Abstract
A common strategy of metabolic engineering is to increase the endogenous supply of precursor metabolites to improve pathway productivity. The ability to further enhance heterologous production of a desired compound may be limited by the inherent capacity of the imported pathway to accommodate high precursor supply. Here, we present engineered diterpenoid biosynthesis as a case where insufficient downstream pathway capacity limits high-level levopimaradiene production in Escherichia coli. To increase levopimaradiene synthesis, we amplified the flux toward isopentenyl diphosphate and dimethylallyl diphosphate precursors and reprogrammed the rate-limiting downstream pathway by generating combinatorial mutations in geranylgeranyl diphosphate synthase and levopimaradiene synthase. The mutant library contained pathway variants that not only increased diterpenoid production but also tuned the selectivity toward levopimaradiene. The most productive pathway, combining precursor flux amplification and mutant synthases, conferred approximately 2,600-fold increase in levopimaradiene levels. A maximum titer of approximately 700 mg/L was subsequently obtained by cultivation in a bench-scale bioreactor. The present study highlights the importance of engineering proteins along with pathways as a key strategy in achieving microbial biosynthesis and overproduction of pharmaceutical and chemical products.
Keywords: GGPP synthase, levopimaradiene synthase, metabolic engineering, Escherichia coli, molecular reprogramming
Metabolic engineering is the enabling technology for the manipulation of organisms to synthesize high-value compounds of both natural and heterologous origin (1–4). In the case of heterologous production, well-characterized microorganisms are used as production hosts because targeted optimization can be performed using widely available genetic tools and synthetic biology frameworks (5, 6). One important application of engineered microbial systems is geared toward the synthesis of terpenoid natural products (7–9). Terpenoids represent one of the largest classes of secondary metabolites that includes pharmaceuticals, cosmetics, and potential biofuels candidates (8, 10–12). Metabolic engineering approaches to produce terpenoids in microbial systems such as Escherichia coli and yeast have commonly focused on increasing the precursor flux into the heterologous terpenoid pathway by rerouting endogenous isoprenoid metabolism (13–15). These engineering strategies have relied heavily on changing the enzyme concentrations in the product pathway.
Many properties of a metabolic pathway, however, are not limited solely by the enzyme concentration, as is particularly true for the terpenoid pathway. In nature, terpenoid biosynthesis is regulated at multiple metabolic branch points to create large structural and functional diversity (16–18). In the major metabolic branch point in terpenoid biosynthesis, the prenyl transferases and terpenoid synthases catalyze the formation of a wide range of structurally diverse acyclic and cyclic terpenoid molecules (17). Sequence analysis of these enzymes showed that they are paralogous proteins evolved through gene duplications that subsequently diverged in functional roles to catalyze the formation of different terpenoid structures (16, 17, 19). Particularly, terpenoid synthases generate enzyme-bound carbocation intermediates that undergo a cascade of rearrangements and quenchings of carbocations to create structural diversity (20). These enzymes are highly promiscuous (21), and the functional promiscuity is often associated with unwanted product formation and poor catalytic properties (22). Thus in an engineered terpenoid pathway, these enzymes lead to low metabolic fluxes and large byproduct losses, limiting yield improvement of the desired product molecules. In some cases, the buildup of intermediate metabolites elicits stress responses detrimental to cell growth (23, 24). Thus, the ability to tune a heterologous terpenoid pathway at regulatory nodes would be a valuable approach both to confer an overproduction phenotype and to minimize toxicity in microorganisms.
