Abstract
ATR kinase is a critical upstream regulator of the checkpoint response to various forms of DNA damage. Previous studies have shown that ATR is recruited via its binding partner ATR-interacting protein (ATRIP) to replication protein A (RPA)-covered single-stranded DNA (RPA-ssDNA) generated at sites of DNA damage where ATR is then activated by TopBP1 to phosphorylate downstream targets including the Chk1 signal transducing kinase. However, this critical feature of the human ATR-initiated DNA damage checkpoint signaling has not been demonstrated in a defined system. Here we describe an in vitro checkpoint system in which RPA-ssDNA and TopBP1 are essential for phosphorylation of Chk1 by the purified ATR-ATRIP complex. Checkpoint defective RPA mutants fail to activate ATR kinase in this system, supporting the conclusion that this system is a faithful representation of the in vivo reaction. Interestingly, we find that an alternative form of RPA (aRPA), which does not support DNA replication, can substitute for the checkpoint function of RPA in vitro, thus revealing a potential role for aRPA in the activation of ATR kinase. We also find that TopBP1 is recruited to RPA-ssDNA in a manner dependent on ATRIP and that the N terminus of TopBP1 is required for efficient recruitment and activation of ATR kinase.
Keywords: DNA damage checkpoint, signal transduction, topoisomerase IIβ binding protein 1
To prevent the potentially catastrophic consequences of DNA damage, eukaryotic cells activate DNA damage checkpoint responses which delay cell cycle progression until the damage is repaired (1, 2). ATR is a member of the phosphoinositide 3-kinase-related protein kinase (PIKK) family and a major regulator of checkpoint responses to incompletely replicated DNA and various forms of damaged DNA, including UV-induced DNA damage (1, 3). ATR forms a complex with ATR-interacting protein (ATRIP) which is essential for the checkpoint function of ATR (3). ATRIP directly interacts with replication protein A (RPA) (4), a heterotrimeric complex consisting of RPA1, RPA2, and RPA3 subunits, which has high binding affinity for single-stranded DNA (ssDNA) (5). Previous work with human, Xenopus, and yeast systems indicates that RPA recruits ATR to ssDNA through an interaction with ATRIP (4, 6–8). ATR activation also requires additional checkpoint components, including the Rad17-RFC complex, the 9-1-1 (Rad9-Hus1-Rad1) checkpoint complex, and TopBP1 (2). Interactions between Rad9 and TopBP1 are important for the activation of ATR (9–11). Notably, TopBP1 has been shown to be a direct activator of ATR and strongly stimulates the kinase activity of ATR even in the absence of other checkpoint components (12, 13). Therefore, the current model for ATR activation is as follows: ssDNA generated at sites of DNA damage during repair, transcription, or replication is bound by RPA, which then recruits ATR through its physical interaction with ATRIP. Independently, the 9-1-1 complex is loaded by Rad17-RFC at ssDNA containing primer/template junctions, and the loaded 9-1-1 complex recruits TopBP1 in the proximity of ATR to activate its kinase function. It has been reported that mutations in RPA1 that disrupt its interaction with ATRIP also abrogate ATR activation in yeast, Xenopus, and humans (4, 6, 8). However, the essential role of the ATRIP-RPA interaction for ATR activation has been questioned by the finding that ATR phosphorylates Chk1 even when the interaction between ATRIP and RPA is disrupted by mutations in ATRIP (7). Furthermore, checkpoint defective mutations in RPA1 also disrupt interactions with other checkpoint proteins, such as Rad9 (14) and Rad17-RFC (15, 16), indicating that RPA may contribute at many levels to the activation of ATR.
Therefore, detailed mechanistic studies on the activation of ATR in a well defined system are necessary to clarify the role of RPA-coated ssDNA in the checkpoint response. Notably, even though RPA-coated ssDNA is considered to be the predominant, if not the sole, signal for activation of the ATR → Chk1 signal transduction pathway, an in vitro system for ATR-initiated checkpoint signaling recapitulating a key feature of the model: (RPA-ssDNA) + (ATR-ATRIP) + TopBP1 → Chk1 phosphorylation has not yet been developed. Here, we describe the reconstitution of such a system. Moreover, we show that mutations in RPA1 that abrogate the checkpoint function also disrupt signaling in our in vitro system, indicating that this system is a reasonable representation of the signal transduction pathway. In addition, we demonstrate that an alternative form of RPA (aRPA), in which the RPA2 subunit is replaced with RPA4, recruits ATRIP to ssDNA and stimulates TopBP1-dependent activation of ATR similar to the canonical RPA. Analysis of the requirements for ATR activation in this system allowed us to discover that the N terminus of TopBP1 is required to form a stable checkpoint complex on RPA-ssDNA required for efficient ATR activation.
