Abstract
Förster resonance energy transfer (FRET) with fluorescent proteins permits high spatial resolution imaging of protein–protein interactions in living cells. However, substantial non-FRET fluorescence background can obscure small FRET signals, making many potential interactions unobservable by conventional FRET techniques. Here we demonstrate time-resolved microscopy of luminescence resonance energy transfer (LRET) for live-cell imaging of protein–protein interactions. A luminescent terbium complex, TMP-Lumi4, was introduced into cultured cells using two methods: (i) osmotic lysis of pinocytic vesicles; and (ii) reversible membrane permeabilization with streptolysin O. Upon intracellular delivery, the complex was observed to bind specifically and stably to transgenically expressed Escherichia coli dihydrofolate reductase (eDHFR) fusion proteins. LRET between the eDHFR-bound terbium complex and green fluorescent protein (GFP) was detected as long-lifetime, sensitized GFP emission. Background signals from cellular autofluorescence and directly excited GFP fluorescence were effectively eliminated by imposing a time delay (10 μs) between excitation and detection. Background elimination made it possible to detect interactions between the first PDZ domain of ZO-1 (fused to eDHFR) and the C-terminal YV motif of claudin-1 (fused to GFP) in single microscope images at subsecond time scales. We observed a highly significant (P < 10-6), six-fold difference between the mean, donor-normalized LRET signal from cells expressing interacting fusion proteins and from control cells expressing noninteracting mutants. The results show that time-resolved LRET microscopy with a selectively targeted, luminescent terbium protein label affords improved speed and sensitivity over conventional FRET methods for a variety of live-cell imaging and screening applications.
Keywords: cellular imaging, dihydrofolate reductase, Forster resonance energy transfer, lanthanide luminescence, protein labeling
Protein–protein interactions, often mediated by modular interaction domains, play a fundamental role in the dynamic organization of cells (1). Various experimental techniques such as immunoprecipitation, affinity chromatography, and yeast two-hybrid analysis have been used to identify putatively interacting proteins and deduce the biomolecular mechanisms of cell function (2, 3). However, cell-free studies and screening assays do not provide information about the spatio-temporal organization of protein networks in the natural environment of the living cell or organism. A variety of optical methods are available for monitoring protein interactions in cells, including fluorescence cross correlation spectroscopy (FCCS) (4, 5), bimolecular fluorescence complementation (6), translocation-based assays (7–9), and methods that detect intermolecular Förster resonance energy transfer (FRET). Among these methods, only FRET allows dynamic and reversible imaging of protein–protein interactions while simultaneously preserving information about their subcellular distribution (10–12).
FRET occurs when a fluorescent donor molecule is brought within close proximity (< 10 nm) to an acceptor whose absorption spectrum overlaps with the donor emission spectrum. FRET can be detected in appropriately configured microscopes as a decrease in the fluorescence intensity, lifetime, or anisotropy of the donor or as an increase in the fluorescence intensity of the acceptor upon donor excitation (if the acceptor is fluorescent). Fluorescent proteins (FPs), especially cyan (CFP) and yellow (YFP) variants, are the most common donors and acceptors used in live-cell FRET imaging (13). Intermolecular FRET between two FP-labeled proteins has been used to visualize receptor oligomerization and interactions between transcription factors and cell signaling components (11, 13–15).
Steady-state monitoring of sensitized acceptor emission upon donor excitation is the most popular FRET microscopy method. This technique involves acquiring three sets of images using different filter combinations: donor excitation/donor emission, acceptor excitation/acceptor emission, and donor excitation/acceptor emission (11, 12, 16, 17). The images are then processed postacquisition to correct for cross-detection of donor fluorescence in the acceptor emission channel (and vice versa), and to normalize for the dependence of FRET on the relative concentrations of donor and acceptor. However, image processing propagates noise and uncertainty from each individual image, lowering the sensitivity and precision of steady-state FRET imaging. Changes in donor emission intensity upon acceptor photobleaching enable direct determination of FRET efficiency (the fraction of the photon energy absorbed by the donor that is transferred to the acceptor), but this method destroys the sample and precludes time-lapse imaging. Alternatively, fluorescence lifetime imaging microscopy (FLIM) allows direct detection of FRET effects on donor excited-state lifetime and, therefore, overcomes artifacts associated with donor/acceptor spectral overlap. However, FLIM image acquisition can take several minutes and requires complicated, expensive instrumentation to accurately resolve changes in nanosecond-scale lifetimes of FPs or organic fluorophores (11). Finally, the maximum FRET dynamic range for nondimerizing fluorescent proteins is < 5-fold (11, 18), and the signal observed by microscopy in living cells may only change ∼10% (14, 15). The small signal variation and considerable noise factors make many interactions difficult or impossible to measure by existing FRET approaches. Therefore, despite numerous reported successes, FRET imaging has significant limitations.
