Abstract
Excitatory amino acid transporters (EAATs) remove glutamate from synapses. They maintain an efficient synaptic transmission and prevent glutamate from reaching neurotoxic levels. Glutamate transporters couple the uptake of one glutamate to the cotransport of three sodium ions and one proton and the countertransport of one potassium ion. The molecular mechanism for this coupled uptake of glutamate and its co- and counter-transported ions is not known. In a crystal structure of the bacterial glutamate transporter homolog, GltPh, only two cations are bound to the transporter, and there is no indication of the location of the third sodium site. In experiments using voltage clamp fluorometry and simulations based on molecular dynamics combined with grand canonical Monte Carlo and free energy simulations performed on different isoforms of GltPh as well on a homology model of EAAT3, we sought to locate the third sodium-binding site in EAAT3. Both experiments and computer simulations suggest that T370 and N451 (T314 and N401 in GltPh) form part of the third sodium-binding site. Interestingly, the sodium bound at T370 forms part of the binding site for the amino acid substrate, perhaps explaining both the strict coupling of sodium transport to uptake of glutamate and the ion selectivity of the affinity for the transported amino acid in EAATs.
Keywords: excitatory amino acid transporters, fluorescence, the sodium/aspartate symporter from Pyrococcus horikoshii (GltPh), simulations
Glutamate, the main excitatory neurotransmitter in the central nervous system, is removed from the extracellular synaptic space by the glutamate excitatory amino acid transporters EAAT1–5 (1, 2). These transporters thereby maintain an efficient synaptic communication between neurons and prevent extracellular glutamate from reaching neurotoxic levels (1, 2). EAATs are trimeric proteins in which each subunit functions as an independent transporter (3, 4). Each subunit has eight transmembrane domains and two membrane inserted hair-pin (HP) loops (5–7). EAATs use the Na+ and K+ gradients in driving the uptake of glutamate against a concentration gradient (8). The uptake of one glutamate is coupled to the cotransport of three Na+ ions and one H+ ion and the countertransport of one K+ ion (9). How the thermodynamically coupled transport of glutamate, H+, Na+, and K+ ions is accomplished by glutamate transporters is not known. Here we present evidence for a binding site for Na+ ions that suggests a mechanism for the coupling of sodium and glutamate transport.
At least one extracellular Na+ ion appears to bind before glutamate can bind, and at least one extracellular Na+ ion appears to bind after glutamate has bound to the transporter (10–12). For example, in the absence of glutamate, a fluorophore attached to a cysteine at position A430 on HP2 in EAAT3 reported voltage- and Na+-dependent fluorescence changes, consistent with Na+ binding to the glutamate-free transporter and inducing a conformational change in HP2. Li+ also supports glutamate uptake in the EAAT3 subtype but with lower affinity than Na+ (11, 13). In addition, the affinity for different transported amino acids was shown to depend on the nature of the cotransported cation (14). For example, the glutamate affinity was 130-fold less in Li+ than in Na+, whereas the aspartate affinity was only 10-fold less in Li+ than in Na+ (14). The molecular mechanism for this cation specificity of the amino acid affinity is not known.
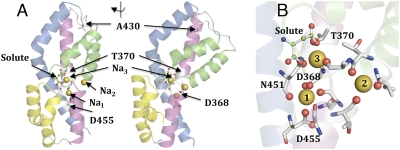
Mutagenesis experiments have identified residues affecting Na+ affinity and/or Li+/Na+ selectivity of glutamate transporters. For example, in EAAT3 the mutation T370S in TM7 lowered the affinity for Na+ and abolished Li+-driven glutamate uptake (13). D368N in TM7 lowered the affinity for Na+ binding to the glutamate-free transporter (15). In the crystal structure of the bacterial glutamate transporter homolog GltPh (7), the side chains of the homologous residues of D368 and T370 are far apart (Fig. 1A). These residues have been suggested to be part of different Na+-binding sites in EAAT3 (7).
Fig. 1.
Putative Na+ ion-binding sites in glutamate transporters. (A) Model with three Na+ sites (Na1-Na2-Na3) suggested by the GCMC simulations. Residues are numbered as in EAAT3. Shown are two views of the C-terminal half of one subunit, rotated 90°. HP1, yellow; HP2, green; TM7, purple; TM8, blue; Na+, yellow balls. (B) The Na+ ion at Na3 interacts with the side chains of T370 and N451 and with the backbone oxygen of A409 (corresponding to T314, N401, and A353 in GltPh) and is in direct contact with the bound substrate. More details are given in SI Appendix, Fig. S3 and S4 and Tables S1–S3.
In a high-resolution crystal structure of GltPh (16), two thallium cations were seen bound to the transporter. These cations were assumed to be binding to two of the Na+ sites, which were called “Na1” and “Na2” (Fig. 1). Na1 was localized close to D455 (EAAT3 numbering) in TM8 (16). Na2 was localized at the tip of HP2 and at break of the alpha helical region in the middle of TM7 (16) and was not located close to any of the residues identified by mutagenesis that affected Na+ binding (Fig. 1). The Na2 site was proposed to be the Na+ site that is occupied after glutamate has bound to the transporter (16).