Results
In the ATR-mediated DNA damage checkpoint, the phosphorylation of Chk1 by ATR in response to ssDNA generated by replication arrest or DNA damage processing is considered a key step (3). Because ssDNA is usually bound to RPA in vivo, it is generally thought that RPA-ssDNA is the checkpoint signaling structure (3). Despite the availability of several in vitro systems for analyzing ATR kinase activity (13, 17–24), this key reaction has not been demonstrated with purified proteins. The discovery of TopBP1 as a potent activator of ATR kinase (12), and the development of a method to purify ATR from a native source (23) provided the means to attempt to reconstitute this reaction in a defined system.
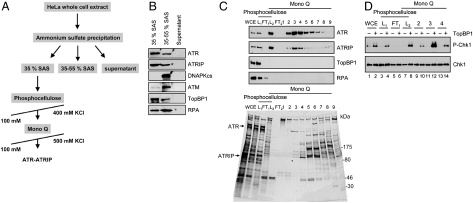
Purification of Native ATR-ATRIP.
Because of the large size of ATR, it has been difficult to purify ectopically expressed ATR-ATRIP using mammalian expression systems. Our efforts to purify ATR-ATIP by immuno-affinity methods have yielded preparations of low activity. Therefore, we decided to carry out our studies with ATR-ATRIP purified from its native source. We previously described a method for the purification of limited amounts of ATR-ATRIP from HeLa cells (23). In this study, we describe an improved purification scheme to obtain highly pure ATR-ATRIP with a better yield (Fig. 1 and Table S1). We have found that in testing for ATR-ATRIP activity in cell extracts or partially purified preparations, some members of the PIKK family, particularly DNA-PK and ATM, are a major source of background activity. Fortuitously, fractionation of HeLa cell extracts with 35% saturated ammonium sulfate (SAS) precipitated 70%–80% of the ATR-ATRIP complex while leaving essentially all DNA-PK and about 90% of ATM in the supernatant (Fig. 1B), thus providing a good source of ATR-ATRIP free of other PIKKs. However, as shown in Fig. 1B, at this stage of the purification, the ATR-ATRIP fraction contains most of the TopBP1 and some of the RPA in the extract. Because TopBP1 is a key component of ATR-mediated DNA damage checkpoint signaling, and we wished to investigate the role of RPA in the checkpoint response, it was necessary to separate TopBP1 and RPA from ATR-ATRIP. Hence, ATR-ATRIP was further purified by chromatography with phosphocellulose and Mono Q resins (Fig. 1C). In the peak ATR-containing fraction from the Mono Q column, ATR was the major band by SDS-PAGE/silver stain analysis, and both ATR and ATRIP were detected by immunoblotting (Fig. 1C, fraction #3). Functional analysis of the Mono Q fractions revealed a close correspondence between the ATR-ATRIP peak identified by immunoblotting and TopBP1-dependent Chk1 phosphorylation activity (Fig. 1D). Importantly, the final purified fraction is free of other known checkpoint proteins examined, including TopBP1 and RPA (Fig. 1C), and its kinase activity on Chk1 is completely dependent on TopBP1 (Fig. 1D, compare lanes 11 and 12).
Fig. 1.
Purification of human ATR-ATRIP from HeLa cells. (A) Purification scheme. (B) Separation of ATR-ATRIP from DNA-PK and ATM by 35% SAS precipitation. ATR was precipitated with 35% SAS, leaving DNA-PK and ATM in the supernatant which then could be precipitated with 55% SAS. The dialyzed fractions were analyzed by Western blotting with the indicated antibodies. (C) Analysis of purified ATR-ATRIP by Western blotting (top) and silver staining (bottom) of 5%–10% discontinuous SDS-PAGE. (WCE), whole cell extract; L1, 35% SAS fraction loaded onto phosphocellulose column; FT1, flow-through of phosphocellulose column; L2, peak ATR-ATRIP fractions from phosphocellulose were applied to the Mono Q column; FT2, flow-through of Mono Q column; 1–9, Mono Q fractions. In the bottom, the band corresponding to ATR and the position of ATRIP (which is inefficiently stained by silver) are marked with asterisks in fraction 3. (D) TopBP1-dependent kinase activity of purified fractions. The indicated fractions (1 μL each) were tested for Chk1 phosphorylation in the absence and presence of TopBP1. The reactions were analyzed by Western blotting for phospho-Chk1 (S345) and Chk1 as indicated.
RPA-ssDNA Dependent Activation of ATR Kinase.
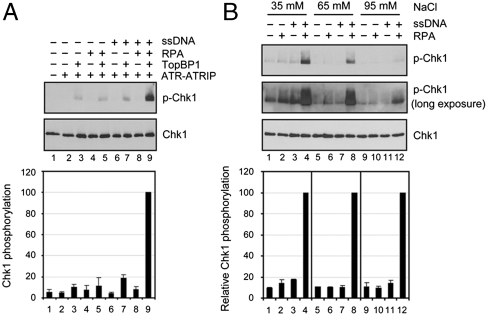
In the ATR-mediated signaling pathway, formation of an RPA-ssDNA complex is considered to be the activator of ATR which phosphorylates Chk1. However, this model, which is largely based on in vivo data and, in part, on data from the Xenopus oocyte extract/sperm chromatin system, has not been tested critically in vitro (3). In fact, previous attempts to activate ATR kinase with either ssDNA or RPA-ssDNA have either failed or yielded marginal stimulation (8, 23, 25), and activation of ATR-mediated Chk1 phosphorylation by RPA-ssDNA has not been demonstrated.