Lanthanide coordination complexes are well established probes for detecting biomolecular interactions in cell-free systems at high signal-to-background ratio (19–22). As lanthanide emission is not technically fluorescence (singlet-to-singlet transition), the term luminescence (or lanthanide-based) resonance energy transfer (LRET) is commonly used to describe transfer from a lanthanide donor to an organic fluorophore or fluorescent protein acceptor. Lanthanide complexes have two distinct properties that enable sensitive LRET measurements. Firstly, large Stokes shifts (separation between excitation and emission wavelengths) and narrow emission lines facilitate efficient spectral separation of donor and acceptor emission signals. Secondly, long-lived (μs-ms) luminescence enables time-resolved detection, where pulsed light excites the sample, and the detector is switched on after decay of short-lived (< 100 ns) autofluorescence and directly excited acceptor fluorescence background signals (21). Direct conjugation of terbium or europium complexes to oligonucleotides, antibodies and proteins has permitted the development of sensitive time-resolved LRET assays of biomolecular interactions in purified biochemical preparations, cellular extracts, and on cell surfaces (22–24). Recently, lanthanide complexes have been developed as probes for live cell imaging applications using time-resolved microscopy with pulsed, near-UV single photon or two photon excitation (25–28). Time-resolved microscopy in the microsecond domain has also been demonstrated using platinum or palladium complexes (29–31). However, to date, lanthanide complexes have not been used as LRET donors for intracellular experiments because: (i) suitably bright and photostable complexes were not available; (ii) methods to deliver membrane-impermeant lanthanide complexes from culture medium to the cytoplasm were lacking; and (iii) novel approaches were needed to specifically attach probes to proteins or subcellular structures.
Herein we report LRET imaging of protein–protein interactions in living cells. We show that a luminescent terbium complex, TMP-Lumi4, can be delivered to the cytoplasm and specifically targeted to Escherichia coli dihydrofolate reductase (eDHFR) fusion proteins. Time-resolved microscopy was used to image LRET between the terbium complex and green fluorescent protein (GFP) by detecting long-lifetime (> 10 μs) terbium-sensitized GFP emission (Fig. 1). Interactions between the first PDZ domain of ZO-1 and the C-terminal YV motif of claudin-1 were detected unambiguously in a single microscope image at time scales (seconds) comparable to steady-state FRET imaging, but without the need for multiple control measurements and associated postimage processing. Whereas steady-state microscopy of intermolecular FRET may yield signal changes of only 10% (14, 15), we observed > 6-fold change in mean, donor-normalized LRET signal between cells expressing interacting proteins and control cells expressing noninteracting mutants. This article shows that time-resolved LRET microscopy enables faster, more sensitive and simpler analyses of molecular interactions in living cells than currently available FRET imaging methods.
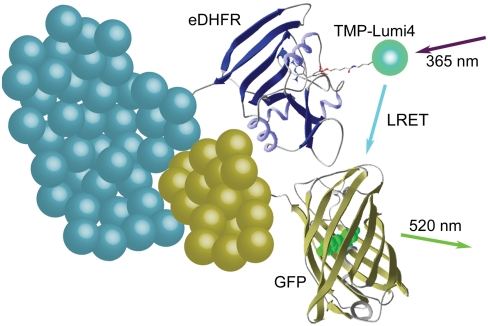
Fig. 1.
Schematic representation of intermolecular terbium-to-GFP LRET. TMP-Lumi4 binds specifically and tightly to eDHFR (KD = ∼ 2 nM). Interaction between eDHFR and GFP fusion proteins and excitation of TMP-Lumi4 (the donor) results in LRET-sensitized emission of GFP (the acceptor). Time-resolved microscopy detects long-lifetime, sensitized GFP emission while eliminating cellular autofluorescence and directly excited GFP fluorescence background signals.
Results and Discussion
Properties of the Selective Terbium Protein Label.
We recently reported a method to impart terbium luminescence to recombinant fusion proteins (32). Heterodimeric conjugates of the common antibiotic trimethoprim (TMP) covalently linked to various terbium chelates were developed that bind noncovalently to eDHFR. TMP and eDHFR interact exclusively with one another in mammalian systems, and this interaction has been previously exploited to develop TMP-fluorophore conjugates that specifically label eDHFR fusion proteins in a variety of wild-type mammalian cell lines (LigandLink™ Universal Labeling Technology, Active Motif, Inc.) (33–35). In our previous study, we found that one particular conjugate, TMP-Lumi4 (Fig. 2A), bound tightly (KD = ∼ 2 nM) to a GFP-eDHFR fusion protein and that intramolecular terbium-to-GFP LRET could be detected with > 100-fold dynamic range as long-lifetime, sensitized GFP emission in a cell-free assay (32).
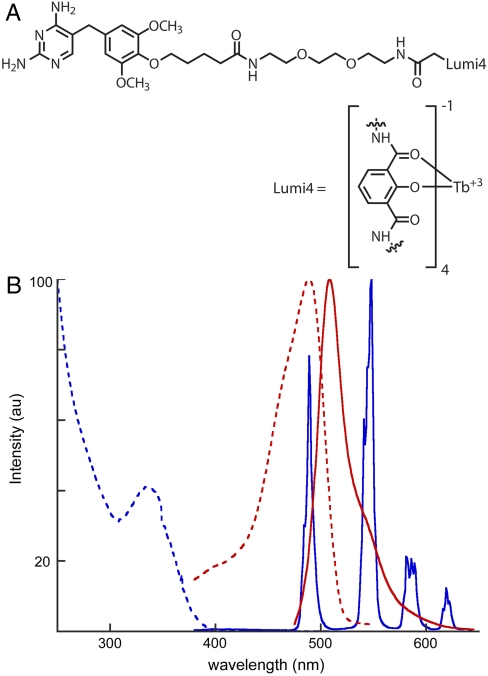
Fig. 2.