Using voltage-clamp fluorometry (VCF), we sought here to determine the effects of different mutations on Na+ binding to the glutamate-free transporter. To guide our experimental studies, Grand Canonical Monte Carlo (GCMC) and molecular dynamics (MD) simulations on GltPh and a homology model of EAAT3 were performed to identify plausible locations of the ion-binding sites. MD and free energy perturbation (FEP) simulations were used to assess the stability of the identified ion-binding sites. The experimental data and the computer simulations suggest that bound amino acid substrate and the side chains of T370 and N451 bind the third Na+ ion essential for the transport to occur.
Results
Computations Suggest an Na Site Close to T370.
We first tried to locate a potential third Na+ site using simulations based on the crystal structures of the bacterial EAAT3 homolog GltPh and homology-modeled EAAT3 (SI Appendix). In contrast to the moderate overall sequence identity (<40%) between GltPh and EAAT3, several functional regions are highly conserved throughout the family (7). For example, the binding pocket for cations and bound amino acid substrate has at least 65% homology. Therefore, we replaced amino acid L328 to D466 of GltPh with the corresponding amino acids of EAAT3, creating a GltPh-EAAT3 chimera. Three Na+ ions are cotransported with one glutamate molecule in both the mammalian EAATs and bacterial GltPh (9, 17). Known crystal structures of GltPh have revealed only two putative sodium-binding sites (16). We used probe particle insertion into GltPh in GCMC simulations (18) to uncover possible locations of the third ion-binding site. Methods and controls regarding stability and selectivity of binding pockets are summarized in SI Appendix.
GCMC/MD Simulations of GltPh
GCMC/MD insertions were run on two different isoforms of GltPh (with either an open or closed extracellular gate, henceforth called the ”open” and “closed” isoforms) with either no bound Na+ ion (‘ion free’) or with two Na+ ions bound to the two earlier proposed Na1 and Na2 sites (‘ion loaded’). We evaluated the electrostatic component of the binding free energy as well as Li+/Na+ selectivity of the proposed ion-binding sites. GCMC/MD simulations of the ion-free closed GltPh isoform identified two sites similar to the crystallographic sites Na1 and Na2 plus thee potential binding sites for a third Na+ ion dubbed “Na3,” “Na3′,” and “Na3′′” (Fig. 1; for Na3′ and Na3′′ (SI Appendix, Fig. S2). Further MD simulation shows that a, Na+ ion at Na3 is stably bound at this site for more than 10 ns, whereas coordination at sites Na3′′ and Na3′ is significantly altered in the transporter with three bound ions. However, an Na+ ion at Na3′ is stable in the absence of an Na+ ion at Na1, and we therefore call Na3′ a “transient initial sodium-binding site” (Discussion). Na3′ is formed by ligands from D312, N401, and N310 residues. The combination of all simulation methods places the third Na+ ion (Na3) close to the side chain of T314 and the backbone oxygen of A353, with possible involvement of N401 side-chain oxygen atoms (homologous to T370, A409, and N451 in EAAT3, respectively) and in direct contact with the charged β-carboxylate group of the bound substrate (Fig. 1). After and MD run of 10 ns, the average rmsd of the heavy backbone atoms of the transporter relative to the initial structure was 2.7, 2.6, and 2.9 Å, for an Na+-free structure (2nwl), a two-ion structure (2nwx; Na1-Na2; SI Appendix, Fig. S5), and our structure with the third Na+ bond (Na1-Na2-Na3; Fig. 1 and SI Appendix, Fig. S3). The average distances from this potential site Na3 to sites Na1 and Na2 are 6.9 and 10.8 Å, respectively. The combination of results from equilibrium MD and FEP data on the ion uncharging in this binding site suggests that this position for the Na3 site might be stable.
GCMC/MD Simulations of EAAT3.
GCMC/MD insertions also were run on ion-free closed and open isoforms of the GltPh-EAAT3 chimera. The results of GCMC simulation for the chimera with the EAAT3-binding pocket are slightly different from the results for GltPh. In the closed isoform of the chimera, only three binding sites were identified initially, and all these binding sites were proven stable in 10-ns MD simulations performed on EAAT3 chimera. Site Na1 is formed by three carbonyl oxygen atoms from G362, N366, and N451 as well as by two carboxylates from D455. Site Na2 is formed by carbonyl oxygen atoms from T364, I365, S405, I406, and A408. Site Na3 is formed by side-chains of T370 and N451, as well as by oxygen atoms from the bound glutamate. The distance between the three bound sodium ions is around 7.1–7.5 Å, indicating an almost ideal triangular arrangement in EAAT3 after MD simulation (SI Appendix, Fig. S3). The stability of the model is encouraging but is insufficient to justify use of this model for further functional studies. This model illustrates similarities in ion/substrate binding in bacterial and mammalian transport systems. Therefore, all ion-selectivity, substrate-binding, and ion-binding computations requiring a high-resolution structure were done on the bacterial GltPh.
