Abstract
Influenza HA is the primary target of neutralizing antibodies during infection, and its sequence undergoes genetic drift and shift in response to immune pressure. The receptor binding HA1 subunit of HA shows much higher sequence variability relative to the metastable, fusion-active HA2 subunit, presumably because neutralizing antibodies are primarily targeted against the former in natural infection. We have designed an HA2-based immunogen using a protein minimization approach that incorporates designed mutations to destabilize the low pH conformation of HA2. The resulting construct (HA6) was expressed in Escherichia coli and refolded from inclusion bodies. Biophysical studies and mutational analysis of the protein indicate that it is folded into the desired neutral pH conformation competent to bind the broadly neutralizing HA2 directed monoclonal 12D1, not the low pH conformation observed in previous studies. HA6 was highly immunogenic in mice and the mice were protected against lethal challenge by the homologous A/HK/68 mouse-adapted virus. An HA6-like construct from another H3 strain (A/Phil/2/82) also protected mice against A/HK/68 challenge. Regions included in HA6 are highly conserved within a subtype and are fairly well conserved within a clade. Targeting the highly conserved HA2 subunit with a bacterially produced immunogen is a vaccine strategy that may aid in pandemic preparedness.
Keywords: hemagglutinin, protein design, bacterial expression
Influenza (flu) virus is the cause of major respiratory illness in humans resulting in 20,000–40,000 deaths annually in the United States alone (1) and killing millions in pandemic years. Viral subtypes are classified on the basis of the sequences of HA and neuraminidase (NA) surface proteins. Currently only strains of the H1N1, H3N2 subtypes and the B-type viruses circulate in human population. In recent years, however, certain strains of highly pathogenic avian influenza (H5N1) have been identified as the causative agents of a severe form of flu in humans, and it has been suggested that these have the potential to cause a pandemic (2).
Existing vaccines are primarily composed of the HA and NA from inactivated virus grown mainly in chicken eggs or the recently approved cold adapted live virus. However, due to antigenic shift and drift, the trivalent vaccine (designed against the circulating H1N1, H3N2, and B-type viral strains) has to be regularly modified based on the newly emergent strains of the virus. A major limitation with current influenza vaccines is that growth of the virus in chicken eggs is time-consuming and the generation of a new vaccine takes six to eight months. Hence, production of large amounts of vaccine at short notice during an epidemic/pandemic is difficult. A recombinant subunit vaccine that is easy to manipulate, produce, and scale up and provides long-lasting protection would be the ideal choice for controlling outbreaks such as the current swine flu pandemic.
HA is the most abundant protein on the viral coat and is highly immunogenic. HA as a stand-alone vaccine candidate has been investigated in several studies (3). HA is synthesized as a precursor (HA0) that trimerizes in the endoplasmic reticulum and is transferred to the cell surface via the Golgi apparatus. HA is cleaved by cellular proteases into HA1 and HA2 subunits, and this cleavage converts the protein into a fusion-active form (4). Entry of virions into target cells is mediated by binding of HA1 to sialic acid on the cell surface. The virion enters the cell by endocytosis and is transported to the endosome, where the resulting acidic pH induces major conformational changes in the HA molecule, leading to exposure of the fusion peptide on the HA2 subunit and subsequent fusion of viral and endosomal membranes. The crystal structures of HA in the precursor, neutral pH, and low pH forms have been solved (5–8) and reveal the changes that occur upon cleavage and after the low pH conformational switch.
Although HA2 is considerably more conserved than HA1 (9), early mapping studies of antigenic regions on HA revealed that neutralizing antibodies are directed only against the receptor binding HA1 subunit (10). Other studies have demonstrated that HA2-directed antibodies can provide protection in mice (11–13). Recently, several neutralizing monoclonal antibodies (Abs) have been isolated that bind the stem region of HA. These Abs have been shown to cross-react and neutralize several subtypes of viruses across clades and thus provide broad range protection (14–18). These Abs act by targeting the HA2 region of the molecule and presumably prevent the conformational change of HA at low pH, thus blocking fusion of viral and host membranes. It is therefore speculated that an engineered antigen that could focus the immune response to these epitopes and elicit protection against viral infection could serve as the basis for a more universal vaccine.
Previous attempts at recombinantly expressing soluble HA2 in the absence of HA1 have exclusively produced the low pH conformation of the protein even though expression and refolding were carried out at a neutral pH (19). However, very recently the stalk region of HA was successfully expressed on the surface of mammalian cells and HIV gag viral-like particles (20). In the present study, we have used a protein design and minimization method for successful expression and refolding of HA2 into its neutral pH conformation. We have previously used related approaches to express fragments of larger proteins in biologically active forms (21). The designed immunogen (HA6) consists largely of the HA2 subunit, is highly immunogenic in mice, and protects them against a lethal challenge with the virus.
Results
Design of the Immunogen.