(A) Structure of TMP-Lumi4, a conjugate of trimethoprim covalently linked to a proprietary analog of a multidentate 2-hydroxyisophthalamide terbium chelate (Lumi4®-Tb) (36). The molecule has a molecular weight of 1879 and a net charge of -1 when complexed with terbium. (B) Normalized absorption (dotted) and emission (solid) spectra of TMP-Lumi4 (blue) and EGFP (red). The characteristically narrow terbium emission bands enable efficient spectral separation of signals.
The effectiveness of TMP-Lumi4 as a LRET donor to GFP results from its overall brightness, long lifetime, and the ability to spectrally discriminate GFP emission from terbium emission. Lumi4®-Tb is an exceptionally bright (ε ≈ 26,000 M-1 cm-1 at 354 nm, quantum yield = 0.59) and long-lived (τ ≈ 2.7 ms) proprietary analog of a multidentate 2-hydroxyisophthalamide terbium chelate previously reported by Raymond and coworkers (36). TMP-Lumi4 exhibits characteristic terbium luminescence upon excitation with near-UV light and absorbs strongly at 365 nm, enabling efficient excitation with commercially available light-emitting diode (LED) sources (Fig. 2B). Both TMP-Lumi4 and unconjugated Lumi4®-Tb were found to exhibit single-exponential decay kinetics with the same apparent lifetime (∼2.3 ms) under identical experimental conditions (Fig. S1). Terbium-sensitized GFP emission from a complex of TMP-Lumi4 bound to GFP-eDHFR occurred with single-exponential kinetics and an apparent lifetime of ∼0.8 ms (Fig. S1). From lifetime data, we calculated an efficiency of ∼67% for energy transfer from terbium to GFP in the TMP-Lumi4/GFP-eDHFR complex (SI Materials and Methods).
Intracellular Probe Delivery and Specific Protein Labeling.
TMP-Lumi4 will not diffuse passively into cells from culture medium (32). Therefore, to perform intracellular LRET imaging, it was first necessary to establish methods of cytoplasmic probe delivery. Microinjection is one possible approach for loading adherent cells, and it has been successfully used for lanthanide complex delivery (25). However, microinjection requires specialized apparatus and can only be used to load relatively few cells at a time. Two techniques were therefore adapted to simultaneously deliver TMP-Lumi4 to the cytoplasm of many cells: (i) reversible plasma membrane permeabilization with streptolysin O (SLO) (37); and (ii) osmotic lysis of pinocytic vesicles (38, 39). Both methods consistently yielded > 50% loading efficiency while maintaining ∼95% cell viability 2 h posttreatment (Table S1).
Whereas both loading techniques have negligible effects on cell viability, the pinocytosis method should be less perturbative to cellular physiology, as the cell membrane is not compromised at any point during the process. With the pinocytic loading method, cells are allowed to undergo pinocytosis in a hypertonic medium containing sucrose, polyethylene glycol, and TMP-Lumi4. Upon transfer to a hypotonic medium, the pinocytic vesicles (containing TMP-Lumi4) burst due to the lowering of osmotic pressure, releasing their contents into the cytoplasm. The overall amount of TMP-Lumi4 delivered into cells can be controlled by varying the concentration in the hypertonic loading medium, or alternatively, by repeated applications of the pinocytosis/lysis process. Given a natural rate of pinocytosis equaling ∼10-16 L/ min for most cell types (39), and assuming typical cell volumes of ∼10-12 L, the estimated cellular concentration of TMP-Lumi4 was < 10-7 M for the experimental conditions used in this study (single loading application, 50 μM TMP-Lumi4, 10 min. pinocytosis) (SI Materials and Methods).
Intracellular delivery of TMP-Lumi4 and specific labeling of eDHFR fusion proteins was visualized using time-resolved microscopy. A conventional epifluorescence microscope was adapted for time-resolved imaging by incorporating a fast-modulated, UV LED (λem = 365 nm) as the excitation source and an intensified CCD camera for image acquisition. The LED circuitry allowed for either continuous wave emission or external on/off modulation with ∼1 μs time resolution. The excitation intensity could be varied; however, we held the intensity constant at a measured value of 1.6 mW at the objective back aperture, yielding an estimated illumination intensity of ∼0.6 W/cm2 at the image plane (see SI Materials and Methods). The image intensifier component of the camera served as both a fast shutter and emission signal amplifier. By synchronizing the LED and intensifier with a digital delay generator, the excitation pulse width, the gate delay (time between pulse and detection), the gate width (intensifier on-time), and the pulse/detection repetition rate could be varied independently. The output from multiple excitation/detection cycles could be integrated on the CCD during a single camera frame, and the camera control software allowed summation of multiple frames (see SI Materials and Methods and Table S2 for microscope operation parameters used in this study).
Madin Darby Canine kidney (MDCKII) epithelial cells were transiently cotransfected with two plasmid DNA vectors; one that expressed nucleus-localized eDHFR and another that expressed nucleus-localized CFP as a positive control for transfection. After SLO-mediated delivery of TMP-Lumi4, a time-resolved image (gate delay = 10 μs) of broadband emission (> 400 nm) revealed specific localization of terbium luminescence in the nucleus of a transfected cell loaded with probe (Fig. 3A). When unconjugated TMP (final conc. ∼ 10 μM) was added to the imaging medium, it diffused into cells, competed with TMP-Lumi4 for eDHFR binding, and markedly diminished nuclear luminescence (Fig. 3A). Specific labeling of nucleus-localized eDHFR was also observed in NIH3T3 fibroblast cells that were loaded with TMP-Lumi4 by SLO-mediated membrane permeabilization (Fig. S2), and results seen with pinocytic delivery were similar.