Mutational Studies Suggest That T370 Binds Sodium Before Glutamate Has Bound.
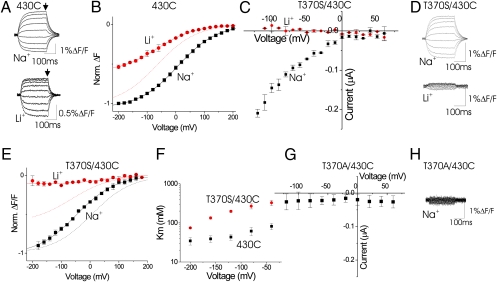
To test experimentally whether cations bind to the proposed Na3 binding site, we used VCF. To attach a fluorescent probe, we introduced a cysteine at position A430 in EAAT3 glutamate transporters. A430C transporters were expressed in Xenopus oocytes and labeled with the thiol-specific Alexa546-maleimide fluorophore. Using VCF, we have shown previously that the fluorescence from Alexa546-labeled A430C transporters reports on a conformational change induced by Na+ binding to the glutamate-free transporter (11). Conformational changes induced by Na+ binding most likely alter the environment around the fluorophore, thereby causing changes in fluorescence. Fluorescence traces from A430C in 100 mM NaCl or 100 mM LiCl are shown in Fig. 2A. As reported earlier, fluorescent-labeled A430C displays voltage-dependent fluorescence changes both in Na+ and Li+ solutions, suggesting that both Na+ and Li+ binding induce conformational changes in HP2 (11). The slopes of the fluorescence versus voltage curve F(V) are similar in Na+ and Li+ [Na+: zδ = 0.41 ± 0.01 (n = 22); Li+: zδ = 0.36 ± 0.01; n = 4]. Na+ and Li+ ions thus bind at closely similar electrical distances into the membrane. However, the F(V) in Li+ solutions is left-shifted by >50 mV relative to the F(V) in Na+ solutions (Na+: V1/2 = −5.6 ± 2.5 mV, n = 22; Li+: V1/2 = −65.2 ± 4.9 mV; n = 4; Fig. 2B). We showed earlier that lowering the Na+ concentration 5-fold shifted the F(V) curve by about the same amount (11), as though Li+ has a 5-fold lower affinity than Na+. In addition, the maximal amplitude of the fluorescence changes is smaller in Li+ than in Na+ (Li+: 3.08% ± 0.5% ΔF/F; Na+: 5.36% ± 0.8% ΔF/F; n = 10), as though Li+ induces a smaller conformational change than Na+ or the efficacy of Li+ to induce conformational change is less than that of Na+.
Fig. 2.
Mutations of T370 affect the affinity for Na+ ions. (A and D) Fluorescence change in Na+ and Li+ for Alexa546-labeled A430C (A) and T370S/A430C (D) transporters in response to voltage steps from −200 mV to +200 mV in glutamate-free solutions. (B and E) Steady-state F(V)s in Na+ (black) and Li+ (red) from A and D (measured at arrows in A). The F(V)s were normalized to −1 at −200 mV in Na+. The data were fit with a Boltzmann curve: F = F0/[1+exp(−zδ(V−V1/2)/kT]. In B, the dashed line shows the fit of the Li+ data normalized to −1. In E, the dashed line shows the fit of the Li+ (red) and Na+ (black) data for A430C from B. (C) Glutamate-activated I/V curves in Na+ (black) and Li+ (red) for Alexa546-labeled T370S/A430C transporters. (F) Voltage dependence of the Km for Na+ for A430C (black) and T370S/A430C (red). The Na+ dependence of the fluorescence for A430C and T370S/A430C was measured at different voltages and fit with the equation F(Na+) = Fmax/(1+Km/[Na+]). Na+ was replaced by equimolar choline. (G) Glutamate-activated I/V curves in Na+ for Alexa546-labeled T370A/A430C. (H) Fluorescence in Na+ for Alexa546-labeled T370A/A430C in response to voltage steps from −200 mV to +200 mV in glutamate-free solutions.
We next studied the mutation T370S in the background of A430C, to test the extent to which the T370S mutation affects the interaction of Na+ with the glutamate-free transporter. Similar to earlier reports for T370S (13), Alexa546-labeled T370S/A430C transporters conduct glutamate-induced transport currents in Na+ but not in Li+ (Fig. 2C). We show here that in glutamate-free solutions Alexa546-labeled T370S/A430C transporters generate fluorescence changes in Na+ but not in Li+ (Fig. 2D). The fluorescence changes in Na+ are similar to the fluorescence changes in 430C. However, the F(V) in Na+ for T370S/A430C is shifted to more hyperpolarized potentials (V1/2 = −60.9 ± 1.5 mV; n = 6), in the same direction as the F(V) for A430C in Li+ (Fig. 2E). Increasing the Na+ concentration from 100 mM to 200 mM for T370S/A430C shifts the V1/2 (V1/2 = −25.4 ± 5.4 mV; n = 4) toward the V1/2 of A430C. The apparent Km for Na+, measured from the amplitude of the Na+-induced fluorescence changes in different Na+ concentrations, increases at all voltages (Fig. 2F). For example at −80 mV, the Km is 61.7 ± 15 mM for 430C and 268 ± 41 mM (n = 4) for the T370S/A430C mutation.