With the identification of TopBP1 as a potent activator of ATR and the availability of ATR-ATRIP isolated from a natural source, we examined the influence of RPA and DNA in our reconstituted system. We incubated purified ATR-ATRIP with the Chk1 substrate in the presence of various combinations of ssDNA, RPA, and TopBP1 (Fig. 2A). Under our experimental conditions, even though TopBP1 with or without RPA or ssDNA has a detectable stimulatory effect, the combination of RPA-ssDNA and TopBP1 causes significant phosphorylation of Chk1 by ATR above that observed with the individual components alone (Fig. 2A and Fig. S1). Even the highest concentration of RPA added marginally stimulated the TopBP1-dependent ATR kinase activity while RPA-ssDNA stimulates Chk1 phosphorylation in a dose-dependent manner (Fig. S1). The effect of RPA-ssDNA is observed even when the ionic strength of the reaction mixture is increased to physiologically relevant concentrations. While at high ionic strength the overall phospho-Chk1 signal intensity decreases (Fig. 2B, compare lanes 4, 8, and 12), the signal-to-noise ratio of RPA-ssDNA vs. RPA or ssDNA alone at 95 mM NaCl becomes apparent after long exposure of the blot (Fig. 2B, bottom inset), indicating the high specificity for stimulation by RPA-ssDNA. Thus, we conclude that our system constitutes a reasonably accurate representation of the in vivo ATR → Chk1 signaling pathway.
Fig. 2.
TopBP1-dependent ATR activation is stimulated by RPA-ssDNA. (A) Effect of RPA-ssDNA on ATR kinase. The reactions were carried out with ATR-ATRTIP, Chk1, TopBP1, RPA, and ϕX174 ssDNA. The DNA (0.6 nanogram = 1.85 pmol nucleotides) was preincubated with RPA (320 fmol) and then ATR-ATRIP (1 fmol), TopBP1 (50 fmol), and Chk1 (100 fmol) were added and the mixture was incubated at 30 °C for 20 min. Reactions were analyzed by immunoblotting for phospho-Chk1 (S345) and Chk1 (top). The levels of Chk1 phosphorylation from three experiments were quantified and are plotted (mean ± standard error) relative to the maximum (bottom). (B) Effect of ionic strength on kinase activity. The reactions were conducted as in (A), in reaction buffers of differing ionic strengths. Short and long exposures of the Western blot are shown to reveal the stimulatory effects of RPA-ssDNA under differing ionic strengths. In the quantitative analysis (bottom) the maximum values obtained with RPA-ssDNA at each ionic strength were set to 100 and the other values within each set are expressed relative to those values (averages of three experiments). Columns 1–8 values were from the short exposure and those in lane 9–12 were from the long exposure.
Specificity of RPA-ssDNA Stimulation of ATR Kinase.
Having developed an in vitro assay for phosphorylation of Chk1 by ATR, we further analyzed in more detail the factors necessary for this stimulation: RPA, ssDNA, and TopBP1. We first determined whether the RPA stimulation required human RPA. To this end, the Saccharomyces cerevisiae ortholog of RPA (scRPA), Escherichia coli SSB (ecSSB), and the newly discovered human ortholog of E. coli SSB, the human SSB1 (hSSB1, which has been implicated in UV-induced DNA damage checkpoint response) (26) were compared to human RPA (hRPA) in the ATR kinase assay. All these ssDNA-binding proteins (Fig. S2A) including scRPA, which is structurally similar to human RPA, failed to replace human RPA in the kinase assay (Fig. S2B). Thus, these data indicate that specific interactions between human ATR-ATRIP and human RPA play an important role in RPA-ssDNA-mediated ATR activation. This point is further analyzed in the following section.
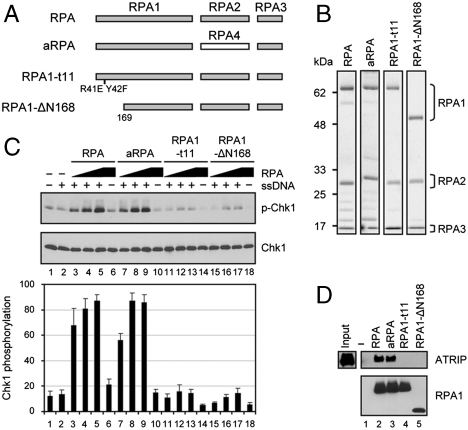
Effect of Alternative RPA and of RPA Checkpoint Mutants on Phosphorylation of Chk1 by ATR.
