DNA is susceptible to diverse types of damage throughout the cell cycle including cross-linking, oxidation, and adduct formation. DNA is especially prone to breaks during S phase, when non-B forms of DNA, such as hairpins or triplexes, can appear in the course of replication or extended regions of single-strand DNA (ssDNA) can arise by polymerase–helicase uncoupling. When genomic DNA is damaged, eukaryotic cells restrain their cell cycle progress while the damage is repaired. This so-called checkpoint mechanism (1), first described in the 1970s (2), is crucial for maintenance of genome integrity. In the ensuing years, genetic studies have contributed dramatically to understanding checkpoint mechanisms to the point where it is believed that most, if not all, proteins involved in the main checkpoint signaling pathways have been discovered. A central question remaining in this area is how the cell recognizes and transduces the diverse noncanonical structures of DNA into a checkpoint response. The paper by Choi et al. (3) in PNAS represents a major advance in this field, and it describes the in vitro assembly of a human checkpoint system from defined components.
The protein kinases ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) are at the apices of the checkpoint pathways and function as sensors to detect damaged DNA, stalled replication forks, or intermediates of DNA repair (4). The ATR checkpoint pathway (Fig. 1) is believed to begin by direct loading of ATR and its essential binding partner ATR interacting protein (ATRIP) onto replication protein A (RPA)-coated ssDNA. On UV-induced DNA damage, stimulation of the ATR pathway results in the phosphorylation of the effector kinase CHK1 and multiple additional substrates that stabilize replication forks and slow the cell cycle. Because RPA-ssDNA is a structural intermediate common to several DNA metabolic processes, the ATR pathway can detect diverse DNA lesions. A distinct set of proteins is responsible for activating ATM, primarily in response to DNA double-strand breaks, although cross-talk exists between the ATR and ATM pathways (4).
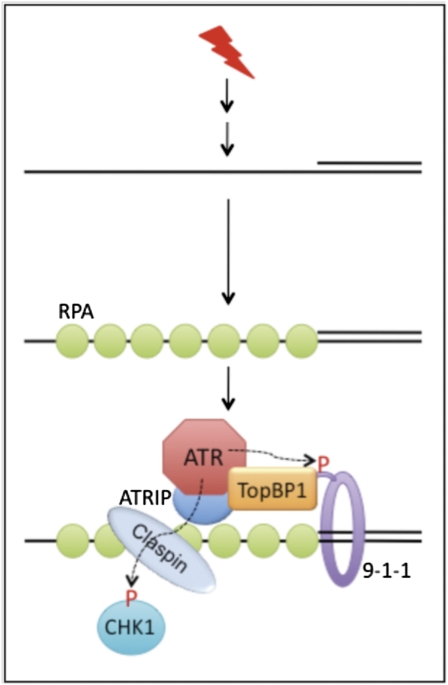
Fig. 1.
The conserved ATR→CHK1 checkpoint signaling pathway in eukaryotes (4, 17). ssDNA can be formed as an intermediate structure during DNA repair or DNA replication. RPA binds to ssDNA, which then recruits ATR-ATRIP by interacting with ATRIP. RPA may also work with RAD17-RFC to load the 9-1-1 (RAD9-RAD1-HUS1) checkpoint clamp to the 5′ recessed junction. TopBP1 interacts with ATRIP-ATR and phosphorylated Rad9 in the 9-1-1 clamp. The ATR-activating domain of TopBP1 stimulates the kinase activity of ATR. ATR can phosphorylate the effector kinase CHK1 and Rad9 of 9-1-1 complex (dashed arrows and P letter in red). Phosphorylation of CHK1 by ATR requires the mediator Claspin that recruits CHK1 to the DNA damage site (18, 19). Note that the in vitro checkpoint system described in PNAS (3) represents a major part of the pathway that does not include 9-1-1 and Claspin.