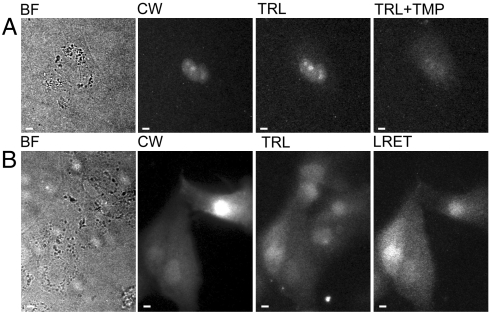
Fig. 3.
Both streptolysin O (SLO)-mediated membrane permeabilization and osmotic lysis of pinocytic vesicles deliver TMP-Lumi4 to the cytoplasm of MDCKII cells, and specific labeling of eDHFR fusion proteins can be visualized by time-resolved luminescence microscopy. (A), (B) Micrographs: BF, bright field; CW, continuous wave fluorescence (λex = 480 ± 20 nm, λem = 535 ± 25 nm); TRL, time-resolved luminescence (λex = 365 nm, λem > 400 nm, gate delay = 10 μs); TRL + TMP, 20 min after addition of TMP (final conc. = 10 μM) to culture medium; LRET, time-resolved luminescence (λex = 365 nm, λem = 520 ± 10 nm, gate delay = 10 μs). TRL and TRL + TMP images were adjusted to identical contrast levels. Complete details of time-resolved microscopy parameters are provided in SI Materials and Methods and Table S2. Scale bars, 10 μm. (A) MDCKII cells transiently cotransfected with DNA encoding nucleus-localized CFP and nucleus-localized eDHFR were incubated with TMP-Lumi4 (15 μM) and streptolysin O (SLO, 50 ng/mL) for 10 min. Time-resolved detection of broadband (> 400 nm) terbium emission reveals localization of TMP-Lumi4 in nucleus of transfected cell (corresponding to continuous wave fluorescence image of CFP emission). A time-resolved image taken 20 min. after addition of TMP (final conc. = 10 μM) to the culture medium shows diminished nuclear luminescence because TMP out-competes TMP-Lumi4 for binding to eDHFR. (B) MDCKII cells transiently transfected with DNA encoding GFP-eDHFR. Cells were incubated in hypertonic medium containing TMP-Lumi4 (50 μM) for 10 min to allow pinocytosis and subsequently exposed to hypotonic medium to effect lysis of pinocytic vesicles and release of probe into the cytoplasm. Time-resolved detection of broadband emission (> 400 nm) reveals terbium luminescence in TMP-Lumi4-loaded cells. Luminescence resonance energy transfer (LRET) is seen only in transfected cells (indicated in continuous wave fluorescence image) coincidentally loaded with TMP-Lumi4 when visualized in time-resolved mode through a narrow-pass filter (520 ± 10 nm), indicating specific labeling of the GFP-eDHFR fusion protein.
Cytoplasmic delivery of TMP-Lumi4 and specific labeling of eDHFR was further assessed by imaging terbium-to-GFP LRET in time-resolved mode. MDCKII cells were transfected with DNA encoding a GFP-eDHFR fusion protein. The cells were loaded with TMP-Lumi4 by pinocytic delivery, and intramolecular, terbium-sensitized GFP emission was seen only in GFP-eDHFR-expressing cells when visualized in time-resolved mode through a narrow-pass filter (λem = 520 ± 10 nm) that blocked terbium emission (Fig. 3B). Intramolecular, terbium-to-GFP LRET was also observed in NIH3T3 fibroblasts that were pinocytically loaded with TMP-Lumi4 (Fig. S2). These results and those above show that both SLO-mediated membrane permeabilization and osmotic lysis of pinocytic vesicles can effectively deliver TMP-Lumi4 into the cytoplasm of at least two cell types (MDCKII and NIH3T3). Upon cellular delivery, TMP-Lumi4 diffuses freely throughout the cytoplasm and nucleus without detectable nonspecific binding and binds selectively to eDHFR, suggesting that other methods of cytoplasmic delivery should prove equally useful. Moreover, time-resolved microscopy can visualize intramolecular terbium-to-GFP LRET as long-lifetime (> 10 μs), sensitized GFP emission.
Quantification of LRET signals.
The concentration of GFP-eDHFR or other fusion proteins expressed from a strong promoter (1–10 μM) (40) likely exceeds the estimated concentration of TMP-Lumi4 (< 100 nM) in pinocytically loaded cells. Given its high affinity for eDHFR (KD = ∼ 2 nM), TMP-Lumi4 was assumed to be mostly bound to fusion protein, and therefore the LRET signal should vary with the amount of TMP-Lumi4 in the cell. To quantify the LRET signal, two time-resolved images were obtained using identical acquisition settings but for different emission wavelengths: (i) the pure sensitized GFP emission signal (λem = 520 ± 10 nm); and (ii) a signal comprising terbium emission plus sensitized GFP emission (λem = 540 ± 10 nm). For each cell in an image, the background-subtracted mean gray value was used as an indicator of emission intensity (SI Materials and Methods and Fig. S3). From these pairs of images, a donor-normalized LRET signal (LRETN) could then be calculated as the 520/540 emission ratio for each measured cell.
TMP-Lumi4 Stability and Photoresistance in cells.
