Abstract
The longevity-promoting NAD+–dependent class III histone deacetylase Sirtuin 1 (SIRT1) is involved in stem cell function by controlling cell fate decision and/or by regulating the p53-dependent expression of NANOG. We show that SIRT1 is down-regulated precisely during human embryonic stem cell differentiation at both mRNA and protein levels and that the decrease in Sirt1 mRNA is mediated by a molecular pathway that involves the RNA-binding protein HuR and the arginine methyltransferase coactivator-associated arginine methyltransferase 1 (CARM1). SIRT1 down-regulation leads to reactivation of key developmental genes such as the neuroretinal morphogenesis effectors DLL4, TBX3, and PAX6, which are epigenetically repressed by this histone deacetylase in pluripotent human embryonic stem cells. Our results indicate that SIRT1 is regulated during stem cell differentiation in the context of a yet-unknown epigenetic pathway that controls specific developmental genes in embryonic stem cells.
Keywords: coactivator-associated arginine methyltransferase 1, HuR, neural differentiation, embryonic stem cells
Sirtuin 1 (SIRT1) is an NAD+-dependent lysine deacetylase involved in multiple cellular events, including chromatin remodeling, transcriptional silencing, mitosis, stress responses, DNA repair, apoptosis, cell cycle, genomic stability, insulin regulation, and control of lifespan (see ref. 1 for a review). In mammals, SIRT1 function is mediated by its deacetylating activity not only on histone tails (mainly K16-H4 and K9-H3 positions; refs. 2–4), but also on key transcription factors such as p53 (p53), forkhead transcription factors (FOXO), p300 histone acetyltransferase, the tumor protein p73 (p73), E2F transcription factor 1 (E2F1), the DNA repair factor Ku antigen, the 70-kDa subunit (Ku70), the nuclear factor κ-B (NF-κB), and the androgen receptor (AR) (see ref. 1 for a review).
Recent studies in mouse models suggest the importance of Sirt1 in stem cell differentiation. Sirt1 influences the neural and glial specification of neural precursors (5), regulates differentiation of skeletal myoblast (6), and inhibits spermatogenesis (7). Independently generated Sirt1-deficient mice are reported to exhibit severe neural defects, including exencephaly and disturbed neuroretinal morphogenesis (8, 9). In contrast to mice, in man the role of SIRT1 in human embryonic stem cell (hESC) differentiation is poorly understood. Here, we report a pathway that down-regulates SIRT1 during stem cell differentiation. In addition, we demonstrate that SIRT1 regulates the expression of specific developmental genes in pluripotent hESC and, thus, that its down-regulation is necessary for correct establishment of specific differentiation programs during stem cell differentiation.
Results
SIRT1 Is Down-Regulated During hESC Differentiation.
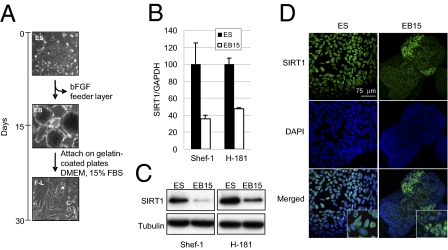
To study the putative role of SIRT1 in hESC differentiation, we first measured SIRT1 mRNA levels during the course of in vitro differentiation of the hESC lines Shef-1 and H-181. Withdrawal of basic fibroblast growth factor (bFGF) and feeder cells causes spontaneous differentiation of both hESC lines into embryonic bodies (EB) (Fig. 1A) (10). We observed that 15 d after induction of differentiation, SIRT1 mRNA levels were 60% lower in Shef-1 and 50% lower in H-181 cells (Fig. 1B). Consistent with this observation, there was a marked reduction in SIRT1 protein levels in 15-d EB compared with hESC (Fig. 1C), as previously shown in mice (11). To further characterize SIRT1 down-regulation during hESC differentiation, we performed immunofluorescence staining of SIRT1 in hESC and 15-d EB. SIRT1 staining in hESC was mostly nuclear except during mitosis, when it was diffused in the cytoplasm (Fig. 1D). In 15-d EB, SIRT1 staining levels were lower and its distribution was heterogeneous in distinct cell populations (Fig. 1D). Costaining of disaggregated EB cells for SIRT1 and for pluripotency/differentiation markers showed that cells with higher SIRT1 levels also had higher levels of the marker of pluripotency OCT4 (Fig. S1A); immunofluorescence analysis corroborated SIRT1 and OCT4 costaining (Fig. S1B). These analyses also showed that MAP2- and TUBB3-expressing neuroectodermal cells did not express high SIRT1 levels (Fig. S1 A and B).
Fig. 1.