We next tested the mutations T370A, which abolishes Na-driven glutamate uptake in EAAT3 (13). Alexa546-labeled T370A/A430C transporters display no detectable glutamate-activated currents (Fig. 2G) or Na+-dependent fluorescence changes (Fig. 2H). Other T370 mutations, such as T370R, display no detectable glutamate-activated currents or Na+-dependent fluorescence changes (SI Appendix, Fig. S7).
A simple interpretation of these observations is that T370 forms part of a Na+-binding site in the glutamate-free transporter and that the conservative mutation T370S retaining a hydroxyl group still allows cation binding (but with a lower affinity and different selectivity), whereas less conservative mutations abolish Na+ binding. The fact that Li+ does not induce fluorescence changes in the T370S mutations suggests that T370S also alters the cation selectivity of this cation-binding site, as expected if T370 forms part of a cation-binding site in EAAT3.
Mutational Studies Suggest That N451 Binds Sodium Before Glutamate Has Bound.
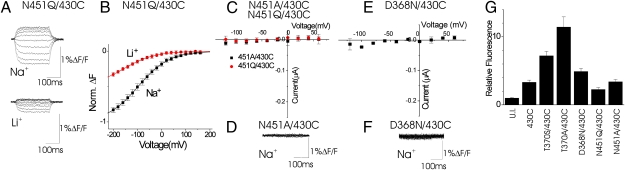
Our computer simulations suggested that residue N451 contributes to the Na3 site in EAAT3 (Fig. 1) and plays an important, if not critical, role in the formation of the transient binding site Na3′ in the open transporter. We therefore tested mutations of N451. Alexa546-labeled N451Q/A430C and N451A/A430C transporters display no detectable glutamate-activated currents (Fig. 3C) or radioactive glutamate uptake (SI Appendix, Fig. S6). Alexa546-labeled N451Q/A430C transporters generate fluorescence changes in Na+ and Li+ (Fig. 3A). The F(V)s were shifted to more hyperpolarized potentials (Na+: V1/2 = −97.0 ± 1.5 mV, n = 12; Li+: V1/2 = −156.9 ± 1.5 mV, n = 9) compared with the F(V) for 430C (Fig. 3B), as if the N451Q mutation lowers the affinity for both Na+ and Li+. In contrast, Alexa546-labeled N451A/A430C transporters display no Na+-dependent fluorescence changes (Fig. 3D). These data are consistent with our computer simulations suggesting that both T370 and N451 contribute to the Na3 site in EAAT3. This result suggests that binding of Na+ to both T370 and N451 is necessary to induce the conformational change reported by a fluorophore attached at 430C.
Fig. 3.
Mutations of N451 and D368 affect the Na+-dependent fluorescence changes. (A) Fluorescence change in Na+ and Li+ for Alexa546-labeled N451Q/A430C transporters in response to voltage steps from −200 mV to +160 mV in glutamate-free solutions. (B) Steady-state F(V)s in Na+ (black) and Li+ (red) from A. (C and E) Glutamate-activated I/V curves in Na+ for N451Q/A430C (C) and D368N/A430C (E). (D and F) Fluorescence in Na+ for Alexa546-labeled N451Q/A430C (D) and D368N/A430C (F) in response to voltage steps from −200 mV to +160 mV in glutamate-free solutions. (G) Alexa546-maleimide fluorescence labeling for oocytes expressing A430C, T370S/A430C, T370A/A430C, D368N/A430C, N451Q/A430C, and N451A/430C. The fluorescence was normalized to the fluorescence of fluorescence-labeled uninjected (UI) oocytes.
Mutational Studies Suggest That D368 Binds Sodium Before Glutamate Has Bound.
We next tested the mutation D368N, which significantly lower the cation affinity of the glutamate-free transporter (15). Alexa546-labeled D368N/A430C transporters display no detectable glutamate-activated currents (Fig. 3E) or Na+-dependent fluorescence changes (Fig. 3F). Oocytes injected with T370A/A430C, N451A/A430C, or D368N/A430C are highly labeled with Alexa488-maleimide compared with uninjected oocytes (Fig. 3G). The amount of fluorescence labeling for these mutations was similar to that of the A430C mutation, showing that the absence of Na+-dependent fluorescence changes in these mutations was not caused by decreased expression in the cell membrane. These fluorescence data, showing that mutations T370A, N451A, or D368N abolish the Na+-dependent fluorescence change, are consistent with our computer simulations suggesting that T370, N451, and D368 contribute to the Na3 (or Na3′) site in EAAT3. This result suggests D368 is necessary for inducing the conformational change reported by a fluorophore attached at 430C. Our simulations suggest that the D368 site (Na3′) plays an important role attracting an Na+ ion in the absence of bound glutamate. However, our simulations show that simultaneous occupancy of both Na1 and Na3′ at D368 is prohibited by the very strong electrostatic repulsion between ions separated by less than 3.5 Å (SI Appendix, Fig. S4B).