To be useful as cellular imaging probes, lanthanide complexes must remain stably luminescent in cells over time and must also be resistant to photobleaching. Long-term cellular stability may be compromised by endogenous chelators that could remove metal from the complex, rendering it nonluminescent. The stability of TMP-Lumi4 in MDCKII cells expressing GFP-eDHFR was determined by imaging terbium-to-GFP LRET. LRETN was observed to be constant over ∼23 h (Fig. S4). However, the unnormalized LRET signal decreased approximately 50% over the same time period, from an initial value of 230 ± 100 to a value of 110 ± 65 at 23 h (mean ± s.d., > 10-cell samples). The results suggest that TMP-Lumi4 remains stably luminescent within cells but may slowly leak out of the cells over time, and therefore the LRET signal remains quantitatively useful over the time course of the experiment.
The photoresistance of TMP-Lumi4 in MDCKII cells expressing GFP-eDHFR was determined by monitoring the LRET signal as a function of accumulated irradiation time. A field of view was exposed to continuous wave LED excitation at the standard illumination intensity (∼0.6 W/cm2). The LRET signal (λem = 520 ± 10 nm) was measured on a cell-by-cell basis at successive timepoints, normalized to each cell’s initial intensity, and fit to a single exponential decay, yielding an estimated photobleaching lifetime of ∼2.0 min (Fig. S4). The measured photobleaching lifetime is considerably longer than typical image exposure times (seconds), and therefore time-lapse LRET imaging should be possible without concern for loss of signal due to photobleaching of the donor.
LRET Imaging of Protein–Protein Interactions.
The ability of this unique LRET system to image protein–protein interactions was next used to measure the association of a PDZ domain with a carboxyl-terminal binding motif in living cells (41, 42). Such PDZ-mediated interactions are fundamental to biological function in species from Drosophila to mammals. In epithelia, a direct, high-affinity (KD = ∼ 10–20 nM) interaction between the C-terminal cytoplasmic tail of claudin-1 and the most N-terminal PDZ domains (PDZ1) of ZO-1 and the related proteins ZO-2 and ZO-3 has been demonstrated using isolated recombinant proteins (43). Moreover, PDZ1 domains of ZO-1, ZO-2, and ZO-3 are recruited to sites of claudin-1 polymerization in transfected fibroblasts (43) but not to sites enriched in mutant claudin-1 lacking the C-terminal YV motif (43). The relevance of this interaction to disease is emphasized by the observations that mutations within the ZO-2 PDZ1 domain are linked to familial hypercholanemia (44), a disease of hepatic bile transport, whereas loss of claudin-1 expression is associated with neonatal sclerosing cholangitis (45), another hereditary cholestatic disease. However, a direct interaction between the C-terminal claudin-1 YV motif and the first PDZ domain of ZO-1 has not been previously demonstrated in live cells.
To examine the interaction between claudin-1 and ZO-1 in living cells, MDCKII cells were transfected with expression vectors encoding a C-terminal fusion of eDHFR to the PDZ1 domain (residues 19–113) of ZO-1 (ZO-1/PDZ1-eDHFR) and an N-terminal fusion of EGFP to the C-terminal cytoplasmic domain (residues 187–211) of claudin-1 (GFP-cldn1/tail). Transfected cells were loaded with TMP-Lumi4 using both SLO-mediated and pinocytic delivery methods. In both cases, LRET imaging revealed terbium-sensitized GFP emission only in transfected cells loaded with TMP-Lumi4, providing an unambiguous image of protein–protein interaction (Fig. 4A and Fig. S2). The LRETN for pinocytically loaded cells equaled 0.38 ± 0.10 (mean ± s.d., Table S3). Addition of excess TMP to growth medium eliminated the LRET signal (Fig. 4A), further confirming that long-lifetime, sensitized GFP emission results from specific binding of TMP-Lumi4 to ZO-1/PDZ1-eDHFR. The predominantly nuclear localization of interactions between ZO-1/PDZ1-eDHFR and the GFP-cldn1/tail is consistent with previous reports showing that GFP-tagged ZO-1 PDZ1 domain accumulates within the nucleus (43) and that ZO-1 directs subcellular claudin trafficking (46).
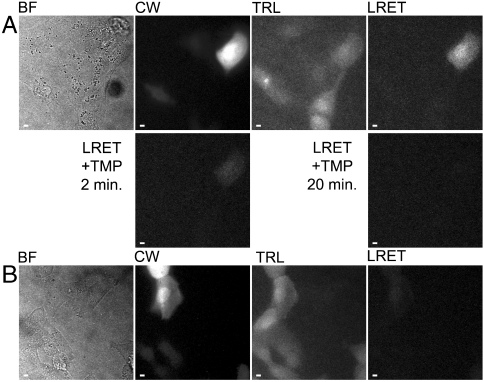
Fig. 4.