Down-regulation of SIRT1 during hESC differentiation. (A) Flowchart of in vitro hESC differentiation. Representative images of hESC (ES), EB, and F-L cells were obtained by phase-contrast microscopy. hESC differentiated to EB after bFGF withdrawal and growth in suspension for 15 d. F-L cells were obtained by EB attachment on gelatin-coated plates and culture in DMEM + 15% FBS. (B) qPCR analyses of SIRT1 mRNA levels in hESC (ES) cells and 15-d EB (EB15) from Shef-1 and H-181 lines. Results are shown as the amount of SIRT1 mRNA relative to control GAPDH mRNA. (C) WB analysis of the samples in B using an anti-SIRT1 and anti-α-tubulin (control) antibodies. (D) Confocal fluorescence microscopy assay showing the cell localization of SIRT1 (green) in Shef-1 undifferentiated hESC and 15-d EB. qPCR values are mean ± SD of three independent experiments.
To characterize precisely the down-regulation of SIRT1 during hESC differentiation, we assessed RNA and proteins levels in a time-course experiment during spontaneous differentiation of Shef-1 cells into EB and subsequently into fibroblast-like (F-L) cells. These cells were obtained by inducing EB attachment, followed by culture in DMEM with 15% FBS for an additional 15 d (Fig. 1A) (10). SIRT1 mRNA levels decreased gradually during hESC differentiation, whereas SIRT1 protein levels dropped markedly only 7 d after induction of differentiation, and remained low throughout the process (30 d) (Fig. S1C); this observation suggests that SIRT1 is regulated during hESC differentiation at more than one level.
SIRT1 mRNA Is Stabilized by Coactivator-Associated Arginine Methyltransferase 1 (CARM1)-Dependent Methylated HuR in Pluripotent hESC.
We next studied the molecular mechanisms involved in SIRT1 down-regulation during hESC differentiation. We tested several possible ways to regulate SIRT1, including SIRT1 promoter epigenetic status and activity (Fig. S2 A and B, respectively), expression of the SIRT1 regulator miR34a (12) (Fig. S2C), and phosphorylation status of SIRT1 (13) (Fig. S2D), but observed no changes that justify SIRT1 down-regulation during differentiation. It was proposed that, in cancer cells, SIRT1 RNA stability can be regulated by the RNA-binding protein HuR (14), which also has a role in cell differentiation (15).
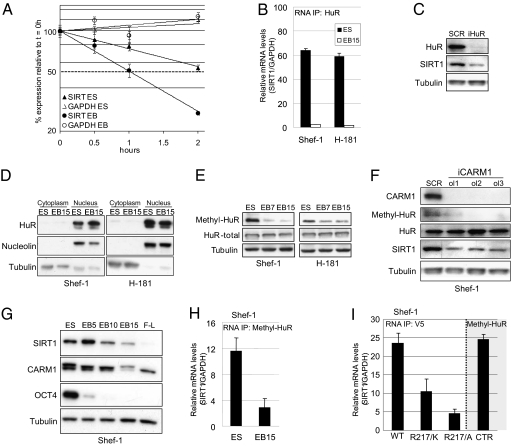
We hypothesized that HuR regulates SIRT1 RNA stability during hESC differentiation. To test this possibility, we examined the stability of SIRT1 RNA by treating cells with actinomycin D to inhibit de novo transcription. We monitored levels of SIRT1 mRNA and the housekeeping control GAPDH mRNA by retrotranscription, followed by quantitative PCR (qPCR) assays, and found that SIRT1 mRNA was notably less stable in differentiating EB than in pluripotent hESC (Fig. 2A). GAPDH mRNA, which is not a HuR target (16), showed no differences between hESC and EB. To determine whether the decrease in SIRT1 stability was due specifically to HuR, we measured the amount of SIRT1 mRNA bound to HuR during hESC differentiation, using immunoprecipitation (IP) assays and by monitoring RNA using qPCR. We observed a clear decrease in the amount of SIRT1 RNA bound to HuR in 15-d EB compared with hESC (Fig. 2B), which suggests that the decrease in SIRT1 RNA during hESC differentiation is mediated by the loss of HuR/SIRT1 mRNA binding. To confirm SIRT1 regulation by HuR, we depleted HuR in the Shef-1 hESC line with siRNA. HuR silencing resulted in a marked reduction in SIRT1 (Fig. 2C), further evidence of HuR-mediated regulation of SIRT1 mRNA stability in hESC.
Fig. 2.