The HA2 subunit is considerably more conserved than HA1 (Table S1, approximately 90% vs. 67% for Influenza A H1 and H3 subtypes in the surface-exposed regions) and might serve as a “universal” influenza vaccine candidate if it were to provide sufficient immunogenicity and protection. Furthermore, since HA2 is not involved in receptor binding, it is much more likely that epidemic or pandemic strains will arise through mutations in the HA1 rather than the HA2 subunit. However, the conformation of HA2 found in the neutral pH HA structure is believed to be metastable. It is known that the cleavage of HA0 and the disassembly of HA1 and HA2 subunits is essential for the low pH-induced conformational change (5, 22). We therefore reasoned that HA2 might be expressed in its neutral pH conformation if regions of HA1 that are in close spatial proximity to HA2 are included in the designed molecule.
The accessibilities of all residues in the HA neutral pH structure [Protein Data Bank (PDB) ID code 1HGD] (7, 8) were calculated, and HA1 fragments that had interactions with HA2 were identified using the program PREDBURASA. The program PREDBURASA calculates accessibilities (23) of all the residues in the protein in presence and absence of the interacting partner. The program was run using the coordinates from the PDB file 1HGD, and the accessibilities of all residues in the HA1 chain in the presence and absence of the HA2 chain were calculated. HA1 residues that have a difference of 5 Å2 or more in absolute accessibility in the above calculation and a 10% or higher accessibility difference were considered to be interacting with HA2. Similar cutoffs and approach were previously used by us to stabilize domain D1 of human CD4 (21). HA2-interacting residues consisted largely of residues (7–46) and (290–321) of HA1 (Fig 1A). Using the software RosettaDesign (24), mutations were chosen to remove exposed hydrophobic patches generated due to interactions lost with the rest of HA. These mutations were V297T, I300E, Y302T, and C305T in HA1. Finally flexible soluble linkers of appropriate length, similar in sequence to a linker used previously in HIV immunogen designs (25), were designed to link the three fragments into a single polypeptide chain. The length of the linker was chosen by taking into account the distance between the C-alpha atoms of the terminal residues of the fragments to be connected in the target structure. The designed protein contains residues (1–172) of HA2, a 7-amino acid linker (GSAGSAG), (7–46) of HA1, a 6-amino acid linker (GSAGSA) followed by residues (290–321) of HA1 with the above-mentioned mutations incorporated in it. The design was based on the sequence of H3 HA (A/HK/68) because only for this strain the crystal structure of both the neutral pH and low pH forms of HA are available (6–8). In addition, H3 viruses show a greater degree of genetic diversity than H1 and B as inferred from the HA sequences of the viruses in the past 10 years (Table S1).
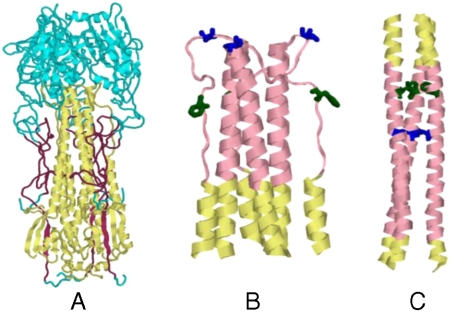
Fig. 1.
Structures of the HA ectodomain. (A) Regions that are included in HA6 are shown in either maroon (HA1) or yellow (HA2). The rest of the molecule is shown in cyan. This figure was derived from the X-ray structure of HA from the (H3N2) isolate A/HK/68 (PDB ID code 1HGD) (7) and was drawn using the program Rasmol (41). Conformation of residues (45–110) HA2 in the neutral pH (B) and low pH (C) structures of HA. The stretch 57–98 is shown in pink and the HA2 residues 63F and 73V, which are buried in the low pH form but are exposed in the neutral pH form, are shown in green and blue, respectively. Only a portion of the HA2 trimer (residues 45–110) is shown here for clarity.
The residues 63F and 73V in the HA2 subunit are buried in the coiled coil of the low pH structure (positions a and d, respectively, in the helical wheel). In the neutral pH conformation, these residues form a part of the loop connecting helices 4 and 5 and are exposed (Fig. 1 B and C). We reasoned that mutation of these hydrophobic residues to Asp would destabilize the coiled coil at neutral pH and introduced two additional mutations, F63D and V73D, in the HA2 chain. The Asp substitution was chosen because we have previously shown that Asp substitutions at buried hydrophobic residues are more destabilizing than substitutions with other charged amino acids (26). The resulting construct, inclusive of all mutations, is denoted as HA6. The final sequence of the protein encoded by the cloned gene is shown in Fig. S1.
A similar immunogen was also designed based on the sequence of a different H3 strain (A/Phil/2/82) that has an overall sequence identity of 85% at exposed residues to HK68 HA. The sequence of HA from A/Phil/2/82 was mapped onto the structure of HA from A/HK/68 and regions corresponding to HA6 were identified. Mutations that remove hydrophobic patches and destabilize the low pH form were also included in this construct as in the case of HA6.
Studies on a Model Peptide System.