To understand the mechanism by which RPA participates in ATR → Chk1 signaling, we analyzed the effects of different forms of human RPA, including an alternative form of RPA (aRPA) in which RPA2 is replaced by RPA4 (27), and of two RPA mutants which have mutations in the N terminus of RPA1 subunit: RPA1-t11 (R41E, Y42F) and RPA1-ΔN168 (deletion of the N-terminal 168 amino acids) (28) (Fig. 3A). Alternative RPA does not support DNA replication although it has biochemical properties similar to the canonical RPA (29, 30), but it does support nucleotide excision repair (31). To determine whether aRPA could function in the DNA damage checkpoint response, we purified aRPA (Fig. 3B) and examined its effect on the activation of the ATR kinase. Interestingly, aRPA significantly stimulates TopBP1-dependent ATR activation in the presence of ssDNA, and the level of stimulation by aRPA is comparable to that of canonical RPA (Fig. S3A). Given this observation and the notion that recruitment of ATR to RPA-ssDNA through an ATRIP-RPA interaction is necessary for activation of ATR, we determined whether aRPA could also recruit ATRIP to ssDNA. To this end, we analyzed the binding of ATRIP to RPA-ssDNA and aRPA-ssDNA complexes by a pull-down assay. We found that ATRIP is efficiently recruited to ssDNA in the presence of either canonical RPA or aRPA (Fig. S3B), suggesting that this alternative RPA can substitute for canonical RPA in the ATR-mediated checkpoint response.
Fig. 3.
TopBP1-dependent ATR activation is stimulated by canonical and alternative RPA but not by checkpoint defective RPA mutants. (A) Schematic of RPA, aRPA, and RPA mutants. (B) Analysis of various forms of RPA by SDS-PAGE and Coomassie Blue staining. (C) Stimulation of TopBP1-dependent ATR kinase by RPA and aRPA, but not by RPA1-t11 and RPA1-Δ168. Where indicated, the reactions contained 80, 240, or 720 fmol each RPA complex. Bottom shows quantitative analysis of three experiments. (D) Recruitment of ATRIP to ssDNA by RPA or aRPA, but not by RPA mutants. Biotinylated 80-mer oligonucleotide (1 pmol)-bound streptavidin beads were incubated with each RPA complex (4 pmol). The beads were retrieved, washed, and incubated with ATRIP (5 pmol). The beads were then isolated and washed, and bound proteins were analyzed by immunoblotting with the corresponding antibodies. The input lane contains 4 pmol of ATRIP.
Previous work has shown that two RPA mutants, RPA1-t11 and RPA1-ΔN168 are defective in checkpoint activation but efficient for DNA replication in vivo (28). Accordingly, we purified the RPA mutants to test in our in vitro checkpoint system (Fig. 3B). In contrast to RPA and aRPA, both RPA1-t11 and RPA1-ΔN168 failed to support Chk1 phosphorylation by ATR (Fig. 3C). DNA recruitment assays for ATRIP revealed that both RPA mutants bind to ssDNA with affinities comparable to that of RPA and aRPA, but fail to recruit ATRIP to the DNA-protein complex (Fig. 3D). These results indicate that aRPA and RPA, both of which contain the RPA1 subunit, can equally function in our checkpoint assay, but RPA with mutations in the RPA1 subunit, RPA1-t11 or RPA1-ΔN168, does not. Therefore, the N terminus of RPA1 is essential for the activation of ATR, and impaired interactions of the RPA mutants with ATRIP cause significant defects in the ATR-mediated phosphorylation of Chk1. This result is in agreement with reports that ATRIP interacts with RPA mainly through the region encompassing the N-terminal 168 amino acids of RPA1 (4, 6, 7, 14).
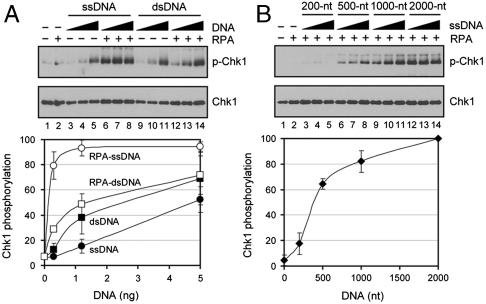
Role of DNA in ATR → Chk1 Signaling.
To further test the model that RPA-ssDNA is the signal for ATR activation, we investigated the effects of DNA secondary structure, DNA damage, and DNA length on the phosphorylation of Chk1 by ATR. In Fig. 4A we compared the effects of ssDNA and dsDNA on ATR activation in the absence and presence of RPA. In agreement with our previous report, both ssDNA and dsDNA stimulated TopBP1-dependent ATR kinase moderately and at comparable levels (23). Importantly, while RPA had no significant effect on the stimulation afforded by dsDNA, it increased the stimulation conferred by ssDNA nearly 5-fold, indicating that the stimulatory effect of RPA is highly specific to ssDNA. These results are consistent with the much higher affinity of RPA to ssDNA relative to dsDNA, and support the model that RPA-ssDNA is a key signal for activating ATR kinase.
Fig. 4.