The report by Choi et al. (3) describes an in vitro checkpoint system consisting of purified ATR-ATRIP, topoisomerase II binding protein 1 (TopBP1), ssDNA, and RPA, the key components in the ATR pathway (3). The readout of this system is the phosphorylation of serine 345 in CHK1, an ATR-specific phosphorylation event (5, 6). To avoid potential complications caused by the kinase activity of activated CHK1, Choi et al. (3) use a mutant form of CHK1 that lacks catalytic function so that the kinase activity observed in the system is solely caused by activated ATR. It is worth noting that Choi et al. (3) purify native ATR-ATRIP, because overexpression may alter the quality of purified proteins, especially protein kinases whose enzymatic activity can be modulated by many factors. As observed by Choi et al. (3), ATR-ATRIP purified by immunoaffinity methods from ectopic expression systems often contains reduced activity. Similarly, overexpression of the human RAD17-replication factor C complex in insect cells (7) resulted in altered activity.
Choi et al. (3) describe a relatively straightforward purification scheme that allows successful separation of ATR-ATRIP from the other PI3K family kinases ATM and DNA-dependent protein kinase (DNA-PK) as well as other components in the ATR → CHK1 pathway. However, purification of the native, biologically active ATR-ATRIP complex is not a meager achievement, simply because of the large size of this complex. The study by Choi et al. (3) finds that, in the presence of TopBP1, RPA-ssDNA significantly stimulates ATR kinase activity under physiologically relevant ionic strength conditions. This result recapitulates much of what has been observed in vivo (i.e., efficient RPA- and TopBP1-dependent phosphorylation of CHK1), indicating that a faithful checkpoint system has been established. Whereas previous attempts to set up such a system were not successful, Choi et al. (3) stress that the activity of the current system is largely because of the availability of highly active ATR-ATRIP and TopBP1 proteins, careful titration of the purified components, and adjustment of reaction conditions to approximate in vivo conditions.
Human TopBP1 is a large protein comprising eight BRCA1 carboxyl-terminal (BRCT) domains distributed across its 1,522 amino acid length. TopBP1 functions in the initiation of DNA replication and in the activation of ATR during the DNA damage response. In Xenopus egg extracts, the N-terminal one-half of TopBP1 (BRCT domains I–IV) is necessary for loading of Cdc45 at origins of replication to form the active replisome (8, 9). The C-terminal portion of TopBP1 contains the ATR-activating domain (AAD) between the sixth and the seventh BRCT domains (10). Previous work showed that, under specific conditions, the TopBP1-AAD alone can activate ATR without the need for the BRCT domains (10, 11). At limiting protein concentrations, however, Choi et al. (3) show that efficient RPA-ssDNA–dependent stimulation of ATR requires the N-terminal portion of TopBP1. Interestingly, under these conditions, RPA coating of ssDNA can exclude TopBP1 binding, probably because of the high abundance and strong affinity of RPA for ssDNA. However, ATRIP bound to RPA-ssDNA acts as an interface between TopBP1 and ATR to overcome this inhibition and increase the local concentration of the kinase and its activator. TopBP1 recruitment requires interaction between ATRIP and multiple BRCT domains of TopBP1. Conversely, TopBP1 binding also enhances the binding of ATR-ATRIP complexes to RPA-ssDNA. Consistent with a previous report (12), the length of ssDNA was found to have a significant effect on the ability of TopBP1 to activate ATR. Although DNA shorter than 200 nt had little stimulatory effect, DNA of 1,000–2,000 nt dramatically enhanced TopBP1-dependent CHK1 phosphorylation, indicating that efficient activation of ATR requires the recruitment of a threshold number of ATR-ATRIP complexes to the same DNA molecule.
Choi et al. (3) propose that binding of ATRIP to RPA-ssDNA is mediated by the large RPA1 subunit of the RPA heterotrimeric complex. This conclusion is based on the observation that Saccharomyces cerevisiae RPA, Escherichia coli ssDNA binding protein (SSB), or human SSB1 failed to replace human RPA in this reaction. Furthermore, two checkpoint defective RPA1 mutants, RPA1-t11 (R41E, Y42F) and RPA1-∆N168, that do not bind ATRIP also failed to stimulate ATR. Corroborating a role for RPA in the DNA damage response, Choi et al. (3) show that a recently identified alternative form of the RPA complex (aRPA), which is active in DNA repair but does not support DNA replication (13), could stimulate ATR with similar efficiency as the canonical RPA.