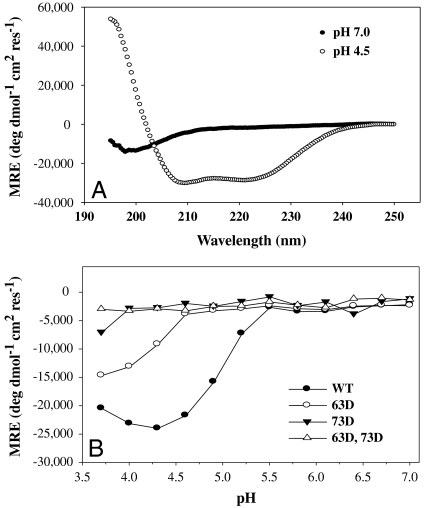
In order to test if the above Asp mutations were capable of destabilizing the extended HA2 coiled coil that is formed at low pH, a small 42-residue peptide (57–98 of HA2) that includes this region was chosen for further study. Previous studies have shown that a similar peptide undergoes a random coil to triple helix transition at low pH, similar to that observed in native HA (27). The gene for this peptide was cloned as a fusion to cytochrome b5 (28), and the mutations 63D and/or 73D were introduced into the gene. The resulting fusion proteins were expressed in Escherichia coli and purified. The peptides were derived from the fusion proteins described above by cleavage with TEV (tobacco etch virus) protease and further purified using reverse phase HPLC. Conformation of WT and mutant peptides was assessed using CD spectroscopy. The molar ellipticity at 222 nm was monitored as a function of pH to estimate alpha-helical content. The WT peptide was a random coil at neutral pH and formed a helical coiled coil at pH 4.5 (Fig. 2A). The decrease in ellipticity observed for 63D and/or 73D in Fig. 2B indicated that the introduced mutations have indeed destabilized the coiled coil. The apparent midpoints of the random coil to helix transitions are 5.3 and 4.7 for the WT and F63D mutant, respectively. The V73D mutation is more destabilizing than the F63D mutation. The mutant V73D and the double mutant F63D,V73D did not form a coiled coil throughout the pH range tested. Both these mutations were therefore included in the designed HA6 molecule as described above with the aim of destabilizing the low pH conformation of HA.
Fig. 2.
CD studies on the WT and mutant (57–98) HA2 peptides. (A) Far UV CD spectra of WT (57–98) HA2 peptide at pH 7.0 (•) and 4.5 (○), 25 °C, depicting the random coil to helix transition upon acidification. (B) pH titration of WT and mutant (57–98) HA2 peptides. The θ222 of the WT (•), 63D (○), 73 D (▾), and 63D, 73D (▵) mutant peptides are plotted as a function of pH. 73D mutant and 63D, 73D double mutant peptides are highly destabilized and do not form a coiled coil at any pH. Lines through the points are only for visual clarity.
Expression, Purification, and Biophysical Characterization of HA6.
An E. coli codon optimized gene for HA6 containing a C-terminal hexa-His tag to facilitate purification was synthesized and cloned into the bacterial expression vector pET-26b(+). The protein was expressed in E. coli BL21(DE3) cells and purified by immobilized metal affinity chromatography after resolubilization from inclusion bodies. The yield was about 2 mg/L of culture. SDS-PAGE with Coomassie staining confirmed that the protein was at least 95% pure.
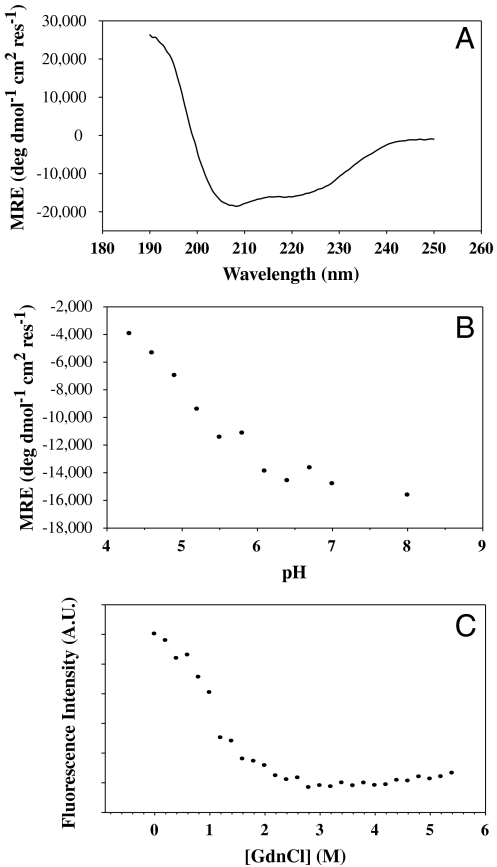
CD spectroscopy of the protein revealed a largely helical structure consistent with the designed target structure (Fig. 3A). In intact HA, each monomer of HA2 has an 18 amino acid helix (residues 38–55) packed against a longer helix (residues 76–125) in an antiparallel fashion. The trimer is stabilized by coiled coil interactions involving the N-terminal region of the longer helix (Fig. 1A). The ellipticity at 222 nm in Fig. 3A corresponds to ∼40% α-helix content, which is consistent with the predicted value (29) of 34% in the target structure. Intrinsic fluorescence emission spectra under native and denaturing conditions also indicate that the molecule is well folded (Fig. S2A) as the peak of the emission spectrum shows the expected red shift upon denaturation. In order to test if the molecule is well folded and compact, we also carried out ANS (1-anilino-8-napthalene-sulphonate) binding studies. In comparison to a molten globule control, HA6 did not bind ANS significantly (Fig. S2B) indicating that it does not have exposed hydrophobic patches. An isothermal denaturation melt using GdnCl revealed a cooperative unfolding transition characteristic of a well-folded protein (Fig. 3C).