Time-resolved LRET imaging reveals interaction between ZO-1/PDZ1-eDHFR and GFP-cldn1/tail in MDCKII cells. (A), (B) Micrographs: BF, bright field; CW, continuous wave fluorescence (λex = 480 ± 20 nm, λem = 535 ± 25 nm); TRL, time-resolved luminescence (λex = 365 nm, λem > 400 nm, gate delay = 10 μs); LRET, time-resolved luminescence (λex = 365 nm, λem = 520 ± 10 nm, gate delay = 10 μs); LRET + TMP, after addition of TMP (final conc. = 10 μM) to growth medium at indicated time-points. LRET and LRET + TMP images were adjusted to identical contrast levels. Complete details of time-resolved microscopy parameters are provided in SI Materials and Methods and Table S2. Scale bars, 10 μm. (A) MDCKII cells coexpressing ZO-1/PDZ1-eDHFR and GFP-cldn1/tail loaded with TMP-Lumi4 by osmotic lysis of pinosomes. Terbium-to-GFP LRET is seen only in transfected cells loaded with TMP-Lumi4. When TMP is added to culture medium, it displaces TMP-Lumi4 from the ZO-1/PDZ1-eDHFR/GFP-cldn1/tail complex, eliminating the LRET signal. (B) MDCKII cells coexpressing ZO-1/PDZ1-eDHFR and GFP-cldn1/tailΔYV, loaded with TMP-Lumi4 as in (A). No LRET signal is seen in expressing cells loaded with TMP-Lumi4.
To assess specificity of the observed interaction, a GFP-cldn1/tail construct lacking the C-terminal YV motif (GFP-cldn1/tailΔYV) was developed and expressed in MDCKII cells with ZO-1/PDZ1-eDHFR. Whereas continuous widefield GFP fluorescence and broadband, time-resolved terbium luminescence signals were easily detected in cells loaded with TMP-Lumi4, only extremely faint LRET was seen (Fig. 4B), verifying that the LRET signal observed between ZO-1/PDZ1-eDHFR and GFP-cldn1/tail reflects a specific PDZ-mediated interaction. There was a highly significant (P < 10-6), > 6-fold difference between the mean LRETN signal from cells expressing ZO-1/PDZ1-eDHFR and GFP-cldn1/tail (0.38 ± 0.10) and that seen from cells expressing ZO-1/PDZ1-eDHFR and GFP-cldn1/tailΔYV (0.06 ± 0.04, mean ± s.d., Table S3).
The data above show that LRET imaging is faster, more sensitive, and in many respects, less complex than widefield, steady-state FRET microscopy. With steady-state FRET, the sensitized emission signal is contaminated by directly excited donor and acceptor fluorescence. Therefore, multiple control images must be collected, requiring acquisition times of at least ∼3 s on optimized systems (12, 16, 17). Even with careful image acquisition and proper analysis, signal changes of only ∼10% are typically observed between FRET-positive samples and negative controls (14, 15). By contrast, LRET imaging only detects signals emanating from interacting molecules. We observed a ∼500% increase in mean LRETN between cells expressing interacting and noninteracting ZO-1/PDZ1-eDHFR and GFP-cldn1/tail pairs (Table S3), and this result represents a ∼50-fold increase in signal-to-background ratio over that typically observed with steady-state FRET imaging of protein–protein interactions.
The dramatic difference in measured signal between LRET-positive cells and negative controls is partly due to the high affinity between ZO-1/PDZ1 and cldn1/tail (in vitro KD = 13 nm) (45). The high affinity makes it likely that most of the TMP-Lumi4 delivered into cells was bound to ZO-1/PDZ1-eDHFR and donated energy to proximal GFP-cldn1/tail. Measurements of lower-affinity interactions should be possible using the LRET method, however, longer image acquisition times may be necessary to generate sufficient signal. Alternatively, increased loading of TMP-Lumi4 (up to saturation of eDHFR binding) would yield a higher measured signal level. For LRETN calculations, we used images that were acquired at relatively long acquisition times (8 s, Table S2), although intermolecular LRET could be detected with acquisition times as short as 0.67 s (Fig. S5). Thus, in addition to enhanced sensitivity, LRET imaging displays increased time resolution that may allow analysis of interaction dynamics that cannot be resolved by traditional steady-state FRET or FRET-FLIM. Whereas other optical methodologies such as induced translocation of fluorescent protein fusions (7–9) and FCCS (4, 5) can monitor interactions in cells with much better time resolution (0.1–1 s for FCCS), these methods do not provide a spatial map of interactions.
Conclusion
Our results show that eDHFR fusion proteins can be specifically and stably labeled with a luminescent terbium complex, TMP-Lumi4, in living, wild-type mammalian cells. The ability to selectively impart terbium luminescence enables dynamic, nondestructive LRET imaging of intracellular interactions between eDHFR and GFP fusion proteins without additional control measurements and mathematical processing. By detecting terbium-sensitized GFP emission at long lifetimes, we effectively eliminate detection of cellular autofluorescence, unbound terbium probe luminescence, and directly excited GFP fluorescence, thereby imaging LRET in living cells at time scales (< 1 s) that are substantially faster than steady-state FRET imaging (∼3 s) and orders of magnitude faster than FLIM imaging of FPs.
Whereas the techniques described here should improve live-cell studies of protein function, there is broad scope for further enhancements and modifications. For instance, intramolecular LRET could be exploited to develop biosensors that incorporate eDHFR and GFP in a single fusion protein, analogous to CFP/YFP sensors designed to detect cellular analytes or enzyme activity (13). Time-resolved microscopy can also be adapted to measure donor or sensitized acceptor lifetimes by capturing and analyzing a series of images at different detector delay intervals (47, 48). The multiple terbium emission peaks of TMP-Lumi4 can serve as a LRET donor to both GFP and red fluorescent proteins, potentially enabling the simultaneous detection of more than one molecular interaction in a single living cell (Fig. 2B). Finally, both SLO and pinocytosis probe delivery methods can be easily adapted for use in multiwell plate format, enabling high-throughput screening assays of intracellular protein–protein interactions using commercially available time-resolved fluorescence plate readers.