CARM1-dependent HuR stabilizing activity in SIRT1 mRNA. (A) mRNA stability assay of SIRT1 and GAPDH in Shef-1 hESC (ES) and 3-d EB (EB) after actinomycin D treatment and collection as indicated. Data are relative to mRNA levels at t = 0 h for each sample. (B) RNA IP of HuR in hESC (ES) and 15-d EB (EB15) of Shef-1 and H-181 hESC lines. SIRT1 levels were detected by qPCR (relative to GAPDH). (C) WB of HuR, SIRT1, and α-tubulin in Shef-1 cells, 3 d after transfection with control (SCR) and HuR-specific siRNA (iHuR). (D) WB of HuR cytoplasmic and nuclear protein fractions from Shef-1 and H-181 hESC (ES) and EB. Nucleolin and α-tubulin were used as nuclear and cytoplasmic markers, respectively. (E) Methylation of HuR analyzed by WB, using an antibody specific for HuR methylated in Arg217 (methyl-HuR), relative to total HuR during EB differentiation of Shef-1 and H-181 hESC lines. (F) WB of CARM1, methyl-HuR, total HuR, SIRT1, and α-tubulin in Shef-1 cells at day 3 after transfection with a control siRNA (SCR) and three specific siRNA oligonucleotides for CARM1 (iCARM1, ol1, ol2, ol3). (G) WB of CARM1 in undifferentiated Shef-1 hESC and EB differentiated for 5, 10, or 15 d (EB5, EB10, and EB15) and fibroblast-like (F-L) cells. SIRT1 and Oct4 (POU5F1) were also tested in the same gel. (H) RNA IP assay of methyl-HuR in hESC (ES) and 15-d EB (EB15) of Shef-1 and H-181 hESC. (I) RNA IP assay with anti-V5 antibody in Shef-1 cells transfected with pCDNA3.3 plasmid expressing WT HuR, R217/K, or R217/A mutant HuR, all with a V5 tag. As control, Shef-1 cells were transfected with an empty plasmid, immunoprecipitated with anti-methyl-HuR (CTR) antibody and tested in the same conditions. qPCR values are expressed as mean ± SD of three independent experiments.
HuR binding to β-catenin, another independent HuR target, also decreased during hESC differentiation (Fig. S2E), which suggests that global HuR activity is also regulated during hESC differentiation. To test this possibility, we measured HuR mRNA levels in hESC and 15-d EB, but only found minor differences between the two cell types (Fig. S2F) and the HuR protein level did not change during hESC differentiation (Fig. 2 D and E). Because HuR activity is reported to be regulated by its nucleocytoplasmic shuttling (17, 18), we examined cellular localization of HuR during hESC differentiation. Western blot (WB) of nuclear and cytoplasmic extracts of hESC and 15-d EB in the Shef-1 and H-181 cell lines showed that, in both cases, HuR was primarily nuclear and its location did not change during hESC differentiation. This result implies that nucleocytoplasmic shuttling of HuR is not the main mechanism that regulates HuR activity in hESC. HuR binding to target mRNA is regulated by Chk2-dependent phosphorylation in cancer cells (14). To determine whether HuR is regulated by phosphorylation during stem cell differentiation, we immunoprecipitated total HuR in Shef-1 cells and EB and analyzed its phosphorylation status by using an antibody to phosphorylated serine. Phosphorylated HuR levels did not change notably during hESC differentiation (Fig. S2G), suggesting that serine phosphorylation is not the primary mechanism of HuR regulation during stem cell differentiation.
HuR is regulated by CARM1-dependent methylation at Arg217 (19). To evaluate whether HuR is regulated by methylation during hESC differentiation, we measured methyl-HuR levels in hESC and 15-d EB and found a marked reduction in methylated HuR in differentiated cells (Fig. 2E). To determine whether this decrease is specifically CARM1-dependent, we used three siRNA to deplete this arginine methyltransferase in Shef-1 hESC. CARM1 knockdown resulted in the loss of methyl-HuR and a marked decrease in SIRT1 (Fig. 2F). Finally, to verify that CARM1-dependent HuR methylation regulates SIRT1 mRNA stability during hESC differentiation, we measured CARM1 levels in hESC and EB over time and determined methyl-HuR binding to SIRT1 during hESC differentiation. The decrease in SIRT1 during hESC differentiation was associated with a decrease in CARM1 (Fig. 2G), and methyl-HuR binding to SIRT1 was much lower in EB than in hESC (Fig. 2H).
To confirm that HuR methylation influences HuR binding to SIRT1 mRNA during hESC differentiation, we compared the amount of SIRT1 mRNA bound to HuR in Shef-1 cells transfected with two Arg217 HuR mutants (Arg217/Lys, Arg217/Ala) resistant to CARM1-dependent methylation to Shef-1 cells transfected with WT HuR. IP assays using an antibody to the V5 epitope tag in our constructs and a control antibody to methyl-HuR showed that CARM1 methylation-resistant HuR mutants bound considerably less SIRT1 mRNA than did WT HuR and methyl-HuR (Fig. 2I). These results suggest that SIRT1 down-regulation during hESC differentiation is mediated by a CARM1-dependent decrease in methyl-HuR/SIRT1 mRNA binding.
SIRT1 Epigenetically Regulates Developmental Genes During Pluripotent hESC Differentiation.