Choi et al. (3) propose a model in which RPA-covered ssDNA recruits ATR-ATRIP to DNA damage sites by interaction between RPA1 and ATRIP. ATRIP also interacts with TopBP1 through both the N- and C-terminal regions of TopBP1, which, in turn, promotes more ATR-ATRIP binding to RPA-ssDNA. The TopBP-AAD domain of bound TopBP1 can then directly activate ATR to phosphorylate target proteins such as CHK1 and initiate checkpoint signaling at the DNA damage site. This model is consistent with the genetic data obtained in vivo that requires TopBP1 and RPA-ssDNA for efficient checkpoint signaling. However, as noted by Choi et al. (3), this experimental system does not yet address the functions of the Rad9-Hus1-Rad1 (9-1-1) clamp or its clamp loader Rad17-RFC complex in checkpoint activation. The orthologous S. cerevisiae 9-1-1 clamp can stimulate Mec1 (ATR) directly (14), whereas the 9-1-1 clamp from higher eukaryotes can stimulate ATR indirectly by recruitment of TopBP1 through its BRCT domains I and II to phosphorylated Rad9. In vivo, ATR-ATRIP and the Rad17-RFC/9-1-1 complexes are thought to bind to DNA independently; however, recent work has indicated that, under certain conditions, 9-1-1 can be recruited to DNA in a TopBP1-dependent manner (15). If ATRIP recruits TopBP1, as shown in the study by Choi et al. (3), then ATR-ATRIP should also recruit 9-1-1. Similarly, in Xenopus egg extracts, the checkpoint mediator Claspin is required specifically for CHK1 phosphorylation by ATR by recruiting CHK1 to DNA damage sites. Elaboration of the current system may clarify the role of TopBP1 as a node for communication between replisome assembly, replisome stabilization, and DNA damage-response pathways.
In a previous study, Choi et al. (11) suggested that human TopBP1 can bind to damaged DNA containing bulky base lesions, recruit ATR-ATRIP to the DNA damage site, and stimulate the ATR kinase activity through its ATR-activating domain (11). In the present study, they show that benzo[a]pyrene diol epoxide-damaged double-stranded DNA can also stimulate ATR activity in a TopBP1-dependent fashion, independent of the ssDNA binding protein RPA. TopBP1 stimulation of ATR bound to damaged dsDNA in vitro may help to reconcile observations that the ATR checkpoint response can be activated in vivo without RPA interaction (16).
As shown by Choi et al. (3), in vitro reconstitution can provide insights that are often unachievable by in vivo methods. However, in vitro reconstitution studies also have limitations that can oversimplify an in vivo system. For example, checkpoint pathways are temporally regulated within the cell during the cell-cycle progression, and in vitro checkpoint systems may not provide a clear picture of how this control is realized or the factors involved in temporal regulation. However, complemented with in vivo genetic studies, in vitro biochemical methods are powerful means of dissecting the molecular details of a complex biological system such as the checkpoint. With a better understanding of the conditions for each reaction step, it should be ultimately possible to reconstitute the checkpoint pathways in vitro to encompass all genetically defined components.
Footnotes
The authors declare no conflict of interest.
See companion article on page 13660.
References
- 1.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 2.Callegari AJ, Kelly TJ. Shedding light on the DNA damage checkpoint. Cell Cycle. 2007;6:660–666. doi: 10.4161/cc.6.6.3984. [DOI] [PubMed] [Google Scholar]
- 3.Choi J-H, et al. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc Natl Acad Sci USA. 2010;107:13660–13665. doi: 10.1073/pnas.1007856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cimprich KA, Cortez D. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Girona A, et al. Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc Natl Acad Sci USA. 2001;98:11289–11294. doi: 10.1073/pnas.191557598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:e33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Hatten RA, et al. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 11.Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc Natl Acad Sci USA. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 13.Kemp MG, et al. An alternative form of replication protein a expressed in normal human tissues supports DNA repair. J Biol Chem. 2010;285:4788–4797. doi: 10.1074/jbc.M109.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou L. Single- and double-stranded DNA: Building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 18.Chini CC, Chen J. Human claspin is required for replication checkpoint control. J Biol Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- 19.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]