Functional Role of the Third Binding Site Assessed by Computations.
Our simulations show that a transporter with only two bound Na+ ions is unable to bind amino acid substrate favorably (Table 1). The introduction of the third ion in direct contact with the cotransported substrate leads to dramatic changes in the binding affinity, stabilizing the amino acid substrate in the binding pocket (Table 1). The removal of the positive charge from direct contact with the substrate (Na1:Na2 occupied state) led to unfavorable binding of the negatively charged aspartate. To assess further the cation binding to the proposed binding sites in GltPh, we compute the free energy of ion binding to the different Na+-binding sites using FEP. We compute the electrostatic component of the free energy of ion binding for both the Na1-Na2 model and Na1-Na2-Na3 model to illustrate the effect of the presence of the third ion on the absolute binding affinity of the other sites (Na1 or Na2). The electrostatic components of the affinities for Na+ are −17.2 and −4.9 kcal/mol for Na1 and Na2, respectively, in the simulations of the two-ion Na1-Na2 model in the presence of aspartate. The computed binding affinities for Na+ are −12.5, −4.7, and −10.1 kcal/mol for Na1, Na2, and Na3, respectively, in simulations of the three-ion Na1-Na2-Na3 model in the presence of aspartate.
Table 1.
Absolute free energy of binding (ΔGtot) for aspartate to the reduced (GSBP) Na1-Na2 and Na1-Na2-Na3 GltPh structure
| aspartate |
||
| Ligand structure | Na1-Na2 | Na1-Na2-Na3 |
| ΔGelec | 9.1 | −29.3 |
| ΔGdisp | −11.3 | −9.3 |
| ΔGrep | −3.3 | 7.2 |
| −ΔGconst(site) | 3.6 | 6.4 |
| − kBTln(Fr) | 6.0 | 6.1 |
| −kBTln(FtC0) | 5.3 | 5.5 |
| ΔGtot | 9.4 | −13.4 |
The absolute binding free energy (ΔGtot) was decomposed into several contributions via a staged protocol, namely, electrostatic component (ΔGelec), dispersive and repulsive parts of van der Waals interactions (ΔGdisp and ΔGrep), configurational constraint component (ΔGconst), and bias from rotational and translational constraining potentials. Units are kcal/mol. Details on the absolute binding free energy computations are found in SI Appendix and our recent publication on the protocol (23).
The ion-binding affinity to GltPh is drastically different in the sites Na1 and Na2. Na1 is a high-affinity binding site for Na+ ions, whereas site Na2 displays only marginally favorable affinity for Na+ ions. The predicted third site (Na3) has relatively high affinity for Na+ as evaluated by the absolute binding free energy from FEP charging/uncharging simulations. As expected, the presence of an ion at site Na3 leads to a small decrease in binding affinity for site Na2. This result is entirely expected, because two ions residing in sites Na2 and Na3 are separated by only about 8 Ǻ, and unfavorable electrostatic repulsion between the two ions will affect ion-binding affinity at these sites. This combination of low-affinity and high-affinity ion-binding sites is similar to that of the leucine transporter LeuT, suggesting different functionality for the high-affinity and low-affinity sites (19).
Discussion
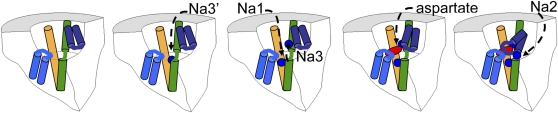
Our results lead us to suggest the following model for extracellular Na+ and glutamate binding to glutamate transporters (Fig. 4).
(i) There are two cation-binding sites in glutamate-free transporters: Na1 contributed by D455 and Na3 contributed by T370 and N451 and transiently by D368. Equilibrium MD simulations suggest that D368 coordinates an early Na+ binding site (Na3′) to the ion- and glutamate-free transporter. However, when an Na+ ion binds to Na1, the Na+ ion at Na3′ quickly (<1 ns) moves over to the Na3 site with T370 (similar results were obtained for D312/T314 sites in the GltPh transporter) because of electrostatic repulsion between ions at the Na1 and Na3′ sites. We therefore conclude that the Na3′ site is only a transient Na+ site and that, once the transporter is fully occupied, the three Na+ ions will bind to Na1, Na2, and Na3 sites. The first two of the three coupled Na+ ions bind to these Na1 and Na3 sites in glutamate-free transporters. It should be noted, however, that D312 may coordinate Na1, because D312 was found to coordinate Na1 in equilibrium MD simulations (SI Appendix, Fig. S4).