SIRT1 was shown to regulate the promoter of the MashI gene in somatic stem cell neural differentiation in mice (5). To identify the downstream effects of SIRT1 down-regulation in hESC differentiation, we performed chromatin IP (ChIP) experiments in Shef-1 hESC by using an anti-SIRT1 antibody and by hybridizing the immunoprecipitated DNA fragments on an Agilent human promoter ChIP-on-chip microarray containing 474,392 probes, which cover almost all described human promoters and many known regulatory regions. These experiments showed significant binding to 428 probes (0.09% of the total), corresponding to 353 gene promoters (because some have more than one probe and some are divergent promoters that regulate two genes), four intergenic regions, and four microRNA promoters; the exact genomic location of each SIRT1-positive probe is presented in Dataset S1. Gene ontology analysis of the SIRT1-bound genes showed nonrandom distribution, largely with respect to molecular function (Table 1 and Datasets S2 and S3). SIRT1-bound genes in hESC are greatly enriched for gene ontology (GO) terms related to development and differentiation, such as developmental process, multicellular organismal development, and cell differentiation (P = 10−10 to 10−4; Table 1). Of the 353 SIRT1-positive genes, 97 are described by the highest-ranking GO term 0032502∼developmental process (1.76-fold enrichment; P = 3.05 × 10−9; Table 1). These observations suggest that SIRT1 is involved in the regulation of specific developmental genes in hESC.
Table 1.
GO terms significantly enriched in SIRT1-bound genes
| GO term∼biological process | No. of genes | P value |
| GO:0032502∼developmental process | 97 | 3.05E-09 |
| GO:0032501∼multicellular organismal process | 104 | 3.01E-08 |
| GO:0007275∼multicellular organismal development | 73 | 1.50E-07 |
| GO:0048856∼anatomical structure development | 68 | 2.75E-07 |
| GO:0048731∼system development | 54 | 1.76E-05 |
| GO:0009653∼anatomical structure morphogenesis | 39 | 3.54E-05 |
| GO:0030154∼cell differentiation | 53 | 1.08E-04 |
| GO:0048869∼cellular developmental process | 53 | 1.08E-04 |
| GO:0035270∼endocrine system development | 6 | 2.45E-04 |
| GO:0003008∼system process | 43 | 4.13E-04 |
| GO:0007154∼cell communication | 94 | 7.75E-04 |
| GO:0048513∼organ development | 38 | 8.78E-04 |
P < 0.001.
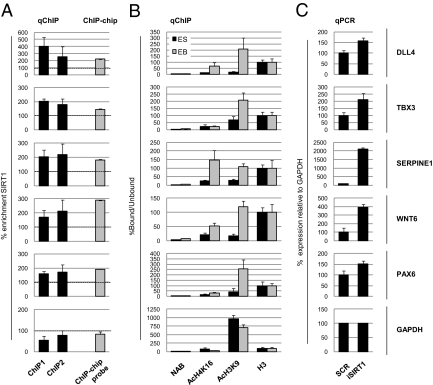
To validate these data, we performed ChIP experiments in Shef-1 hESC and EB by using antibodies to SIRT1 and to AcK16-H4 and AcK9-H3, two of its known histone targets (2, 5). The relative abundance of specific DNA fragments within the immunoprecipitated chromatin was assessed by quantitative PCR (qChIP). We randomly selected 10 of the 97 SIRT1-positive genes classified within the GO term “developmental process” (DLL4, LHX1, PAX6, WNT6, BMP1, HES7, TBX3, SERPINE1, HOXA5, TIMP1; for more information, see SI Materials and Methods). As controls, we used GAPDH and rDNA. These experiments showed that all 10 genes were enriched in SIRT1 in hESC (significant in most cases, P < 0.05; Fig. 3A and Fig. S3A), confirming the consistency of the whole ChIP-on-chip data. In 15-d EB (which have very low SIRT1 levels relative to hESC), we also observed a marked increase for the best-known SIRT1 histone targets, AcK16-H4 and AcK9-H3, in most target genes analyzed (Fig. 3B and Fig. S3B). The GAPDH and rDNA controls showed no SIRT1 enrichment or significant changes in AcK16-H4 or AcK9-H3. To verify direct regulation of these genes by SIRT1, we depleted its activity by siRNA in Shef-1 cells (Fig. S3 C and D) and used qPCR to measure the mRNA levels of five target genes (DLL4, TBX3, SERPINE1, WNT6, PAX6) with GAPDH as control. SIRT1 knockdown resulted in significant up-regulation of all five targets (P < 0.05; Fig. 3C), supporting its role in regulating selected genes in hESC. We also found that mRNA levels of eight genes were clearly up-regulated in 7- and 14-d EB, which are characterized by low SIRT1 levels (Fig. S3E). These data suggest that SIRT1 regulation of specific developmental genes during hESC differentiation is mediated by epigenetic mechanisms that involve its histone deacetylase activity.
Fig. 3.