Fig. 3.
Spectroscopic analysis of HA6. (A) Far UV CD spectrum of 5 μM HA6 in PBS, pH 7.4 at 25 °C. (B) Mean residue ellipticity of HA6 as a function of pH in 5 mM citrate glycine HEPES buffer, 25 °C. The molecule loses its helicity as the pH is lowered, confirming that HA6 does not form the extended coiled coil structure observed at the low pH for WT HA2 (6). (C) Isothermal Chemical Denaturation curve at pH 8.0, 25 °C for 2 μM HA6. The denaturation of the protein was monitored by the fluorescence intensity at 338 nm as a function of denaturant concentration.
The four Cys residues in the molecule (residues 14 HA1, 137 HA2, 144 HA2, and 148 HA2) appeared to be oxidized as judged from a 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) assay. These residues are involved in two disulfide bridges (14 HA1-137 HA2 and 144 HA2-148 HA2) in the native HA molecule. The lack of free thiols in the protein suggests that it is properly folded.
HA6 Adopts the Desired Neutral pH Conformation.
In order to examine if HA6 was indeed folded into the desired neutral pH conformation and not the low pH conformation, the following additional studies were carried out. First, engineered disulfide bridges were used to confirm that the target fold was achieved. Comparing the structures of the neutral pH (PDB ID code 1HGD) and low pH conformations of HA (PDB ID code 1HTM) and using the program MODIP (30) to pick possible positions for engineering disulfide bonds, we chose two pairs of residues in the molecule (3,116 of HA2 and 40,118 of HA2). Residues in each pair are spatially close and predicted to form disulfide bonds when mutated to Cys if the molecule were in the neutral pH-like conformation. In the low pH conformation, these pairs of residues would be separated by greater than 90 Å (Fig. S3). The mutations were introduced into HA6 and the two mutant proteins were expressed and purified. Both these proteins were well folded and had CD spectra similar to that of the WT protein (Fig. S4A). The proteins were tested for their free thiol content under denaturing conditions in a DTNB assay. In both the mutant proteins, the engineered disulfide bond was formed. There were no free Cys residues in the absence of DTT and the expected six Cys residues were detected after reduction with DTT (and removal of DTT on a desalting column). WT and mutant HA6 proteins were characterized by RP-HPLC on a C5 HPLC column. Under reducing conditions, all three proteins eluted at a different acetonitrile concentration relative to the corresponding protein prior to reduction and as compared to each other (Fig. S4B). Formation of the additional disulfide bonds in the (3C, 116C) and (40C, 118C) mutants results in a more compact unfolded conformation as evidenced by the decreased retention times relative to WT HA6. The formation of additional disulfide bonds in the (3C,116C) and (40C,118C) mutants indicates that HA6 is in the desired neutral pH-like conformation and not the low pH conformation.
To further confirm this assertion, the helical content of the molecule was measured as a function of pH using CD spectroscopy. The θ222 of HA6 is plotted as a function of pH in Fig. 3B. The results show that the protein loses its helicity as the pH is lowered contrary to what would be expected if it were in the low pH conformation.
Immunization and Challenge Studies in Mice.
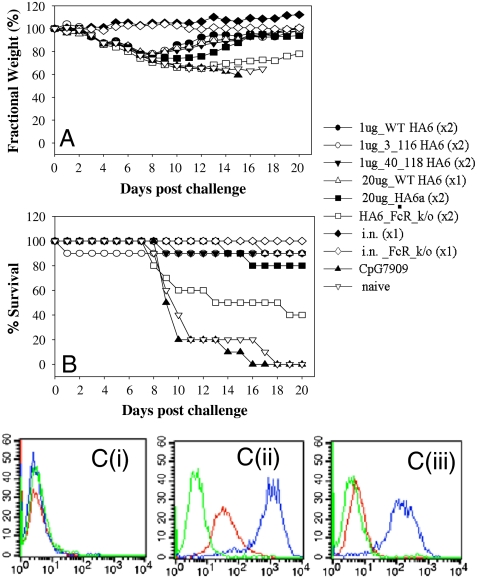
HA6 [WT as well as (3C, 116C) and (40C, 118C) disulfide mutant proteins] were used to immunize mice before challenging them with one LD90 of homologous H3N2(A/HK/68) virus. The antibody titers in the immunized mice were determined by ELISA. It was found that all three immunogens tested were highly immunogenic and resulted in high antibody titers (Table S2). All the animals immunized with the HA6 proteins (WT or disulfide mutants) were protected from a lethal challenge with the homologous virus (Fig. 4B and Table S2). The same results were reproduced with different adjuvants in a repeat study, and protection was observed even with a single dose of 1 μg of HA6 (Table S2). The disulfide mutants also conferred protection in mice. Due to a lack of available mouse adapted H3 strains, it was not possible to examine protection against other H3 strains. Weight measurement studies showed weight loss in the mice following pathogenic challenge before they fully recovered (Fig. 4A).