(ii) The presence of a bound Na+ ion at Na3 induces a conformational change of HP2 that creates the binding site for glutamate. These conformational changes are reported by the fluorophore in Figs. 2 and 3. The Na+ ion bound at Na3 forms part of the glutamate-binding site.
(iii) Glutamate binding induces a conformational change that closes HP2 around the bound glutamate, bringing HP2 and TM7 close together as seen in the GltPh crystal structure (16).
(iv) The glutamate-induced movement of HP2 creates the binding site (Na2) for the last Na+ ion, which is found between TM7 and HP2 in the crystal structure (Fig. 1A) (16).
Fig. 4.
Third Na+ ion at T370 interacts directly with substrate. In this model of the binding order of the three Na+ ions and glutamate: First one Na+ ion binds to the transient Na3′ site contributed by D368 in the outward-facing state of the transporter. When the second Na+ ion binds to Na1, the Na+ ion at Na3′ is repelled and moves to Na3 contributed by T370 and N451; the binding of the two Na+ ions induces a conformational change in HP2 that creates the glutamate-binding site. Glutamate binds and HP2 closes further around the binding site, creating the last Na+ binding site (Na2) at TM7. Then a Na+ ion binds to Na2.
Experimentally, mutations of T370 (T314) and N451 (N401) altered the ability of Na+ ions to induce conformational changes in the absence of glutamate, and mutations of T370 also changed the cation selectivity for these conformational changes, consistent with our computer simulations suggesting that both of these two residues contribute to the Na3 sites in glutamate-free EAAT3 (Fig. 1B). Mutations of D368 also prevented Na+ ions from inducing the conformational changes underlying the fluorescence changes in the absence of glutamate, in accordance with earlier studies showing that D368N altered the affinity for Na+-induced leak currents in the glutamate-free transporter (15). In addition, our simulations suggest that the Na+ ions bound at T370 interacts directly with the bound amino acid and affects the affinity of the bound amino acid substrate.
Earlier theoretical studies also have suggested that the third Na+ ion binds in proximity to the bound amino acid substrate (20, 21). Holley and Kavanaugh (21), using the knowledge-based method VALE, identified a putative third ion-binding site in which a Na+ ion was coordinated by the bound amino acid substrate and three backbone oxygen atoms from HP2. T370 was not predicted to form part of this Na+-binding site (21). Shrivastava et al. (20) noted that T370 (T314 in GltPh) is able to bind water and that this water, in combination with the bound glutamate, could participate in coordinating a Na+ ion. Taken together, both thermodynamic and steric considerations point toward the potential location of an ion-binding site in this region, in good agreement with our results from GCMC/MD modeling.
The Na3 site, formed by the side chains of T370 and N451 and the backbone oxygen atom of A409, is reminiscent of that found in another Na+-coupled transporter, LeuT (Fig. 1) (22). In the Na3 site in EAATs the cation interacts directly with the bound substrate, as does one cation in LeuT (22). Another similarity with LeuT is the direct structural coupling from the Na3 cation site to one of the other Na+ sites (Na1) in the original crystal structure, because the backbone of N451 forms part of the binding site for Na1. Our MD simulations suggest that the Na+ ion bound at T370 interacts directly with the carboxyl oxygen of the transported amino acid, so that the amino acid binding is coordinated by the Na+ ion at site Na3 (Fig. 1). Previously, it has been shown that the affinity of the amino acid substrate depends critically on the type of cotransported cation (14). For example, the affinity for l-glutamate is 130-fold lower in the presence of Li+ ions than in the presence of Na+ ions, whereas the affinity for aspartate is only 10-fold lower in the presence of Li+ ions than in the presence of Na+ ions (14). Here, we found that the direct interaction between Na+ ions and transported amino acid is critical for favorable substrate binding to the transporter (Table 1). The difference in the binding affinity of aspartate in the two- and three-ions bound transporter is more than 20 kcal/mol, as evaluated with the staged free energy simulations protocol developed previously (23). The analysis of the path-dependent free energy decomposition into components suggests that the key reason for this favorable gain in binding energy is a change in the electrostatic component (Table 1). While in the binding pocket of the GltPh transporter, the negatively charged aspartate is unable to compensate for the loss of the favorable electrostatic interaction with bulk water, amounting to as much as −140 kcal/mol (24), without direct coupling to the positively charged cation.