SIRT1 binding and regulation of developmental gene promoters. (A) qChIP of SIRT1 in Shef-1 hESC. Enrichment relative to a chromatin sample immunoprecipitated without antibody (NAB) for the SIRT1-bound regions of DLL4, TBX3, SERPINE1, WNT6, and PAX6 was studied by qPCR. Results are expressed as the percent enrichment of the bound/unbound ratio (DNA copy number) of SIRT1 relative to the NAB immunoprecipitate in two sets of ChIP experiments (ChIP1, ChIP2). The enrichment value of the Agilent Human Promoters Array Probe (ChIP-chip probe) is also shown as the percentage of the normalized IP signal divided by the normalized input signal for the probe giving the highest signal for each gene represented (Dataset S1). qChIP primers were designed around the positive probe in the ChIP-on-chip array. The GAPDH promoter was included as a negative control for SIRT1 binding and histone modifications. (B) qChIP of acetyl-lysine 16 of histone H4 (AcH4K16) and acetyl-lysine 9 of histone H3 (AcH3K9) in Shef-1 hESC and EB cells. qPCR corresponded to the same genomic regions described above. Results are expressed as the bound/unbound percentage ratio (DNA copy number) for each IP and further normalized for the total H3 enrichment considered constant in chromatin. (C) Expression of these genes was measured by qPCR in Shef-1 hESC 3 d after transfection with a control siRNA (SCR) and an siRNA oligo for SIRT1 (iSIRT1). Data are normalized with respect to GAPDH expression, relative to the undifferentiated hESC sample (GAPDH expression set to 100%). qPCR values are mean ± SD of three independent experiments.
SIRT1 Is Involved in Lineage Specification During hESC Differentiation.
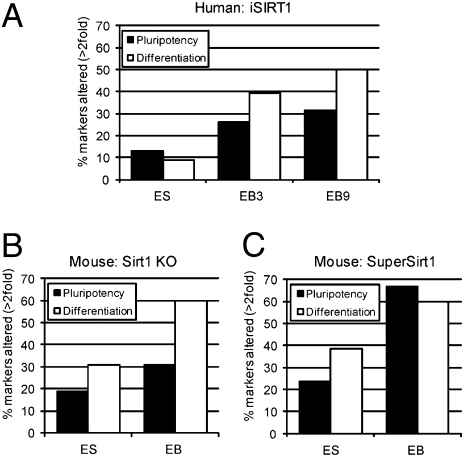
To study the functional role of SIRT1 down-regulation during hESC differentiation, we used the TaqMan Human Stem Cell Pluripotency Array and WB to compare the relative expression of stemness and differentiation markers during in vitro differentiation of siRNA-mediated SIRT1-depleted Shef-1 cells, mouse SIRT1 knockout ES cells, and mouse ES cells genetically modified to overexpress SIRT1 (20) with their respective wild-type controls. siRNA down-regulation of SIRT1 resulted in increased acetylation of the SIRT1 target p53 (Fig. S3F), consistent with previous reports (21). Global acetylation of H4K16 remained unchanged (Fig. S3F), further supporting the idea that SIRT1 histone deacetylase activity is largely restricted to gene promoters (Fig. 3 A and B). This expression array showed that siRNA-mediated depletion of SIRT1 in hESC altered the expression of only 12% of pluripotency and 9% of differentiation markers (Fig. 4A). Accordingly, protein expression of pluripotency markers OCT4, NANOG, SOX2, and E-cadherin was not notably altered (Fig. S3G). The impact of SIRT1 silencing on the expression of pluripotency and differentiation markers was much greater after induction of differentiation (Fig. 4A); the effect was clearly stronger on expression of developmental markers (50% in 9-d EB) than on that of pluripotency markers (31% in 9-d EB) (Fig. 4A).
Fig. 4.
SIRT1 knockdown in hESC and its regulation of the differentiation program. (A) A TaqMan Human Stem Cell Pluripotency Array that tests expression of 98 genes was carried out on Shef-1 hESC transfected with control (SCR) and SIRT1 siRNA (iSIRT1) at 2 d after transfection and after 3 (EB3) and 9 (EB9) d of differentiation of control and knocked-down samples. Results are shown as the percentage of genes cataloged as pluripotency- or differentiation-related, whose expression changed >2-fold between control and SIRT1 knocked-down samples at each differentiation stage. (B) A TaqMan Mouse Stem Cell Pluripotency Array was performed on TC1 mouse ES cells, the same cell line knocked out for SIRT1, and both lines differentiated in vitro to 15-d EB. Results are expressed as in A. (C) A TaqMan Array as in B was carried out on SuperSirt1 and the corresponding WT mESC, and both lines differentiated in vitro to 15-d EB. Results are expressed as in A. qPCR values are mean ± SD of three independent experiments.
To study SIRT1 function during ES differentiation in greater detail, we used several mouse ES cell (mESC) lines: Sirt1 KO, in which Sirt1 was either inactivated by deletion of exon 4 containing the catalytic domain (9), and SuperSirt1, in which Sirt1 was increased by insertion of an additional copy of the Sirt1 gene (20). Sirt1 expression was verified by WB in these mESC lines (Fig. S4A) and during in vitro differentiation (Fig. S4B). As in human cells, the effect of Sirt1 deficiency on expression of pluripotency and developmental markers was more pronounced after induction of differentiation (Fig. 4 B and C); moreover, the impact on developmental marker expression was greater than on that of pluripotency markers (Fig. 4 B and C). Sirt1 overexpression also resulted in greater alterations in pluripotency and developmental marker levels when differentiation was induced (Fig. 4C); in contrast to Sirt1 down-regulation, however, the number of pluripotency and developmental markers affected in cells overexpressing Sirt1 during differentiation was similar (≈60%) (Fig. 4C).