Effect of DNA secondary structure and DNA length on TopBP1-dependent activation of ATR. (A) Effect of DNA secondary structure. ATR and TopBP1 were incubated with 0.3, 1.2, or 5 ng of ϕX174 ssDNA or dsDNA in the absence or presence of RPA (320 fmol) and the phosphorylation of Chk1 was detected by immunoblot (top) and quantitative analysis plotted (bottom). (B) DNA length-dependent ATR activation in the presence of RPA. The reactions were carried out with various sizes of ssDNA fragments (0.3, 1.2, or 5 ng) in the presence of RPA (320 fmol). Bottom shows averages of Chk1 phosphorylation obtained with 5 ng ssDNA.
Next, we compared the effect of DNA damage on ATR kinase activity in the absence and presence of RPA, because of previous findings that bulky base adducts in dsDNA stimulate TopBP1-dependent ATR kinase activity (23, 32). At the salt concentration used in these experiments where no significant stimulation is seen by dsDNA, the BPDE (benzo[a]pyrene diol epoxide)-damaged DNA stimulates TopBP1-dependent phosphorylation of Chk1 4–5-fold and this stimulation is not significantly affected by RPA (Fig. S4). These results are in agreement with the proposal that recruitment of ATR to DNA by either RPA or directly by DNA damage enhances its TopBP1-dependent kinase activity on downstream targets.
The results shown in Fig. 4B reveal a strong effect of DNA length. At equimolar nucleotide concentrations (and hence 10-fold molar excess of 200 nt-long DNA) the 2,000 nt-long RPA-DNA was at least 5-fold more efficient than the 200 nt-long RPA-DNA in stimulating ATR kinase. These findings suggest a cooperative mechanism in RPA-ssDNA-stimulated and TopBP1-dependent ATR activation. However, at this point we cannot propose a physical model for this cooperative reaction. We note, however, that we previously observed a similar behavior with stimulation of TopBP1-dependent ATR kinase by damaged DNA in the absence of RPA (32). Thus, it appears that the activation of the ATR kinase requires the presence of a critical number of ATR-ATRIP molecules on the same DNA molecule.
Role of TopBP1 in ATR → Chk1 Signaling.
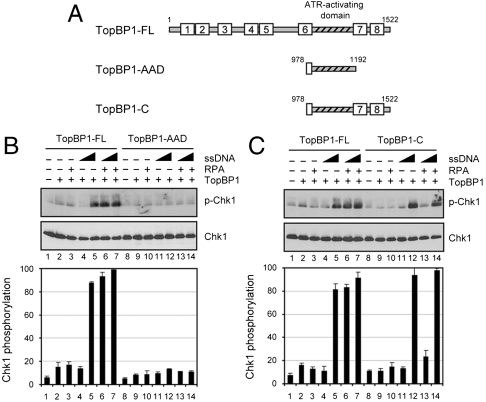
TopBP1 is a multifunctional protein that plays important roles during both DNA replication and the DNA damage checkpoint response (33). TopBP1 contains 1,522 amino acids with eight BRCT (BRCA1 carboxyl-terminal) domains located over the entire length of the protein (33) (Fig. 5A). A region spanning amino acids 978–1,192 (ATR-Activating Domain (AAD), TopBP1-AAD in Fig. 5A) has been previously demonstrated to be sufficient to activate ATR kinase under reaction conditions that were independent of DNA or RPA (8, 12, 23). However, under stringent reaction conditions (higher ionic strength), where ATR activation was both TopBP1- and DNA-dependent, TopBP1-AAD was unable to stimulate ATR kinase due to lack of DNA binding affinity (23, 32). Hence, we wished to determine if TopBP1-AAD is able to stimulate ATR activation in the presence of RPA-coated ssDNA. To this end, we tested TopBP1-AAD and full-length TopBP1 in the presence of ssDNA or RPA-ssDNA (Fig. 5B). In agreement with our earlier reports, at the high concentration of DNA, full-length TopBP1 was capable of stimulating ATR kinase in the absence of RPA (lane 5) (23, 32). With the combination of RPA + ssDNA, full-length TopBP1 stimulated ATR kinase at the low DNA concentration (lane 6). In contrast, neither ssDNA, RPA, nor the combination of the two (lane 8–14) was capable of conferring ATR kinase activation in the presence of TopBP1-AAD, implying that the AAD of TopBP1 is not sufficient for RPA-ssDNA-mediated ATR activation. It should be noted that these concentrations of full-length TopBP1 and TopBP1-AAD produced similar levels of basal ATR-stimulatory activity with higher amounts of ATR kinase (23, 32).
Fig. 5.