The direct contact of the cation at site Na3 to the transported amino acid could underlie the Na+/Li+ selectivity of the apparent affinity for the transported amino acid in EAAT3. Other possibilities suggested by our fluorescence data (Fig. 2B) are that Li+ is less efficient than Na+ in inducing the conformational change once it is bound to the transporter or that Li+ binding does not induce as large conformational change as Na+. This latter possibility also would explain why the affinity of cysteine, which lacks the γ-carboxylate group of glutamate that would interact directly with the cation bound at Na3, also is dramatically different in Li+ and Na+ (14). We therefore propose that there is both a cation specific effect on the conformational change of HP2 and a direct interaction of the bound cation with substrates that contain a β- or γ-carboxylate group, such as aspartate and glutamate. This intimate link between the cation and the amino acid substrate in the glutamate transporter is similar to the interaction shown in the crystal structure for the LeuT transporter (22), where the Na+ ion and the substrate are bound together, and suggests a simple mechanism for how the transport of Na+ and cotransported substrate are tightly coupled in glutamate transporters.
Materials and Methods
Molecular Biology.
Site-directed mutagenesis of the human EAAT3, in vitro synthesis of RNA, and RNA injection into Xenopus laevis oocytes were performed as described previously (11).
Electrophysiology.
Glutamate-induced currents were calculated by subtracting the current measured in glutamate-free Na+ Ringer's solution from the two-electrode voltage-clamp current measured in Na+ Ringer's solution containing 1 mM glutamate as described previously (11). For Li+ experiments, Na+ was replaced by Li+.
Voltage-Clamp Fluorometry.
VCF experiments were performed as described previously (11). Oocytes were labeled for 60 min with 10 μM Alexa-546 maleimide (Molecular Probes) in Na+ Ringer's solution. Fluorescence was monitored through a rhodamine filter cube (HQ545/x30, Q570LP, and HQ620/60m), low-pass filtered at 200–500 Hz, and digitized at 1 kHz.
Grand Canonical Monte Carlo Simulations.
For computations of GCMC simulations and for computations of the free energy of binding, only a sphere of 15 Å around the bound substrate was treated explicitly; the rest of the system was accounted for via generalized solvent boundary potential developed earlier (24). The GCMC simulation was run for 300 cycles, each with 20,000 MC steps. The structures with inserted water molecules were relaxed for 50 ps of Langevin dynamics for every GCMC cycle. These two approaches combined provide a comprehensive description of the electrostatic map and accessibility, thus helping identify buried metal sites in proteins. A similar methodology has been used by Beck's group (25).
Molecular Dynamics Simulations.
MD simulations were run on optimized structures of GltPh using the CHARMM c33b2 (26). Na+ ions were introduced at the binding pockets identified in GCMC calculations. The simulation box contained one GltPh transporter (trimer), bound Na+, three cotransported substrates, and 298 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) lipid molecules, solvated by a 100-mM NaCl aqueous salt solution, comprising a total of ~120,000 atoms. The lipid bilayer was built as described previously (27). All simulations were carried at constant pressure (1 atm) and constant temperature with periodic boundary conditions (28). Electrostatic interactions were treated using a particle mesh Ewald (PME) algorithm (29). All systems were equilibrated for 1.5 ns and then subjected to 5- to 15-ns runs to test stability.
Free Energy Simulations: Evaluation of Absolute Ion and Substrate-Binding Affinities.
FEP simulations were carried out using the CHARMM PERT command for GltPh containing two and three Na+ ions. The ligand-binding computations were performed with the staged protocol of Deng and Roux (30). A similar scheme had been used previously for studies of substrate and antidepressant binding to the neurotransmitter transporter LeuT (23). The absolute binding free energies for ions were evaluated from FEP experiments with periodic boundary conditions using parameters developed earlier (19). The weighted histogram analysis method was used to postprocess the FEP data and evaluate ion-binding affinities for the binding sites (Na1, Na2, and Na3). The free energy of Na+ uncharging (Gbulkelec) in the bulk water was estimated to be −98.6 kcal/mol (27). The absolute binding affinity (electrostatic component) was calculated as ΔGelec = Gsiteelec − Gbulkelec.
Supplementary Material
Acknowledgments
We thank Drs. B. Roux and Y. Deng for their help with the staged free energy protocol and Drs. F. Elinder and W. Nonner for helpful comments on the manuscript. This work was funded by Grant NS051169 from the National Institute of Neurological Disorders and Stroke (to H.P.L.) and by Grant 200804MOP-186232 from the Canadian Institute for Health Research (to S.Y.N). S.Y.N. is an Alberta Heritage Foundation for Medical Research Scholar and Canadian Institute for Health Research New Investigator. B.L. is an Alberta Ingenuity Graduate Student.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006289107/-/DCSupplemental.