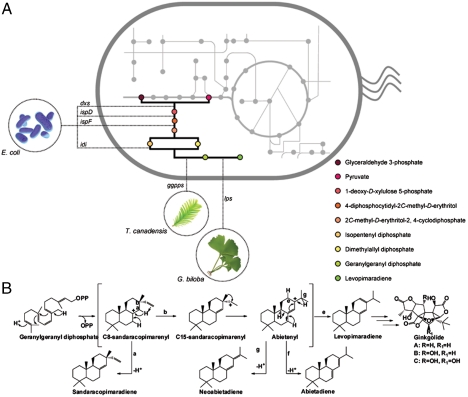
In the present work, we engineered Escherichia coli to produce levopimaradiene, the diterpenoid gateway precursor of the pharmaceutically important plant-derived ginkgolides (25–28). The biosynthesis of levopimaradiene from simple carbon sources (glucose or glycerol) starts from the formation of the precursors isopentenyl diphosphate (IPP), and dimethylallyl diphosphate (DMAPP) derived from the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway in E. coli. Geranylgeranyl diphosphate synthase (GGPPS) then catalyzes the condensation of IPP and DMAPP to the linear diphosphate intermediate geranylgeranyl diphosphate (GGPP). In the final step, levopimaradiene synthase (LPS) catalyzes the conversion of GGPP to levopimaradiene via a complex reaction cascade of cyclization, rearrangement, and proton transfers (Fig. 1A). The promiscuous function of LPS also results in the formation of isomeric side products such as abietadiene, sandaracopimaradiene, and neoabietadiene (29, 30) (Fig. 1B). The functional expression of codon-optimized genes encoding for GGPPS and LPS in E. coli only generated minute quantities of levopimaradiene. Levopimaradiene synthesis was increased when GGPPS–LPS expression was coupled with the systematic amplification of genes in the upstream MEP pathway to elevate flux toward IPP and DMAPP; however, titers remained low. We postulated that the threshold of levopimaradiene production was limited by inherent GGPPS–LPS capacity. To overcome this constraint, we adopted the principle of molecular reprogramming through engineering combinatorial mutations of the GGPPS–LPS pathway. This approach was inspired by natural systems, in which biosynthetic pathways undergo molecular reprogramming processes (e.g. via mutations of transcription regulators and enzymes) to accommodate important changes in metabolite concentrations (31–34). By combining protein and metabolic engineering, we achieved approximately 2,600-fold improvements in levopimaradiene productivity and demonstrated a strategy to harness the potential of engineered biosynthetic pathway for large scale microbial production of valuable molecules.
Fig. 1.
(A) Engineering levopimaradiene synthesis in E. coli. A plant-derived pathway was constructed by introducing T. chinensis ggpps and G. biloba lps codon-optimized genes. To amplify the endogenous precursor pools of GGPPS substrates (IPP and DMAPP), copy numbers of rate-limiting steps (dxs, idi, ispD, ispF) in the MEP pathway were amplified by additional episomal expression. (B) General reaction mechanism of “LPS-type” enzymes. Levopimaradiene, the major product of G. biloba LPS is the gateway precursor of ginkgolides. Coproducts of LPS include abietadiene, neoabiatadiene, and sandaracopimaradiene that stem from the different deprotonation patterns throughout intermediates in the reaction cascade.
Results and Discussion
Levopimaradiene Production Improvement via Precursor Pathway Amplification.
The initial attempt to synthesize levopimaradiene from E. coli by coexpressing codon-optimized genes encoding for GGPPS and LPS resulted in only a small amount of levopimaradiene, 0.15 mg/L (Table S1), with no detectable amount of the related isomers produced by LPS. The first step taken to improve productivity was a metabolic engineering approach via incremental overexpression of the bottleneck enzymatic steps in the MEP pathway, namely dxs, idi, ispD, and ispF (Fig. 1A). Indeed, this approach improved levopimaradiene production (Table S1). The highest production of approximately 92 mg/L was achieved by inclusion of approximately 5 additional copies of the MEP pathway genes. At this level, the levopimaradiene isomers abietadiene, sandaracopimaradiene, and neoabietadiene (Fig. 1B) were also identified in the culture, at product fractions of 11%, 2%, and trace amounts, respectively (Fig. S1). However, further increasing the MEP pathway from approximately 5 to approximately 10 copies was not accompanied by additional increases in levopimaradiene production. In fact, the titer was lower (approximately 23 mg/L) than when MEP pathway was amplified by approximately 5-fold (Table S1). These results demonstrated that stepwise improvement of precursor flux, a strategy commonly employed in metabolic engineering, did not yield an incremental amplification of terpenoid molecules. Hence, we postulated that the efficiency of the regulatory node in terpenoid biosynthesis, the GGPPS–LPS (prenyltrasnferase–terpenoid synthase) portion of the pathway, is rate-limiting under high IPP and DMAPP precursor flux (16–18, 20–22). Quantitative RT-PCR confirmed that higher MEP pathway transcript levels were achieved with 10-fold amplification of the MEP pathway, likely resulting in a buildup of toxic intermediates that negatively affected overall pathway flux (23). Thus, we set out to reprogram GGPPS and LPS to develop mutant pathways that confer high-level levopimaradiene production in the 10-copy MEP pathway background.