The N terminus of TopBP1 is required for efficient ATR activation in the presence of RPA-ssDNA. (A) Schematic of full-length TopBP1 and its fragments. The boxes indicate the BRCT regions, and the AAD is indicated. (B) No stimulatory effect of DNA or RPA-ssDNA on ATR activation in the presence of the AAD of TopBP1. The reactions were carried out with 50 fmol of full-length TopBP1 (TopBP1-FL) or 500 fmol of TopBP1-AAD in the presence of RPA (240 fmol) and different amounts of ϕX174 ssDNA (1 or 10 ng). The levels of Chk1 phosphorylation were quantified and averages of three experiments are presented (bottom). (C) Effect of DNA or RPA-ssDNA on ATR activation in the presence of TopBP1 C terminus. The reactions were performed as in (B), except with 200 fmol of the C-terminal 1/3rd of TopBP1 (TopBP1-C) compared with 50 fmol of full-length TopBP1.
We have previously shown that the C terminus of TopBP1, encompassing the AAD and the last two BRCT domains (TopBP1-C), efficiently stimulates the activation of ATR kinase in a manner dependent on the presence of DNA (32). Thus, we examined this fragment of TopBP1 for ATR activation in the presence of RPA-ssDNA (Fig. 5C). Interestingly, the results reveal that even though TopBP1-C can activate ATR at a high DNA concentration (lane 12), no effect of RPA was observed (lane 13). Using different concentrations of TopBP1-C, DNA, and RPA, we confirmed that TopBP1-C is sufficient for DNA-dependent ATR activation, but defective for RPA-ssDNA-dependent activation of ATR (Fig. S5). These results indicate that the N terminus of TopBP1 is required for efficient activation of ATR kinase by RPA-ssDNA.
Recruitment of TopBP1 to ssDNA.
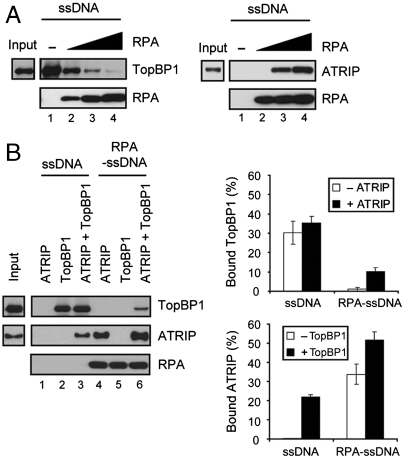
As mentioned above, previous studies have shown that the chromatin-bound 9-1-1 complex recruits TopBP1 proximate to ATR, leading to efficient ATR activation (3). However, it has also been reported that TopBP1 can directly interact with ATR in a manner dependent on ATRIP (12, 34). Therefore, we reasoned that under our experimental reaction conditions, where the 9-1-1 complex is not present, TopBP1 could be recruited to RPA-ssDNA through interactions with ATRIP. To address this hypothesis, we first tested whether TopBP1 could directly interact with RPA, but observed no significant interaction. Moreover, we found that DNA-bound RPA strongly inhibits binding of TopBP1 to DNA (Fig. 6A, left), whereas ATRIP binding to ssDNA is strongly dependent on the presence of RPA under these reaction conditions (right). Next, we examined whether TopBP1 could recruit ATRIP to DNA and whether ATRIP could recruit TopBP1 to RPA-ssDNA (Fig. 6B). Although ATRIP does not bind ssDNA directly (lane 1), we observed that DNA-bound TopBP1 recruits ATRIP (lane 3). In the presence of RPA-ssDNA, TopBP1 no longer binds to DNA (lane 5), but importantly we observed that TopBP1 is recruited to RPA-ssDNA through interaction with ATRIP (lane 6). Interestingly, we consistently observed cooperative binding of ATRIP and TopBP1 to RPA-ssDNA (compare lane 6 with lane 4 for ATRIP binding).
Fig. 6.
TopBP1 is recruited to RPA-ssDNA in a manner dependent on ATRIP. (A) Binding of TopBP1 or ATRIP to ssDNA in the absence or presence of RPA. DNA pull-down assays were performed with biotinylated 80-mer oligonucleotide (1 pmol)-bound streptavidin beads, RPA (0, 1, 2, 4 pmol), and 1 pmol of TopBP1 (left) or ATRIP (right). The input lane contains 0.2 pmol of TopBP1 or ATRIP. (B) Recruitment of ATRIP and TopBP1 to ssDNA and RPA-ssDNA. Biotinylated 80-mer oligonucleotide (1 pmol)-bound streptavidin beads were incubated with no protein (lane 1–3) or 4 pmol of RPA (lane 4–6). The beads were retrieved, washed, and incubated with 1 pmol of ATRIP (lane 1 and 4) or TopBP1 (lane 2 and 5), or both (lane 3 and 6). The bound proteins were separated on SDS-PAGE and analyzed by immunoblotting with the corresponding antibodies. The input lane contains 0.25 pmol of TopBP1 or ATRIP. Levels of the bound proteins from three independent experiments are presented (left).