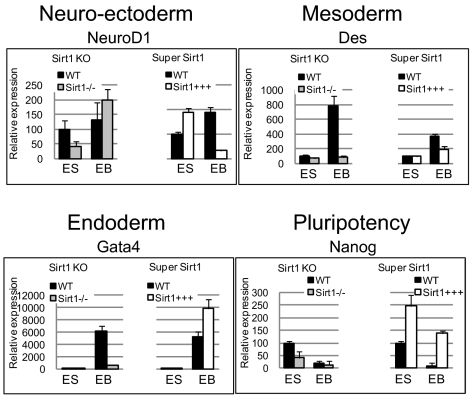
We used qPCR to analyze the marker expression in differentiating Sirt1 KO and SuperSirt1 mESC; we tested six pluripotency (Fgf4, Tdgf1, Nanog, Nodal, Rest, Tdfcp2l1), six neuroectodermal (NeuroD1, Syp, Nes, Isl1, Sfrp2, Hlxb9), three mesodermal (Col2a1, Brachyury, Des), and three endodermal markers (Fn1, Gata4, Lama1) (Fig. 5 and Fig. S4C). Sirt1 knockout or overexpression in undifferentiated mESC had little effect on developmental marker expression (Fig. 5 and Fig. S4C), in accordance with the expression array data. During differentiation, both Sirt1 deficiency and overexpression resulted in a clear alteration of most of developmental markers analyzed (Fig. 5 and Fig. S4C), as in hESC. The greatest changes were observed in neuroectodermal markers; most were overexpressed in Sirt1 KO EB and down-regulated in SuperSirt1 EB. These findings coincide with our data for hESC and indicate that, in humans and in mice, SIRT1 might have an important role in the establishment of the neuroectodermal layer.
Fig. 5.
Expression of pluripotency and differentiation markers in differentiation of Sirt1 KO (Sirt1−/−) or SuperSirt1 (Sirt1+++) mESC. A marker each for neuroectodermal (NeuroD1), mesodermal (Des), endodermal (Gata4), and undifferentiated (Nanog) cells was measured by qPCR in mESC (ES) and 15-d EB. Data are normalized to Gapdh expression, relative to the undifferentiated ES sample for each line. qPCR values are mean ± SD of three independent experiments.
Sirt1 KO mESC showed minor changes in expression of the pluripotency markers tested; SuperSirt1 mESC markedly overexpressed these markers (Fig. 5 and Fig. S4C). Although Sirt1 KO EB showed no clear changes in pluripotency marker expression during differentiation, SuperSirt1 EB retained notable expression of these markers; in most cases, it was comparable to their expression in WT ES cells.
Our data indicate that Sirt1 affects the expression of developmental genes and, to a lesser extent, of pluripotency genes during differentiation of human and mouse ESC. hESC ChIP-on-chip data indicated that the effect on developmental genes is direct, whereas the effect on pluripotency genes is mainly indirect. To determine whether this case is also true for mESC, we used ChIP experiments to analyze Sirt1 occupancy at the promoters of two pluripotency (Nanog, Oct4) and three developmental genes (NeuroD1, Nes, Sfrp2). Expression of the latter is affected by Sirt1 deficiency and overexpression during ES differentiation; Sfrp2 has an important role in neuroectodermal differentiation (22) and is directly regulated by SIRT1 (23). These experiments showed that Sirt1 bound Sfrp2 and Nes, but none of the pluripotency gene promoters, suggesting that, as in hESC, Sirt1 preferentially binds developmental gene promoters in mESC (Fig. S4D). Because not all developmental genes with altered expression in Sirt1-engineered cells were bound by Sirt1 in ESC, our data suggest that Sirt1 has a direct or indirect effect on developmental genes, whereas its effect on pluripotency genes is mainly indirect.
Discussion
Our studies show that SIRT1 regulation of specific developmental genes is important in hESC differentiation. Compelling evidence from mouse models indicates that Sirt1 is a central regulator of embryonic (21) and somatic (5) stem cell function. In mESC, Sirt1 is associated to the DNA damage response after mild oxidative stress (24). Our results extend these findings by showing that SIRT1 might be an important regulator of hESC differentiation, and also suggest a molecular pathway for SIRT1 regulation in stem cells. We show that SIRT1 is down-regulated during hESC differentiation at mRNA and protein levels and that the mRNA reduction depends on CARM1-dependent HuR Arg217 methylation. It was suggested that Carm1 helps to maintain mESC pluripotency by regulating histone methylation at the promoters of specific pluripotency genes (25, 26). In addition, Wu et al. (26) showed that developmental factors are overexpressed after iCarm1, but they did not address how these genes are up-regulated. Because there is some overlap of the iCarm1-overexpressed genes in mESC with those whose promoters are bound by SIRT1 in hESC (Datasets S4 and S5), we consider that our data and those of Wu et al. help to explain the central role of CARM1 in pluripotency. CARM1 down-regulation can have a direct effect on pluripotency by regulating chromatin structure and an indirect effect on priming developmental genes through SIRT1 (Fig. 6). This hypothesis would explain our observation that, at difference from iCARM1, iSIRT1 alone cannot induce hESC differentiation. CARM1 acts on both pluripotency and differentiation genes, and its knockdown is thus able to force differentiation. Repression of pluripotency genes is absolutely necessary to induce ES cell differentiation and, in hESC, SIRT1 appears to act preferentially on developmental genes.