Probing the Putative Binding Pocket in LPS to Design Improved Variants.
Fully combinatorial strategies to search for important mutations in LPS are impractical due to the lack of a suitable high-throughput screen. To circumvent this limitation, we first constructed a homology structure for the second active site of LPS that was used as a guide to probe residues important for catalytic function. In the second active site of an “LPS-type” enzyme, the bicylic (+)-copalyl diphosphate (CPP) intermediate (derived from the deprotonation of GGPP in the first active site) undergoes a diphosphate-ionization cyclization. The resulting C8-sandaracopimarenyl cation intermediate is further deprotonated at two alternative sites to release isopimaradiene or sandaracopimaradiene end products. However, this intermediate can also undergo intramolecular proton transfer and 1,2-methyl migration to yield abietenyl cation. Subsequent deprotonation of abietenyl cation at four possible sites then produces abietadiene, levopimaradiene, neoabietadiene, and palustradiene (29, 30).
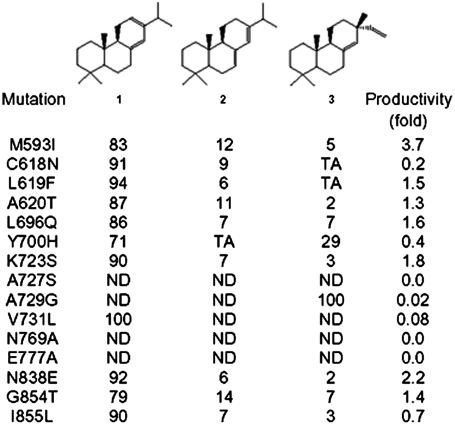
Following the construction of the LPS model, fifteen residues constituting the binding pocket were selected (Fig. S2A). We perturbed these residues using phylogeny-based mutation (35, 36) to determine if they conferred a different production phenotype. Specifically, residue replacements were determined from paralogous LPS-type enzymes that are functionally different from LPS (Fig. S2B), namely Abies grandis abietadiene synthase (AS) (37), Picea abies AS, and P. abies isopimardiene synthase (ISO) (38). Using this information, we created single mutations of M593I, C618N, L619F, A620T, L696Q, K723S, A729G, N838E, G854T, and I855L based on residues in A. grandis and P. abies AS; whereas Y700H, A727S and V731L were created based on P. abies ISO. Alanine was used to replace Asn769 and Glu777 because these amino acids are conserved throughout LPS, AS, and ISO (Fig. 2).
Fig. 2.
Summary of LPS mutations and the impact with respect to product distribution and productivity of the engineered pathway. Trace amount (TA), not detected (ND). Numbers indicate percentage of each isomer (1-levopimaradiene; 2-abietadiene; 3-sandaracopimaradiene).