Fig. 4.
Immunogenicity and protection against lethal challenge (A/HK/68) by HA6 in mice. Mice of 10 per group were immunized with indicated vaccines either twice (× 2) or once (× 1) and challenged intranasally with an LD90 of mouse adapted A/HK/68. Weight changes (A) and mortality (B) were monitored for 20 days post challenge. Convalescent mice from intranasal A/HK/68 inoculation were included as a positive control, and mice receiving CpG7909 alone as adjuvant control. (C) Binding of HA6 antiserum to viral infected cells. HA6 antiserum (red), sera from CpG7909 adjuvant control group (green), and convalescent sera from A/HK/68 infected mice (blue) were tested by FACS for binding to mock infected (i), A/HK/68 infected (ii), or A/PR/8/34 infected (iii) MDCK cells.
However, sera were not able to prevent viral entry when assayed in a standard microneutralization assay at the lowest dilution (1∶4) of the serum tested. Attempts to concentrate the HA6 binding fraction of the serum using an HA6-affinity column were unsuccessful as the majority of the antibodies bound tightly and could not be eluted. Hence we did not attempt neutralization assays with this material.
In order to probe the mechanism of protection conferred by HA6, FcRγ knockout (ko) mice were immunized with HA6 and subjected to viral challenge with one LD90 of A/HK/68 virus (Fig. 4). FcRγ ko mice were only partially protected (40% protection) indicating that antibody dependent effector functions such as antibody-dependent cellular cytotoxicity (ADCC) play an important role in HA6-induced protection in mice. However, 40% of ko mice still survived. This indicates that other Ab mediated mechanism(s) such as neutralizing antibodies or complement-dependent cellular cytotoxicity are also responsible for protection.
Cross-Strain Protection.
In order to test if HA6 or a similar molecule designed from another strain could provide cross-strain protection, we made an HA6-like construct (HA6a) from the HA sequence of A/Phil/2/82. Mice that were immunized with HA6a were protected (80% protection) against an infection with A/HK/68 (Fig. 4).
However, the HA6 protein failed to confer protection against a heterosubtypic H1 strain (A/PR/8/34). Consistent with the protection data, FACS analysis showed that HA6 antisera bind to A/HK/68 but not to mock or A/PR/8/34-infected Mardin Darby canine kidney (MDCK) cells (Fig. 4C).This was not unexpected as a sequence comparison of HA from the H3N2 (A/HK/68) and H1N1 (A/PR/8/34) revealed a large number of differences between the proteins [identity in the exposed regions (> 20% accessibility) of the corresponding HA6 molecules being only 45%]. The sequence identity in the exposed regions of HA6 for all vaccine strains used over the past 10 years, on the other hand, is 91% and 96%, respectively, for H3 and H1 subtypes (Table S1 and Fig. S5). In comparison, the fraction of conserved residues in the exposed regions of the HA1 subunit for vaccine strains over the past 10 years is 80% and 86% for H3 and H1 strains, respectively. For all H3 and H1 strains of the past 10 years, the numbers are 64% and 71%, respectively. As the degree of conservation is considerably higher within a given subtype for HA6 relative to HA1, an HA6-like immunogen might confer subtype specific protection (Table S1 and Fig. S5).
Binding Studies with Recombinant HA.
In order to test if HA6-induced antibodies could bind to HA from a recent H3 strain that is very different from A/HK/68 (75% identity in exposed regions), binding studies were carried out with recombinant HA of A/Brisbane/16/07. ELISA studies using anti-HA6 sera and convalescent sera from A/HK/68-infected mice showed that anti-HA6 sera bound immobilized recombinant HA from A/Brisbane/16/07 with approximately 100 fold greater half maximal titers than convalescent sera (Fig. S6). The latter contain Abs primarily directed against the exposed, globular head of HA. A sequence analysis revealed that the identity in exposed regions in the HA6 fragment of the two HA molecules (A/HK/68 and A/Brisbane/10/07) was 91%, whereas in the HA1 fragment it was 66%. These results indicate that HA6 designed from HA of HK/68 strain could possibly provide protection against recent strains of H3 viruses also owing to the higher degree of conservation in this region of the molecule. This was confirmed by the protection conferred by HA6a (based on A/Phil/2/82) against A/HK/68 pathogenic challenge.
Binding of HA6 to the Broadly Neutralizing mAb 12D1.