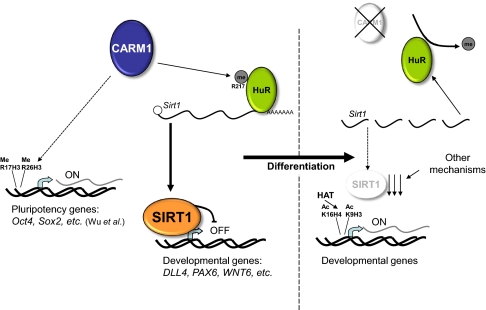
Fig. 6.
Model for SIRT1 action on developmental gene promoters during hESC differentiation. As described by Wu et al. (26), Carm1 regulates pluripotency gene promoters by histone methylation in mESC. In pluripotent hESC, CARM1 methylation of HuR increases HuR/SIRT1 binding and, consequently, SIRT1 mRNA stability and the SIRT1 protein level. In these conditions, SIRT1 binds to the promoter and epigenetically represses specific developmental genes such as DLL4, PAX6, and WNT6. During differentiation, the decrease in CARM1 is associated with a decrease in HuR methylation and, consequently, of HuR/SIRT1 binding, resulting in less SIRT1 mRNA and protein and the epigenetic reactivation of developmental genes targeted by SIRT1.
Because the principal mechanism thought to regulate HuR function is a change in its cytoplasmic levels (17), we would have predicted this as the cause of reduced association between HuR and the SIRT1 transcript. Our results nonetheless suggest that HuR/SIRT1 mRNA binding relies on CARM1-dependent HuR methylation, in accordance with the ability of CARM1 to methylate HuR and the increased HuR/RNA binding activity mediated by HuR methylation at Arg217 (19). HuR regulation of SIRT1 mRNA is not a new finding; HuR regulates SIRT1 in cancer cells (14). Here, we show that HuR also regulates SIRT1 during hESC differentiation, consistent with the reported role of HuR in myogenic differentiation (15), suggesting that HuR regulation of SIRT1 is a common mechanism of stem cell differentiation. It should nonetheless be stressed that although the CARM1/HuR pathway described here appears to be the main mechanism in SIRT1 mRNA down-regulation during hESC differentiation, it is not alone responsible for SIRT1 control in hESC. The more rapid down-regulation of SIRT1 protein than of mRNA suggests that other mechanisms are involved in controlling SIRT1 protein levels.
Our results indicate that elimination of Sirt1 in ES cells had little impact on the expression of pluripotency and developmental factors. This observation concurs with previous findings (27) and suggests that mammalian ES cells have redundant activities that mask Sirt1 function. Our data nonetheless show that SIRT1 down-regulation is needed to establish correct, specific differentiation programs during human and mouse ESC differentiation for two reasons: (i) SIRT1 binds to and epigenetically regulates specific developmental genes in pluripotent hESC, and (ii) expression of pluripotency and, above all, of differentiation markers is clearly altered in differentiating SIRT1-knocked-down hESC and Sirt1 KO/SuperSirt1 mESC. We show that some downstream effects of SIRT1 down-regulation are mediated by epigenetic reactivation of specific developmental genes, consistent with the role of Sirt1 in cell differentiation as a component of the polycomb repressive complex 4 (PRC4) (11). SIRT1 function in hESC maintenance must be more complex, however, because its down-regulation is also associated with p53 hyperacetylation (this study and refs. 9 and 21). Of the 10 SIRT1 target genes selected for ChIP validation, three (LHX1, PAX6, WNT6) are associated with neural development (28–30); another three (DLL4, TBX3, PAX6) are thought to have a role in retinal morphogenesis (29, 31). These data suggest a SIRT1 contribution to neural fate determination and retina formation during embryonic development. This possibility is supported by the findings that (i) expression of the neuroectodermal markers tested was clearly up-regulated during Sirt1 KO mESC differentiation and down-regulated during SuperSirt1 mESC differentiation (Fig. 5 and Fig. S4C), (ii) Sirt1 is involved in determining neural progenitor fate (5), and (iii) Sirt1 KO mice have notable retinal defects (8, 9).
