Influenza A virus causes seasonal flu epidemics and periodic worldwide pandemics (e.g., the 1918 Spanish flu, which caused ≈50 million deaths). The viral surface protein HA is the primary target of neutralizing Abs in natural infections (1). At any given time there are a limited number of viral strains circulating in humans, restricting widespread immunity to a small subset of potential viruses. Yearly seasonal epidemics arise from antigenic drift in the sequence of HA of currently circulating viruses, whereas pandemics are caused by the emergence of new, antigenically divergent viruses to which there is little to no immunity in the population (i.e., antigenic shift). The worldwide spread of a new H1N1 virus in 2009 caused the first recorded pandemic in more than 40 years. Current influenza vaccines are primarily produced from killed virus and mimic natural infection, inducing strain-specific, mainly HA-based, neutralization. Vaccine is produced from representative circulating strains grown in chicken eggs in a months-long process. Recent efforts aim to produce a broader influenza vaccine that focuses on common neutralization epitopes shared by multiple influenza strains. Such a vaccine should target a variety of influenza strains and better combat pandemics. A study published in PNAS describes an exciting strategy toward developing a broader influenza vaccine (2).
HA is trimeric and comprises a receptor-binding surface subunit (HA1) and a transmembrane subunit (HA2) that mediates entry after exposure to low pH in the endosome. Decades of HA structural characterization have defined the steps of membrane fusion and the epitopes of neutralizing Abs (Fig. 1) (1). HA is produced as a single-chain precursor with an HA1 cap covering an HA2 stalk. Cleavage leads to burial of the N-terminal HA2 fusion peptide and primes HA for fusion. Exposure to low pH induces large conformational changes in HA2 that propel the fusion peptide to the host endosomal membrane and lead to membrane fusion. The epitopes for most strain-specific neutralizing Abs are on the variable HA1 cap, and these Abs typically prevent receptor binding (3, 4). However, a handful of more broadly neutralizing monoclonal Abs have recently been identified that bind to the more conserved HA2 stalk (5–8). Designing an HA2-based immunogen to elicit these broadly neutralizing Abs is a high priority in the field.
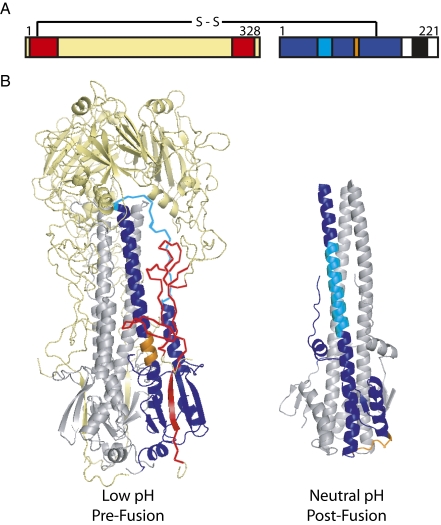
Fig. 1.
Structures defining the conformational changes of HA-induced membrane fusion. (A) The primary structure of cleaved HA. Color indicates the regions defined in B. (B) Two high-resolution structures of influenza HA representing the prefusion neutral-pH (Left) (16) and postfusion low-pH conformations (Right) (17). Trimers are depicted with HA1 (yellow) and HA2 (gray). One monomer is colored to indicate residues present in the HA2-stalk antigen, HA6 (2) (HA1 7–46 and 290–321 are red; HA2 1–172 is blue). Two regions within HA2 (and HA6) that change conformation between the neutral-pH and low-pH structures are indicated (55–76 in cyan and 106–112 in orange). Figure drawn with PyMOL.
Without HA1, HA2 folds into its most stable postfusion (low pH) conformation (9), making it challenging to produce HA2 antigen in its prefusion (neutral pH) conformation in the absence of HA1. Using structures of pre- and postfusion HA as a guide, Bommakanti et al. designed and produced an Escherichia coli-expressed single-chain antigen, HA6 (2). HA6 comprises the majority of the HA2 stalk and two smaller segments of HA1 that stabilize the stalk in the prefusion structure (Fig. 1). Importantly, introduced mutations (i) remove hydrophobic residues that become exposed upon deletion of the remainder of HA to improve solubility, and (ii) place charged residues at buried sites in the low-pH conformation of HA2 to destabilize this state. Standard biochemical and biophysical techniques confirm that HA6 adopts the prefusion structure.
After immunization with as little as 1 μg of HA6, >90% of mice were protected from lethal challenge of a homologous influenza strain (1968 H3N2). Surface-exposed regions of HA6 across a wide range of strains within the H3 subtype demonstrate high conservation of this region (91% vs. 66% for surface HA1 residues, for representative strains from 1968 and 2007), which supports the hypothesis that HA6-like immunogens will provide broader protection than traditional flu vaccines. Indeed, mice immunized with an HA6 antigen from a 1982 H3 strain were 80% protected from challenge by the 1968 H3 strain.