Binding of HA6 to a recently isolated, broadly neutralizing mAb 12D1 that cross-reacts with several H3 viruses spanning approximately 40 drift years 1968–2003 (31) was tested by ELISA. The epitope to this mAb is believed to be in the region (76–106) of HA2 from binding studies to various truncated HA proteins. However, residues (99–106) of HA2 are largely buried in the neutral pH structure of HA. Hence, as a control, a peptide consisting of residues (57–98) of HA2 was used. This peptide is unstructured at neutral pH (Fig 2A). Although the mAb failed to bind to the peptide, it bound well to HA6 as well as to other recombinant HA proteins expressed in baculovirus (Fig. S7A). These data demonstrate that 12D1 recognizes a conformational epitope that is well presented on the HA6 immunogen and that the HA2 region in HA6 is folded in a similar fashion to the corresponding region in the neutral pH structure of HA. Further, anti-HA6 antisera compete with 12D1 binding to HA (Fig. S7B) indicating that 12D1-like Abs are elicited by HA6.
Discussion
In natural influenza infection, neutralizing antibodies elicited against HA lead to clearance of the virus. Four major antigenic sites have been identified on the HA molecule, all of which are on the receptor binding HA1 subunit (10). The HA2 subunit of influenza HA is typically considered not to play a major role in recovery from natural infection. However, recent studies have shown that antibodies against HA2 may help in improving recovery from viral infection (11). Several studies have also shown that cross-reactive neutralizing antibodies are directed to HA2 (14–18, 32). Until the present work, production of a soluble, native (neutral pH-like) HA2 immunogen has proved to be difficult, owing to the metastable nature of HA (19). Here we report the successful design and characterization of a soluble immunogen that largely consists of HA2 in its neutral pH conformation. This was achieved by introducing regions of HA1 that are interacting with the HA2 subunit and by destabilizing the low pH conformation. Since this immunogen is expressed in E. coli, it can be rapidly and cheaply produced in large quantities.
A sequence analysis of vaccine strains of influenza virus used over the past decade revealed very few changes in the region of the molecule included in the present HA6 immunogen. The present immunogen is based on an H3 (A/HK/68) HA sequence. Phylogenetic analysis of HA sequences from various subtypes has revealed two groups (H1 and H3 groups) and four clades (H1, H9, H3, and H7—the former two clades belong to the H1 group and the latter two clades belong to the H3 group) (14–16, 33). H1, H2, and H5 HAs belong to the H1 clade and are closely related. A monoclonal antibody C179 has previously been shown to neutralize viruses from these three subtypes (12, 13). mAbs directed to closely related epitopes on the stem of HA have been shown to neutralize several viruses belonging to the homologous H1 clade, and other Abs have recently been shown to neutralize several H3 subtype viruses indicating the possibility that an HA2-based antigen might elicit broadly neutralizing Abs (14–16, 31). Binding studies of HA6 with a broadly neutralizing mAb 12D1 indicate that epitope(s) that are capable of eliciting a broadly neutralizing Ab response are present on the HA6 immunogen. mAb 12D1 binds to both HA6 and recombinant HA expressed in insect cells with similar affinities (Fig. S7A). Further, competition binding experiments indicate that 12D1-like Abs are present in anti-HA6 guinea pig serum, though not at high enough levels to show neutralization in vitro. Unlike receptor binding site Abs that have IC50’s as low as 60 ng/mL (34), HA2 binding neutralizing mAbs such as 12D1, CR6261, and F10 have IC50s ∼5 to 100-fold higher, in the range of 0.3–5 μg/mL (15, 16, 31).
Owing to the lack of mouse-adapted versions of more recent H3 strains, we designed an immunogen from the A/Phil/2/82 HA and tested for protection against HK/68 virus instead. Mice immunized with HA6a (based on A/Phil/2/82) were protected against a challenge with HA from HK/68 strain indicating successful cross-protection. Binding studies with recombinant HA of a recently circulating H3 strain (A/Brisbane/16/07), which is closely related to the H3 strain in the 2009–2010 flu vaccine, showed good binding with HA6 antisera while convalescent sera showed 100-fold lower binding (Fig. S6). These results indicate that HA6 elicits high titer cross-reactive Abs, by focusing the immune response to the conserved stem region of the molecule. Even relatively few mutations (one or two mutations) in the HA globular domain are known to abolish neutralization of the virus by antibodies (35, 36). The A/Phil/2/82 HA that has been used in this study to show cross-strain protection has only 85% identity in the exposed regions with the challenge strain A/HK/68. Hence it is unlikely that immunization with inactivated A/Phil/2/82 virus would confer protection to an A/HK/68 challenge. Recent studies with human mAbs generated against HAs from different years have indicated that there are three classes of such Abs (those that react with viruses from 1968–1973, 1977–1993, or 1997–2003, respectively). There is substantial antigenic drift between viruses of these groups (37). This finding also supports the notion that immunization with a 1982 virus (A/Phil/2/82) would not have protected against challenge with a 1968 virus (A/HK/68), in contrast to protection conferred by the HA6a immunogen.