References
- 1.Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 2.Tzingounis AV, Wadiche JI. Glutamate transporters: Confining runaway excitation by shaping synaptic transmission. Nat Rev Neurosci. 2007;8:935–947. doi: 10.1038/nrn2274. [DOI] [PubMed] [Google Scholar]
- 3.Grewer C, et al. Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry. 2005;44:11913–11923. doi: 10.1021/bi050987n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch HP, Larsson HP. Small-scale molecular motions accomplish glutamate uptake in human glutamate transporters. J Neurosci. 2005;25:1730–1736. doi: 10.1523/JNEUROSCI.4138-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grunewald M, Bendahan A, Kanner BI. Biotinylation of single cysteine mutants of the glutamate transporter GLT-1 from rat brain reveals its unusual topology. Neuron. 1998;21:623–632. doi: 10.1016/s0896-6273(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 6.Yernool D, Boudker O, Folta-Stogniew E, Gouaux E. Trimeric subunit stoichiometry of the glutamate transporters from Bacillus caldotenax and Bacillus stearothermophilus. Biochemistry. 2003;42:12981–12988. doi: 10.1021/bi030161q. [DOI] [PubMed] [Google Scholar]
- 7.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 8.Kanner BI, Bendahan A. Binding order of substrates to the sodium and potassium ion coupled L-glutamic acid transporter from rat brain. Biochemistry. 1982;21:6327–6330. doi: 10.1021/bi00267a044. [DOI] [PubMed] [Google Scholar]
- 9.Zerangue N, Kavanaugh MP. Flux coupling in a neuronal glutamate transporter. Nature. 1996;383:634–637. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]
- 10.Wadiche JI, Arriza JL, Amara SG, Kavanaugh MP. Kinetics of a human glutamate transporter. Neuron. 1995;14:1019–1027. doi: 10.1016/0896-6273(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 11.Larsson HP, Tzingounis AV, Koch HP, Kavanaugh MP. Fluorometric measurements of conformational changes in glutamate transporters. Proc Natl Acad Sci U S A. 2004;101:3951–3956. doi: 10.1073/pnas.0306737101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watzke N, Bamberg E, Grewer C. Early intermediates in the transport cycle of the neuronal excitatory amino acid carrier EAAC1. J Gen Physiol. 2001;117:547–562. doi: 10.1085/jgp.117.6.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borre L, Kanner BI. Coupled, but not uncoupled, fluxes in a neuronal glutamate transporter can be activated by lithium ions. J Biol Chem. 2001;276:40396–40401. doi: 10.1074/jbc.M104926200. [DOI] [PubMed] [Google Scholar]
- 14.Menaker D, Bendahan A, Kanner BI. The substrate specificity of a neuronal glutamate transporter is determined by the nature of the coupling ion. J Neurochem. 2006;99:20–28. doi: 10.1111/j.1471-4159.2006.04003.x. [DOI] [PubMed] [Google Scholar]
- 15.Tao Z, Zhang Z, Grewer C. Neutralization of the aspartic acid residue Asp-367, but not Asp-454, inhibits binding of Na+ to the glutamate-free form and cycling of the glutamate transporter EAAC1. J Biol Chem. 2006;281:10263–10272. doi: 10.1074/jbc.M510739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–393. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 17.Groeneveld M, Slotboom DJ. Na(+): Aspartate coupling stoichiometry in the glutamate transporter homologue Glt(Ph) Biochemistry. 2010;49:3511–3513. doi: 10.1021/bi100430s. [DOI] [PubMed] [Google Scholar]
- 18.Woo HJ, Dinner AR, Roux B. Grand canonical Monte Carlo simulations of water in protein environments. J Chem Phys. 2004;121:6392–6400. doi: 10.1063/1.1784436. [DOI] [PubMed] [Google Scholar]
- 19.Caplan DA, Subbotina JO, Noskov SY. Molecular mechanism of ion-ion and ion-substrate coupling in the Na+-dependent leucine transporter LeuT. Biophys J. 2008;95:4613–4621. doi: 10.1529/biophysj.108.139741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrivastava IH, Jiang J, Amara SG, Bahar I. Time-resolved mechanism of extracellular gate opening and substrate binding in a glutamate transporter. J Biol Chem. 2008;283:28680–28690. doi: 10.1074/jbc.M800889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holley DC, Kavanaugh MP. Interactions of alkali cations with glutamate transporters. Philos Trans R Soc Lond B Biol Sci. 2009;364:155–161. doi: 10.1098/rstb.2008.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl-dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 23.Zhao CF, Caplan DA, Noskov SY. Evaluations of the absolute and relative free energies for antidepressant binding to the amino acid membrane transporter LeuT with free energy simulations. J Chem Theor Comp. 2010;6:1900–1914. doi: 10.1021/ct9006597. [DOI] [PubMed] [Google Scholar]
- 24.Banavali NK, Im W, Roux B. Electrostatic free energy calculations using the generalized solvent boundary potential method. J Chem Phys. 2002;117:7381–7388. [Google Scholar]
- 25.Kuang ZF, Liu AP, Beck TL. TransPath: A computational method for locating ion transit pathways through membrane proteins. Proteins. 2008;71:1349–1359. doi: 10.1002/prot.21822. [DOI] [PubMed] [Google Scholar]
- 26.Brooks BR, et al. CHARMM: A program for macromolecular energy minimization and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 27.Noskov SY, Bernèche S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 28.Feller S, Zhang Y, Pastor R, Brooks B. Constant pressure molecular dynamics simulation—the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 29.Essman U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 30.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J Phys Chem B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.