Given the observation that the C terminus of TopBP1 is defective in RPA-ssDNA activated ATR → Chk1 signaling, we also tested whether TopBP1-C is recruited by ATRIP to RPA-ssDNA. While we consistently observed significant recruitment of full-length TopBP1 by ATRIP to RPA-ssDNA, there is only weak binding of the C terminus of TopBP1 (Fig. S6). Moreover, we confirmed that the N terminus of TopBP1 also interacts weakly with ATRIP and is recruited to RPA-ssDNA, indicating that both the C- and N-terminal domains of TopBP1 are required to form a stable checkpoint complex on ssDNA, leading to efficient ATR → Chk1 signaling (Fig. S6). Collectively, these data indicate that TopBP1 is recruited to RPA-ssDNA through an interaction with ATRIP, and this recruitment of TopBP1 might be potentiated by other checkpoint proteins, such as the Rad17-RFC/9-1-1 complexes (9, 10).
Discussion
Phosphorylation of Chk1 by ATR is possibly the most important reaction in the DNA damage checkpoint response to genotoxic stress by UV and UV-mimetic agents. Despite the extensive in vivo data, it has not been demonstrated that RPA-ssDNA is a particularly strong signal for activating ATR in a defined system. In fact, it was found that both in Xenopus and human cell-free systems the multifunctional replication/repair protein TopBP1 is a potent activator of ATR kinase in the absence of DNA and any other proteins (12).
However, initial attempts to establish an in vitro system in which the ATR kinase activity was dependent on RPA-ssDNA and TopBP1 were not successful. In one study, using the AAD of TopBP1, no effect of RPA-ssDNA on ATR kinase activity was observed (8). However, our data show that the AAD of TopBP1 is unable to confer ssDNA-RPA stimulation. In another study, ATR kinase reactions containing full-length TopBP1 were stimulated to comparable levels by ssDNA and dsDNA, but addition of RPA had only a marginal effect (23). In contrast, unexpectedly, it was found that TopBP1-dependent ATR activation was strongly stimulated by dsDNA damaged by either BPDE or N-Aco-AAF (N-acetoxy-2-acetylaminofluorene) above and beyond the stimulation afforded by dsDNA (23, 32). In yeast, it has been shown that Dpb11 (TopBP1 ortholog) also directly activates the Mec1-Ddc2 kinase (ATR-ATRIP ortholog) to phosphorylate its substrate (25, 35). However, in vitro studies with purified yeast proteins show that DNA or RPA-ssDNA have no or marginal stimulatory effect on Mec1-Ddc2 kinase activity (25). Finally, in the current study we have succeeded in establishing an in vitro system in which phosphorylation of Chk1 by ATR is dependent on both TopBP1 and RPA-ssDNA.
We believe that the system we have developed approximates the in vivo reaction for the following reasons. First, as stated above, it exhibits a strict requirement for TopBP1 and RPA-ssDNA. Second, it depends on ATRIP-RPA interactions: RPA mutants that fail to interact with ATRIP and are defective in the checkpoint response in vivo also fail to support TopBP1-mediated Chk1 phosphorylation by ATR in our system. However, our in vitro system differs from the in vivo signaling pathway in some significant aspects as well. In particular, there appears to be a strict requirement for the 9-1-1 complex in vivo which is not present in our in vitro system. Based on the experience we have gained from establishing an RPA-ssDNA-dependent system, we expect it should be possible to develop an in vitro system dependent on the 9-1-1 complex as well by careful titration of the multiple components of the reaction (ATR-ATRIP, TopBP1, Rad17-RFC/9-1-1 complex, RPA, Chk1, and DNA) and using appropriate reaction conditions to achieve higher Chk1 phosphorylation than our current system in which only about 10% of Chk1 is phosphorylated (24). Indeed, we have found that the reaction of Chk1 phosphorylation by ATR is rather sensitive to the reaction conditions, suggesting that with a better understanding of the conditions that affect the reaction it should eventually be possible to reconstitute the checkpoint system encompassing all of the genetically defined components.
Finally, we wish to comment on the mechanistic significance of ATR activation under different experimental conditions that do not encompass the full component of elements necessary for ATR activation in vivo (Fig. S7). First, the activation by TopBP1 in the absence of DNA or any additional proteins indicates that TopBP1 might be considered an ATR coactivator which at high concentrations and in buffers of low ionic strength can bypass the requirements for DNA or other factors needed for ATR activation in vivo. Second, under conditions of limiting checkpoint factors and in low ionic strength buffers, TopBP1 can interact with DNA and thus form a DNA-TopBP1-(ATR-ATRIP) complex in which ATR is now active. Under more stringent conditions of limiting checkpoint factors and high ionic strength, DNA with bulky adducts, but not undamaged DNA, binds TopBP1 which in turn binds to and activates ATR. Under still higher stringency of limiting concentrations of DNA and high ionic strength, which perhaps most closely approximates the in vivo conditions, the recruitment of ATR-ATRIP to DNA by ATRIP-RPA interactions predominates. As a consequence, under these conditions, ATR, in the presence of TopBP1, is activated by the RPA + ssDNA combination but not by RPA + dsDNA. It should be noted, however, that even though the RPA-ATRIP interaction is sufficient to recruit ATR to DNA, this recruitment is not enough to enable the AAD or C-terminal one-third of TopBP1 (which carries two BRCT repeats as well as the AAD) to activate ATR kinase. For activation under these conditions the N terminus of TopBP1 is also required to form a stable checkpoint complex on RPA-ssDNA.