The HA6 protein is highly immunogenic in mice, provides protection against a homologous viral challenge, and provides cross-strain protection within the subtype. It also elicits Abs that can compete with broadly neutralizing mAb12D1. Lack of protection against an H1 virus (A/PR/8/34) is not surprising as A/HK/68 (an H3 clade virus) is very distantly related to H1 clade viruses. It is known that broadly neutralizing Abs against H1 clade viruses are incapable of neutralizing H3 clade viruses (14–16).
Immunity to natural influenza infection is attributed to anti-HA antibodies and immunity is lost if Abs fail to recognize HA (38). HA-directed Abs protect from infection primarily by preventing attachment of the virus to the host cell and also by preventing fusion of the viral and host cell membranes (36). The HA6 immunogen described here is devoid of the globular head domain of HA that contains the receptor binding site. Hence receptor binding site directed Abs (that would show HI inhibition activity) would not be elicited using this protein. Studies with FcRγ ko mice demonstrate that ADCC contributes significantly to protection observed by HA6-elicited antibodies. However, the fact that HA6 was capable of eliciting mAb12D1-like responses suggests that neutralizing Abs are also likely to contribute to protection by mechanisms similar to 12D1. The mAb 12D1 confers protection by inhibition of viral fusion to the host cell membrane, unlike HA1-directed Abs that prevent infection by blocking receptor binding.
Broadly neutralizing HA2 stem-specific monoclonal antibodies with neutralizing activity for H1 clade viruses (14, 15) and recently for H3 subtype viruses (31) have been isolated. These studies indicated that epitopes in the stem of HA are accessible to antibodies. Further improvements in the design of the HA6 immunogen and those of other subtypes can lead to constructs that possibly elicit high titers of cross-reactive antibodies capable of neutralizing multiple subtypes of the virus. The H1 subtype of influenza (including pandemic H1N1) also contains hydrophobic residues in the loop contacting helices 4 and 5 that are exposed at neutral pH and buried in the low pH structure. These are 63F and 73L (HA2). Asp mutations at these sites are likely to stabilize a corresponding HA6-like construct derived from H1N1 sequence.
Materials and Methods
Cloning and Purification of HA Proteins and Model HA Peptide.
E. coli codon optimized genes corresponding to the designed proteins (HA6 and HA6a) were synthesized and cloned into pET-26b(+) vector. The proteins were overexpressed in BL21(DE3) cells and purified from inclusion bodies using Ni-NTA affinity chromatography. They were finally refolded by desalting into water. The gene corresponding to residues (75–98) of HA2 was cloned into the vector pET-21a(+) as a cyt.b5 fusion (28). Mutations were introduced by PCR using complementary primers. The fusion proteins were expressed in BL21(DE3) cells, purified on a DEAE-Sephacel column and cleaved using TEV protease. The peptides were obtained after RP-HPLC purification.
Biophysical Characterization.
CD, fluorescence, and ANS binding studies were carried out as described previously (39). Free thiol content was determined using a standard DTNB assay (40).
Immunization and Challenge Studies.
Female BALB/c mice were immunized intramuscularly with 20 μg or less of either of the immunogens [HA6, HA6a, (3C, 116C)HA6, or (40C, 118C)HA6] along with 100 μg of adjuvant CpG7909 and boosted four weeks later. At week 7, the mice were challenged with 1LD90 of A/HK/68 virus, and their survival and weight was monitored for 20 days. Naive mice and adjuvant-treated mice were used as controls. FcRγ ko mice were immunized with WT HA6 in a similar manner. Guinea pigs were immunized with 100 μg HA6 i.m. thrice in four-week intervals, and sera were collected two weeks after the last immunization.
FACS and ELISA.
MDCK cells were infected with A/HK/68 or A/PR/8/34 virus at an moi of 1. After overnight incubation, test sera (1∶200 diluted) were incubated with infected cells for 1 h. After washing, the cells were treated with FITC-labeled goat anti-mouse Ab, fixed with 1% formaldehyde in PBS, and analyzed by flow cytometry. For ELISA, peptide (57–98), HA6, and recombinant HA proteins were immobilized (250 ng of Ag per well) and probed with different concentrations of 12D1. After washing, Alkaline Phosphatase (ALP)-conjugated anti-mouse Ab was added. After 2 h of incubation, ALP substrate was added and the absorbance at 405 nm was measured. Anti-HA6 titers were also determined in a similar assay by testing for binding to immobilized HA6.
Details of the materials and methods are given in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Siddhartha. P. Sarma for the pET-21a(+)cytb5-tev fusion vector and Dr. C. Ramakrishnan for the PREDBURASA program. We thank Dr. Peter Palese for the mAb 12D1. We acknowledge the Council of Scientific and Industrial Research for financial support.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 13563.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007465107/-/DCSupplemental.