Influenza A is classified into 16 subtypes based on the sequences of HA, each belonging to one of two phylogenetic groups (10). Although HA2-stalk Abs, such as C179 and CR6261, are capable of neutralizing several related subtypes within a group (5, 7), it is unlikely that one HA2-stalk antigen will provide universal protection because of sequence variation between groups. Indeed, HA6 (H3N2) immunized mice were not protected from challenge by a distantly related H1N1 strain (2). Multiple HA6 immunogens will likely be required to combat divergent evolutionary groups.
How does HA6 immunization protect against influenza? By design, HA6 is intended to induce neutralizing Abs like 12D1 and CR6261 (6–8) that bind to prefusogenic HA and block the low pH-induced conformational change of HA2. As intended, HA6 is highly immunogenic, and the induced Abs compete with 12D1 for HA binding. However, sera from HA6 immunized mice fail to neutralize virus in vitro, and Fc receptor effector functions seem to play a significant role in protection. Taken together, these data are suggestive of a neutralizing, albeit weak, Ab response. As pointed out by the authors (2), anti-HA2 Abs generally have weaker neutralizing activity than the more common anti-HA1 Abs. Possible explanations include steric or kinetic restrictions and weakened binding affinity (e.g., due to the low pH of the endosome or the relatively small binding surface area). This loss of potency may be the cost of broader neutralizing activity.
Bommakanti et al. designed and produced an Escherichia coli-expressed single-chain antigen.
A similar “headless” antigen designed by Steel et al. (11) lacks most of the HA1 region. The HA1 “head” domain is enclosed by a disulfide bond and was replaced with a short linker peptide to expose the HA2 stalk in its prefusion conformation. This immunogen protected against homologous influenza challenge and produced antisera cross-reactive to heterologous HAs from within the same group. As with HA6, in vitro neutralizing activity could not be demonstrated.
Other approaches attempt an even more ambitious goal of a truly universal influenza vaccine that would cover all subtypes. One promising approach targets the highly conserved influenza M2 protein, which is not typically antigenic during natural infection (12). An intriguing nonimmune strategy is to remove sialic acid on target cells with a sialidase (Fludase; NexBio), which blocks entry of diverse influenza strains (13).
In addition to providing broader protection, a recombinant protein vaccine like HA6 would be more rapid and cost-effective to produce than traditional egg or cell culture vaccines. Future versions of this antigen will need to be optimized to enhance immunogenicity and focus on the most conserved regions of the HA2 stalk. For example, additional trimming of HA1 and HA2 residues could lead to a more minimized construct, and distracting immunogenic regions that are required to maintain the neutral pH structure could be masked by glycosylation or PEGylation.
This protein minimization approach may have broad utility in immunogen design. The same group designed a minimized single-domain version of the HIV receptor, CD4, that binds to and exposes CD4-induced epitopes on HIV gp120 (14) and a minimized gp120 outer domain, which induced HIV-neutralizing sera in rabbits (15). In principle, this design strategy could be broadly applied to the production of soluble domains of proteins for which structural information is available for all relevant conformations. This requirement for detailed structural information on pre- and postfusion conformations may hamper application to HIV, for which the prefusion structures of several key epitopes (e.g., gp41 prefusion conformation, trimeric gp41/gp120 complexes) remain unknown. Finally, this research emphasizes the importance of identifying rare, broadly neutralizing Abs using high-throughput methods. Beyond demonstrating that broad neutralization is achievable, mapping epitopes of such Abs, especially in costructures, greatly informs the design of immunogens capable of eliciting such Abs.
Acknowledgments
Work on viral entry in our laboratory is supported by National Institutes of Health Research Grant R01 AI076168.
Footnotes
The authors declare no conflict of interest.
See companion article on page 13701.
References
- 1.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Bommakanti G, et al. Design of an HA2 based E. coli expressed influenza immunogen that protects mice from lethal challenge. Proc Natl Acad Sci USA. 2010;107:13701–13706. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119:1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 5.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang TT, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, et al. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc Natl Acad Sci USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki Y, Nei M. Origin and evolution of influenza virus hemagglutinin genes. Mol Biol Evol. 2002;19:501–509. doi: 10.1093/oxfordjournals.molbev.a004105. [DOI] [PubMed] [Google Scholar]
- 11.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010 doi: 10.1128/mBio.00018-10. 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du L, Zhou Y, Jiang S. Research and development of universal influenza vaccines. Microbes Infect. 2010;12:280–286. doi: 10.1016/j.micinf.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Malakhov MP, et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob Agents Chemother. 2006;50:1470–1479. doi: 10.1128/AAC.50.4.1470-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma D, et al. Protein minimization of the gp120 binding region of human CD4. Biochemistry. 2005;44:16192–16202. doi: 10.1021/bi051120s. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharyya S, et al. Design of a non-glycosylated outer domain derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J Biol Chem. doi: 10.1074/jbc.M110.152272. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauter NK, et al. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: Analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry. 1992;31:9609–9621. doi: 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- 17.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]