Materials and Methods
Chemical Synthesis.
Details describing the synthesis and characterization of TMP-Lumi4 can be found in SI Materials and Methods and in Rajapakse et al. (32).
Cell Culture, Transfection, and Probe Delivery Methods.
NIH3T3 and MDCKII cells were cultured and and transfected with plasmid DNA according to standard protocols. Complete details of these as well as methods for pinocytic and SLO-mediated probe delivery and cell viability assay protocols are provided in SI Materials and Methods.
Live Cell Imaging and Data Analysis.
Comprehensive descriptions of the time-resolved microscope system, image acquisition parameters and methods of image processing and data analysis are offered in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank K.N. Raymond (University of California) and N.G. Butlin (Lumiphore, Inc.) for material and intellectual support. Lumi4®-Tb is a registered trademark of Lumiphore, Inc. We thank V.W. Cornish (Columbia University) for providing GFP-eDHFR plasmid. This study was supported by the National Institutes of Health (National Institute of General Medical Sciences Grant R01GM081030 and National Institute of Diabetes and Digestive and Kidney Diseases Grant R01DK061931) and by the Chicago Biomedical Consortium (C-008, with support from The Searle Funds at The Chicago Community Trust).
Footnotes
Conflict of interest statement: L.W.M. is a coinventor on a patent relevant to this work (US Patent 7575866) and inventor on a relevant provisional patent application filed by the University of Illinois at Chicago.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002025107/-/DCSupplemental.
References
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300(5618):445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 3.Phizicky EM, Fields S. Protein–protein interactions: Methods for detection and analysis. Microbiol Rev. 1995;59(1):94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudendistel N, Muller G, Waldeck W, Angel P, Langowski J. Two-hybrid fluorescence cross-correlation spectroscopy detects protein–protein interactions in vivo. Chemphyschem. 2005;6(5):984–990. doi: 10.1002/cphc.200400639. [DOI] [PubMed] [Google Scholar]
- 5.Kim SA, Heinze KG, Waxham MN, Schwille P. Intracellular calmodulin availability accessed with two-photon cross-correlation. Proc Natl Acad Sci USA. 2004;101(1):105–110. doi: 10.1073/pnas.2436461100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7(6):449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heydorn A, Lundholt BK, Praestegaard M, Pagliaro L. Protein translocation assays: Key tools for accessing new biological information with high-throughput microscopy. Methods Enzymol. 2006;414:513–530. doi: 10.1016/S0076-6879(06)14027-6. [DOI] [PubMed] [Google Scholar]
- 8.Knauer SK, Stauber RH. Development of an autofluorescent translocation biosensor system to investigate protein–protein interactions in living cells. Anal Chem. 2005;77(15):4815–4820. doi: 10.1021/ac050413o. [DOI] [PubMed] [Google Scholar]
- 9.Piljic A, Schultz C. Analysis of protein complex hierarchy in living cells. ACS Chem Biol. 2008;3(12):749–755. doi: 10.1021/cb8002539. [DOI] [PubMed] [Google Scholar]
- 10.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21(11):1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 11.Piston DW, Kremers GJ. Fluorescent protein FRET: The good, the bad, and the ugly. Trends Biochem Sci. 2007;32(9):407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Vogel SS, Thaler C, Koushik SV. Fanciful FRET. Sci STKE. 2006;(331):re2. doi: 10.1126/stke.3312006re2. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3(12):906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel MT, Garcia EL, Kajimoto T, Hall RA, Newton AC. The protein scaffold NHERF-1 controls the amplitude and duration of localized protein kinase D activity. J Biol Chem. 2009;284(36):24653–24661. doi: 10.1074/jbc.M109.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llopis J, et al. Ligand-dependent interactions of coactivators steroid receptor coactivator-1 and peroxisome proliferator-activated receptor binding protein with nuclear hormone receptors can be imaged in live cells and are required for transcription. Proc Natl Acad Sci USA. 2000;97(8):4363–4368. doi: 10.1073/pnas.97.8.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berney C, Danuser G. FRET or no FRET: A quantitative comparison. Biophys J. 2003;84(6):3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74(5):2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23(3):355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 19.Mathis G. Rare earth cryptates and homogeneous fluoroimmunoassays with human sera. Clin Chem. 1993;39(9):1953–1959. [PubMed] [Google Scholar]
- 20.Prat O, Lopez E, Mathis G. Europium(III) cryptate: A fluorescent label for the detection of DNA hybrids on solid support. Anal Biochem. 1991;195(2):283–289. doi: 10.1016/0003-2697(91)90331-m. [DOI] [PubMed] [Google Scholar]
- 21.Selvin PR. Principles and biophysical applications of lanthanide-based probes. Annu Rev Biophys Biomol Struct. 2002;31:275–302. doi: 10.1146/annurev.biophys.31.101101.140927. [DOI] [PubMed] [Google Scholar]