With respect to the mechanistic aspect of ATR activation, one more point deserves some consideration: There is a DNA length dependence for activation, whether ATR is activated by TopBP1 binding to damaged DNA or whether it is recruited by an ATRIP-RPA interaction to RPA-ssDNA. We suspect that damaged DNAs or ssDNA shorter than 200 bp (nt) that can only accommodate a few TopBP1s or RPAs cannot activate ATR, indicating that a one-dimensional ATR array of certain length is required to activate the kinase. At present, other than stating this striking effect, we cannot offer a mechanistic model. However, we note that a similar observation was made with ATM in vitro (36, 37) and moreover that ATM was activated in vivo when it was artificially recruited to DNA by an array of 256 lac operator-lac repressor interactions (38).
Finally, in this paper we show that aRPA, which is unable to support DNA replication (29, 30), can function similarly to canonical RPA in the activation of ATR. Moreover, since recent work has shown that aRPA can function in nucleotide excision repair and homologous recombination as well (31), it appears that this form of RPA is specialized for DNA damage response pathways.
Materials and Methods
Antibodies and Preparation of Checkpoint Proteins.
See SI Text for details.
Purification of ATR-ATRIP from HeLa Cells.
See SI Text for details.
Preparation of DNA Substrates.
See SI Text for details.
ATR Kinase Assay.
The kinase activity of the ATR-containing fractions was analyzed using Chk1 as a substrate as described previously (23, 32, 39), with some modifications. See SI Text for details.
DNA Pull-Down Assays.
The binding of checkpoint proteins to DNA was analyzed by a DNA pull-down assay using the biotin-streptavidin affinity system as previously described (4), with some modifications. See SI Text for details.
Supplementary Material
Acknowledgments.
We thank S. Bereketoglu and Dr. J. D. Griffith for providing hSSB1 and E. coli SSB, respectively. We also thank Drs. J. Hurwitz, M. Leffak, P. Modrich, Z.-Q. Pan, and D. Wang for helpful comments on the manuscript. This work was supported by National Institutes of Health (NIH) Grant GM32833 (to A.S.) and GM44721 (to M.S.W.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 13561.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007856107/-/DCSupplemental.
References
- 1.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 5.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 6.Kim SM, Kumagai A, Lee J, Dunphy WG. Phosphorylation of Chk1 by ATM- and Rad3-related (ATR) in Xenopus egg extracts requires binding of ATRIP to ATR but not the stable DNA-binding or coiled-coil domains of ATRIP. J Biol Chem. 2005;280:38355–38364. doi: 10.1074/jbc.M508673200. [DOI] [PubMed] [Google Scholar]
- 7.Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball HL, et al. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 11.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto Y, Tsujimura T, Sugino A, Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, et al. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol. 2008;28:7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HS, Brill SJ. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 17.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan S, Lindsay HD, Michael WM. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J Cell Biol. 2006;173:181–186. doi: 10.1083/jcb.200601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke CA, Clarke PR. DNA-dependent phosphorylation of Chk1 and Claspin in a human cell-free system. Biochem J. 2005;388:705–712. doi: 10.1042/BJ20041966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc Natl Acad Sci USA. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsey-Boltz LA, Sercin O, Choi JH, Sancar A. Reconstitution of human claspin-mediated phosphorylation of Chk1 by the ATR (ataxia telangiectasia-mutated and rad3-related) checkpoint kinase. J Biol Chem. 2009;284:33107–33114. doi: 10.1074/jbc.M109.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richard DJ, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 27.Keshav KF, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol Cell Biol. 1995;15:3119–3128. doi: 10.1128/mcb.15.6.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haring SJ, Mason AC, Binz SK, Wold MS. Cellular functions of human RPA1 Multiple roles of domains in replication, repair, and checkpoints. J Biol Chem. 2008;283:19095–19111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haring SJ, Humphreys TD, Wold MS. A naturally occurring human RPA subunit homolog does not support DNA replication or cell-cycle progression. Nucleic Acids Res. 2010;38:846–858. doi: 10.1093/nar/gkp1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason AC, et al. An alternative form of replication protein A prevents viral replication in vitro. J Biol Chem. 2009;284:5324–5331. doi: 10.1074/jbc.M808963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp MG, et al. An alternative form of replication protein A expressed in normal human tissues supports DNA repair. J Biol Chem. 2010;285:4788–4797. doi: 10.1074/jbc.M109.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi JH, Lindsey-Boltz LA, Sancar A. Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res. 2009;37:1501–1509. doi: 10.1093/nar/gkn1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci USA. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 37.You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat Cell Biol. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- 38.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi JH, Sancar A, Lindsey-Boltz LA. The human ATR-mediated DNA damage checkpoint in a reconstituted system. Methods. 2009;48:3–7. doi: 10.1016/j.ymeth.2009.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.