The preengineered E. coli (with the addition of approximately 10 copies of the MEP pathway genes) expressing the wild-type GGPPS provided an in vivo screening system for titer and product distribution changes by the LPS mutations. We observed that the profiles of diterpenoid product distribution resulting from expressing LPS mutants M593I, C618N, L619F, A620T, L696Q, K723S, V731L, N838E, G854T, and I855L were similar to expression of wild-type LPS (Fig. 2). Hence, with respect to LPS product selectivity, these mutations were neutral or close to neutral. Diterpenoid productivities resulting from expressing LPS mutants L619F, A620T, and G854T were also not significantly changed compared to wild-type LPS (within 50%). However, diterpenoid production levels were notably altered by expressing mutants M593I, C618N, L696Q, K723S, V731L, N838E, and I855L (Fig. 2 and Table S2). The highest total diterpenoid production increase (approximately 3.7-fold) was mediated by expressing LPS M593I. In all cases, expression of these mutants did not significantly affect product distribution. Expression of LPS mutant Y700H, however, resulted in significant alteration of diterpenoid product distribution by abolishing abietadiene synthesis and increasing sandaracopimaradiene proportion (Fig. 2). Although a single mutation in P. abies AS, Y686H (corresponding to Tyr700 in LPS) did not result in product selectivity changes in vitro, it promoted isopimaradiene synthesis when combined with another mutation (Y686H/A713S) (38); hence Tyr700 may play an important role in mediating the evolvability of LPS-type enzymes. Additionally, the expression of the A729G mutant resulted in the exclusive production of sandaracopimaradiene; however, it was concomitant with a reduction in productivity of approximately 98% (Fig. 2). Finally, diterpenoid production was not observed in systems expressing LPS A727S, N769A, and E777A variants. These residues fell within approximately 4.7 Å from substrate in the homology model; therefore, it was not surprising that the mutations were deleterious to LPS activity given the close proximity to the substrate.
Mutational Enrichment of Tunable LPS Residues to Identify Variants Conferring Increased Productivity.
The previous results pointed to mutations in LPS that significantly affected production phenotype, namely M593I and Y700H. Although the preliminary mutation of Ala729 imparted product selectivity changes, it was excluded from further analysis because even a conservative replacement such as glycine was deleterious. From analyzing the structural model, we observed that Met593 is located at the posterior of the binding pocket, whereas Tyr700 is positioned at the entrance (in close vicinity of the DDXXD magnesium binding motif). To obtain the complete LPS evolvability profile by these residues, we sampled all amino acids through saturation mutagenesis. Additionally, we also explored the effects of expressing the saturation mutagenesis library of Ala620 because a mutation at this position in A. grandis AS changed its product selectivity in vitro (37).
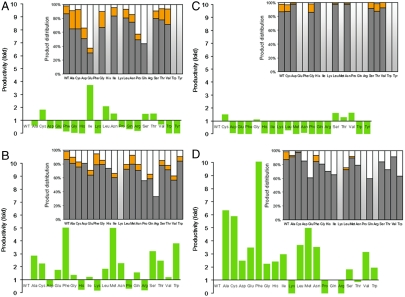
From the saturation mutagenesis library of Met593, we found two substitutions that conferred significant productivity improvement (Fig. 3A and Table S3). In addition to isoleucine, which was discovered in the phylogenetic-based mutation, the replacement with leucine, another hydrophobic residue similar in size as methionine, increased diterpenoid productivity by approximately 2-fold without significantly changing product distribution (Fig. 3A). Based on the structural significance of this position, this productivity improvement appeared to be caused by the disruption of H-bonding at the end of the binding pocket, thus increasing the flexibility of the cavity to better fit the CPP substrate. Therefore, the M593I mutation likely resulted in the highest production increase (approximately 3.7-fold) because isoleucine is the most hydrophobic amino acid. Substitutions with smaller residues than methionine only yielded moderate production improvement (< 2-fold in the case of cysteine, serine, and threonine), and were disruptive in the case of alanine, glycine, and valine. Furthermore, substitutions with amino acids longer than five heavy atoms and those with bulky rings such as phenylalanine, tyrosine, and tryptophan also consistently decreased or abolished activity. These trends are consistent with the requirement for the substrate to have an unobstructed cyclization pocket, as the center of the bend is proximal to this residue. Moreover, replacements with hydrophilic amino acids such as aspartic acid, glutamic acid, lysine, and arginine also generally reduced productivity possibly because their ability to form their own H-bonding may reduce the capacity of the binding pocket.
Fig. 3.