- 22.Selvin PR, Hearst JE. Luminescence energy transfer using a terbium chelate: Improvements on fluorescence energy transfer. Proc Natl Acad Sci USA. 1994;91(21):10024–10028. doi: 10.1073/pnas.91.21.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leblanc V, et al. Homogeneous time-resolved fluorescence assay for identifying p53 interactions with its protein partners, directly in a cellular extract. Anal Biochem. 2002;308(2):247–254. doi: 10.1016/s0003-2697(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 24.Maurel D, et al. Cell-surface protein–protein interaction analysis with time-resolved FRET and snap-tag technologies: Application to GPCR oligomerization. Nat Methods. 2008;5(6):561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanaoka K, Kikuchi K, Kobayashi S, Nagano T. Time-resolved long-lived luminescence imaging method employing luminescent lanthanide probes with a new microscopy system. J Am Chem Soc. 2007;129(44):13502–13509. doi: 10.1021/ja073392j. [DOI] [PubMed] [Google Scholar]
- 26.Law GL, et al. Emissive terbium probe for multiphoton in vitro cell imaging. J Am Chem Soc. 2008;130(12):3714–3715. doi: 10.1021/ja710418d. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery CP, Murray BS, New EJ, Pal R, Parker D. Cell-penetrating metal complex optical probes: Targeted and responsive systems based on lanthanide luminescence. Acc Chem Res. 2009;42(7):925–937. doi: 10.1021/ar800174z. [DOI] [PubMed] [Google Scholar]
- 28.Song B, Vandevyver CD, Chauvin AS, Bunzli JC. Time-resolved luminescence microscopy of bimetallic lanthanide helicates in living cells. Org Biomol Chem. 2008;6(22):4125–4133. doi: 10.1039/b811427g. [DOI] [PubMed] [Google Scholar]
- 29.Botchway SW, et al. Time-resolved and two-photon emission imaging microscopy of live cells with inert platinum complexes. Proc Natl Acad Sci USA. 2008;105(42):16071–16076. doi: 10.1073/pnas.0804071105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Haas RR, et al. Phosphorescent platinum/palladium coproporphyrins for time-resolved luminescence microscopy. J Histochem Cytochem. 1999;47(2):183–196. doi: 10.1177/002215549904700207. [DOI] [PubMed] [Google Scholar]
- 31.de Haas RR, et al. Platinum porphyrins as phosphorescent label for time-resolved microscopy. J Histochem Cytochem. 1997;45(9):1279–1292. doi: 10.1177/002215549704500911. [DOI] [PubMed] [Google Scholar]
- 32.Rajapakse HE, Reddy DR, Mohandessi S, Butlin NG, Miller LW. Luminescent terbium protein labels for time-resolved microscopy and screening. Angew Chem Int Ed Engl. 2009;48(27):4990–4992. doi: 10.1002/anie.200900858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calloway NT, et al. Optimized fluorescent trimethoprim derivatives for in vivo protein labeling. Chembiochem. 2007;8(7):767–774. doi: 10.1002/cbic.200600414. [DOI] [PubMed] [Google Scholar]
- 34.Gallagher SS, Sable JE, Sheetz MP, Cornish VW. An in vivo covalent TMP-tag based on proximity-induced reactivity. ACS Chem Biol. 2009;4(7):547–556. doi: 10.1021/cb900062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller LW, Cai Y, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: A flexible chemical tag. Nat Methods. 2005;2(4):255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 36.Petoud S, Cohen SM, Bunzli JC, Raymond KN. Stable lanthanide luminescence agents highly emissive in aqueous solution: multidentate 2-hydroxyisophthalamide complexes of Sm(3 + ), Eu(3 + ), Tb(3 + ), Dy(3 + ) J Am Chem Soc. 2003;125(44):13324–13325. doi: 10.1021/ja0379363. [DOI] [PubMed] [Google Scholar]
- 37.Ahnert-Hilger G, Bader MF, Bhakdi S, Gratzl M. Introduction of macromolecules into bovine adrenal medullary chromaffin cells and rat pheochromocytoma cells (PC12) by permeabilization with streptolysin O: Inhibitory effect of tetanus toxin on catecholamine secretion. J Neurochem. 1989;52(6):1751–1758. doi: 10.1111/j.1471-4159.1989.tb07253.x. [DOI] [PubMed] [Google Scholar]
- 38.Okada CY, Rechsteiner M. Introduction of macromolecules into cultured mammalian cells by osmotic lysis of pinocytic vesicles. Cell. 1982;29(1):33–41. doi: 10.1016/0092-8674(82)90087-3. [DOI] [PubMed] [Google Scholar]
- 39.Rechsteiner M. Osmotic lysis of pinosomes. Methods Enzymol. 1987;149:42–48. doi: 10.1016/0076-6879(87)49042-3. [DOI] [PubMed] [Google Scholar]
- 40.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 41.Songyang Z, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275(5296):73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 42.Doyle DA, et al. Crystal structures of a complexed and peptide-free membrane protein-binding domain: Molecular basis of peptide recognition by PDZ. Cell. 1996;85(7):1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 43.Itoh M, et al. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147(6):1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlton VE, et al. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34(1):91–96. doi: 10.1038/ng1147. [DOI] [PubMed] [Google Scholar]
- 45.Hadj-Rabia S, et al. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: A tight junction disease. Gastroenterology. 2004;127(5):1386–1390. doi: 10.1053/j.gastro.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 46.Umeda K, et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126(4):741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 47.Elangovan M, Day RN, Periasamy A. Nanosecond fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy to localize the protein interactions in a single living cell. J Microsc. 2002;205(1):3–14. doi: 10.1046/j.0022-2720.2001.00984.x. [DOI] [PubMed] [Google Scholar]
- 48.Heyduk T, Heyduk E. Luminescence energy transfer with lanthanide chelates: interpretation of sensitized acceptor decay amplitudes. Anal Biochem. 2001;289(1):60–67. doi: 10.1006/abio.2000.4925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.