References
- 1.Fan J, et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine. 2004;22:2993–3003. doi: 10.1016/j.vaccine.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 2.Horimoto T, Kawaoka Y. Influenza: Lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 3.Cox MM. Cell-based protein vaccines for influenza. Curr Opin Mol Ther. 2005;7:24–9. [PubMed] [Google Scholar]
- 4.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, et al. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 6.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Sauter NK, et al. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: Analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry. 1992;31:9609–9621. doi: 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- 8.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–73. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 9.Both GW, Sleigh MJ, Cox NJ, Kendal AP. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: Multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983;48:52–60. doi: 10.1128/jvi.48.1.52-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 11.Gocnik M, et al. Antibodies induced by the HA2 glycopolypeptide of influenza virus haemagglutinin improve recovery from influenza A virus infection. J Gen Virol. 2008;89:958–967. doi: 10.1099/vir.0.83524-0. [DOI] [PubMed] [Google Scholar]
- 12.Smirnov YA, Lipatov AS, Gitelman AK, Class EC, Osterhaus AD. Prevention and treatment of bronchopneumonia in mice caused by mouse-adapted variant of avian H5N2 influenza A virus using monoclonal antibody against conserved epitope in the HA stem region. Arch Virol. 2000;145:1733–1741. doi: 10.1007/s007050070088. [DOI] [PubMed] [Google Scholar]
- 13.Okuno Y, Matsumoto K, Isegawa Y, Ueda S. Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J Virol. 1994;68:517–520. doi: 10.1128/jvi.68.1.517-520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Fauquier A, Villanueva N, Melero JA. Isolation of cross-reactive, subtype-specific monoclonal antibodies against influenza virus HA1 and HA2 hemagglutinin subunits. Arch Virol. 1987;97:251–265. doi: 10.1007/BF01314425. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, et al. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc Natl Acad Sci USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010;1:e00018. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma D, et al. Protein minimization of the gp120 binding region of human CD4. Biochemistry. 2005;44:16192–16202. doi: 10.1021/bi051120s. [DOI] [PubMed] [Google Scholar]
- 22.Godley L, et al. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 23.Lee B, Richards FM. The interpretation of protein structures: Estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 24.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 25.Varadarajan R, et al. Characterization of gp120 and its single-chain derivatives, gp120-CD4D12 and gp120-M9: implications for targeting the CD4i epitope in human immunodeficiency virus vaccine design. J Virol. 2005;79:1713–1723. doi: 10.1128/JVI.79.3.1713-1723.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajaj K, Chakrabarti P, Varadarajan R. Mutagenesis-based definitions and probes of residue burial in proteins. Proc Natl Acad Sci USA. 2005;102:16221–16226. doi: 10.1073/pnas.0505089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 28.Mitra A, et al. High level expression of peptides and proteins using cytochrome b5 as a fusion host. Protein Expr Purif. 2005;41:84–97. doi: 10.1016/j.pep.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 29.Joyce JG, et al. Enhancement of alpha -helicity in the HIV-1 inhibitory peptide DP178 leads to an increased affinity for human monoclonal antibody 2F5 but does not elicit neutralizing responses in vitro. Implications for vaccine design. J Biol Chem. 2002;277:45811–45820. doi: 10.1074/jbc.M205862200. [DOI] [PubMed] [Google Scholar]
- 30.Dani VS, Ramakrishnan C, Varadarajan R. MODIP revisited: Re-evaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Eng. 2003;16:187–193. doi: 10.1093/proeng/gzg024. [DOI] [PubMed] [Google Scholar]
- 31.Wang TT, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim AP, et al. Neutralizing human monoclonal antibody against H5N1 influenza HA selected from a Fab-phage display library. Virol J. 2008;5:130. doi: 10.1186/1743-422X-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci USA. 1981;78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knossow M, et al. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology. 2002;302:294–298. doi: 10.1006/viro.2002.1625. [DOI] [PubMed] [Google Scholar]
- 35.Hensley SE, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119:1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada J, et al. Monoclonal antibodies in man that neutralized H3N2 influenza viruses were classified into three groups with distinct strain specificity: 1968–1973, 1977–1993 and 1997–2003. Virology. 2010;397:322–330. doi: 10.1016/j.virol.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Gerhard W. The role of the antibody response in influenza virus infection. Curr Top Microbiol Immunol. 2001;260:171–190. doi: 10.1007/978-3-662-05783-4_9. [DOI] [PubMed] [Google Scholar]
- 39.Prajapati RS, Indu S, Varadarajan R. Identification and thermodynamic characterization of molten globule states of periplasmic binding proteins. Biochemistry. 2007;46:10339–10352. doi: 10.1021/bi700577m. [DOI] [PubMed] [Google Scholar]
- 40.Ramachandran S, Udgaonkar JB. Stabilization of barstar by chemical modification of the buried cysteines. Biochemistry. 1996;35:8776–8785. doi: 10.1021/bi9600759. [DOI] [PubMed] [Google Scholar]
- 41.Sayle RA, Milner-White EJ. RASMOL: Biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.