometric analysis, reveal a novel CD marker for exclusion of pluripotent stem cells. Stem Cell Res (Amst) 2009;2:113–124. doi: 10.1016/j.scr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Lee EJ, et al. Novel embryoid body-based method to derive mesenchymal stem cells from human embryonic stem cells. Tissue Eng Part A. 2010;16:705–715. doi: 10.1089/ten.tea.2008.0596. [DOI] [PubMed] [Google Scholar]
- 22.Tada S, et al. Characterization of mesendoderm: A diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.