Sirt1 interference and knockout, respectively, during human and mouse ESC differentiation showed that Sirt1 can also affect pluripotency gene expression. Because the number of pluripotency genes affected is much lower than that of developmental genes and this regulation is not direct (because Sirt1 rarely binds pluripotency gene promoters in ESC), the role of Sirt1 as a potential regulator of pluripotency genes is unclear. Although Sirt1 is implicated in pluripotency maintenance in stem cells, it is not strictly necessary, because Sirt1 KO mESC do not differentiate spontaneously, Sirt1 KO mice are viable (8, 9), and iSIRT1 in Shef-1 cells does not in itself induce differentiation (Fig. S3 D and G). Sirt1 overexpression during mESC differentiation altered expression of many more pluripotency genes than did its depletion in human or mouse ESC, indicating that abnormally high Sirt1 levels might have an impact on pluripotency programs. Although we found altered expression patterns for developmental and pluripotency genes in SuperSirt1 mESC, these mice do not exhibit apparent developmental problems (20). This discrepancy suggests that, although Sirt1 down-regulation is delayed during SuperSirt1 mESC differentiation, with immediate effects on developmental/pluripotency gene expression, this alteration is probably compensated in the long term.
In conclusion, our data provide evidence for a previously unreported epigenetic pathway in hESC differentiation, in which SIRT1 regulates hESC differentiation, partly via a CARM1/HuR pathway (Fig. 6). This pathway involves epigenetic regulation of key developmental genes such as the neuroretinal morphogenesis effectors DLL4, TBX3, and PAX6. In conjunction with the phenotype of various Sirt1-deficient mouse strains, our results indicate that whereas SIRT1 has a minor role in promoting or impairing hESC differentiation, it contributes to the establishment of specific developmental/differentiation programs of particular relevance for neuroectodermal fates.
Materials and Methods
Full details are provided in SI Materials and Methods. Briefly, hESC lines were obtained from the HUCA and the CABIMER, mouse TC1 ES cells and the knockout ES line (SIRT1Δex4/Δex4) were provided by F. Alt (Harvard Medical School, Boston), and Sirt1 transgenic ES cells were derived by the CNIO Transgenic Mouse Unit. Expression of SIRT1, HuR, CARM1, and all pluripotency and developmental markers was determined by quantitative RT-PCR, WB, immunofluorescence, and/or flow cytometry analysis. RNA interference was performed with a modification of a reported protocol (32). HuR-bound mRNA was immunoprecipitated as described (14). The ChIP assay was carried out as described (33); for the ChIP-on-chip assay, we used the Agilent Human Promoter Array. For qChIP, PCRs were done with SYBR-green PCR master mix and analyzed by using the 7900HT Fast Real-Time PCR System (all from Applied Biosystems).
Supplementary Material
Acknowledgments
We thank Dr. Ite Laird-Offringa (University of Southern California, Los Angeles) for the kind gift of antimethylated HuR antibody, Dr. Frederick Alt (Harvard Medical School) for mouse TC1 ESC and the Sirt1 knockout ES line (SIRT1Δex4/Δex4), and Catherine Mark for editorial assistance. V.C. received a Formación de Profesorado Universitario Spanish Research Programme Fellowship. A. Horrillo, A. Hmadcha, and B.S. are supported by Red TerCel and the Consejeria de Salud Junta de Andalucía (FPS). M.F.F. is funded by Spanish Ministry of Health Grants PI061267 and PS09/02454 and Spanish National Research Council (Consejo Superior de Investigaciones Cientificas) Grant 200820I172. This work was supported primarily by European Union Grant (ESTOOLS) LSHG-CT-2006-018739. The Instituto Universitario de Oncología is supported by Obra Social Cajastur, Spain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001399107/-/DCSupplemental.
References
- 1.Guarente L, Picard F. Calorie restriction–the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 3.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 4.Pruitt K, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prozorovski T, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 6.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One. 2008;3:e1571. doi: 10.1371/journal.pone.0001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBurney MW, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li SS, et al. Target identification of microRNAs expressed highly in human embryonic stem cells. J Cell Biochem. 2009;106:1020–1030. doi: 10.1002/jcb.22084. [DOI] [PubMed] [Google Scholar]
- 11.Kuzmichev A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 14.Abdelmohsen K, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa A, et al. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HH, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, et al. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 20.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han MK, et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubert J, Dunstan H, Chambers I, Smith A. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 2002;20:1240–1245. doi: 10.1038/nbt763. [DOI] [PubMed] [Google Scholar]
- 23.Pruitt K, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:0344–0352. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q, et al. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–2645. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBurney MW, et al. The absence of SIR2α protein has no effect on global gene silencing in mouse embryonic stem cells. Mol Cancer Res. 2003;1:402–409. [PubMed] [Google Scholar]
- 28.Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–173. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- 29.Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt C, McGonnell IM, Allen S, Otto A, Patel K. Wnt6 controls amniote neural crest induction through the non-canonical signaling pathway. Dev Dyn. 2007;236:2502–2511. doi: 10.1002/dvdy.21260. [DOI] [PubMed] [Google Scholar]
- 31.Lobov IB, et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci USA. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braam SR, et al. Feeder-free culture of human embryonic stem cells in conditioned medium for efficient genetic modification. Nat Protoc. 2008;3:1435–1443. doi: 10.1038/nprot.2008.140. [DOI] [PubMed] [Google Scholar]
- 33.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.