Characteristics (productivity and product distribution) of the preengineered strains expressing wild-type ggpps and lps saturation mutagenesis library of: (A) Met593, (B) Tyr700, (C) Ala620, and (D) Tyr700 using lps M593I. In the product distribution charts, gray bars, orange bars, white bars represent proportion of levopimaradiene, abietadiene, and sandaracopimaradiene in the product mixture, respectively. Two toned (white and light gray) bars represent nil production. WT represents the preengineered strain expressing wild-type ggpps and lps.
We also found that the replacement of Tyr700 with phenylalanine, methionine, and tryptophan improved productivity up to approximately 5-fold (Fig. 3B and Table S4). The reaction cascade toward the formation of abietenyl cation requires an energetically unfavorable transition from a tertiary to a secondary carbocation (30). Therefore it was postulated that the latter species is stabilized by the ionic interaction with the paired diphosphate anion that is chelated by the magnesium ion (37). Tyr700 is located within close proximity to the magnesium binding site, thus the absence of the hydroxyl group in amino acids that are similar to tyrosine may allow the repositioning of the magnesium closer to the aspartate-rich region, hence increasing reaction efficiency by improving the chelation of the diphosphate group. A few mutations, i.e. replacements with aspartic acid, histidine, proline, arginine, and lysine, abolished abietadiene synthesis in the product mixture, and conferred a decrease in productivity (Fig. 3B). The replacement with positively charged residues or a helix breaker (proline) might cause a misalignment of the diphosphate anion that impaired catalysis or prevented the deprotonation of abietenyl cation at carbon position f (Fig. 1B) to create abietadiene.
Finally, the sampling of all amino acid substitutions of Ala620 revealed that only replacement with residues similar to alanine (small or hydrophilic) (cysteine, glycine, serine, and threonine) as well as valine retained LPS activity; whereas other substitutions were destructive or deleterious (Fig. 3C and Table S5). A few destructive mutations (replacements with aspartic acid, leucine, asparagine) also destabilized abietenyl deprotonation to yield abietadiene. Therefore, Ala620 in LPS did not appear to control product selectivity and productivity in LPS, yet it was important for catalysis.
Combining Beneficial Mutations in LPS to Further Improve Productivity.
In laboratory experiments, the beneficial effects of single mutations are often additive (39, 40). Therefore, the production improvement resulting from expressing the LPS M593I variant encouraged us to explore the effect of this beneficial mutation in combination with saturation mutagenesis of Tyr700. As shown in Fig. 3D (Table S6), this combination successfully integrated the advantageous properties of the individual mutations, resulting in one variant (M593I/Y700F) that increased diterpenoid titer by approximately 10-fold over the expression of the wild-type enzyme without significant changes in product distribution. Interestingly, the expression of most LPS variants carrying the double mutations resulted in the reduction of abietadiene proportion (Fig. 3D). In two variants, M593I/Y700A and M593I/Y700C, the reduction of abietadiene was complemented by an increase in levopimaradiene, resulting in mixtures that contained up to 97% levopimaradiene. The combinations of M593I with Y700A and Y700C also conferred productivity improvements of approximately 6.3- and approximately 5.8-fold, respectively. Thus, it appeared that these mutations facilitated the stabilization of the intermediates that led to favoring abietenyl intermediate formation yet prevented proton extraction from route f (Fig. 1B). It is noteworthy that using the M593I mutant LPS as a parent for the subsequent saturation mutagenesis only produced three variants that decreased or abolished diterpenoid productivity (Fig. 3D). In contrast, previous saturation mutagenesis of Tyr700 resulted in five pathway variants that decreased/abolished diterpenoid production (Fig. 3B). Although a single M593I mutation was rendered neutral with respect to product selectivity, this mutation restored LPS activity upon subsequent defective mutations by aspartic acid and histidine replacement at residue 700. Thus M593I appeared to facilitate subsequent mutational robustness, a paradigm of neutral genetic drift (39).
Engineering Functional Mutations in GGPPS to Further Increase Diterpenoid Production.
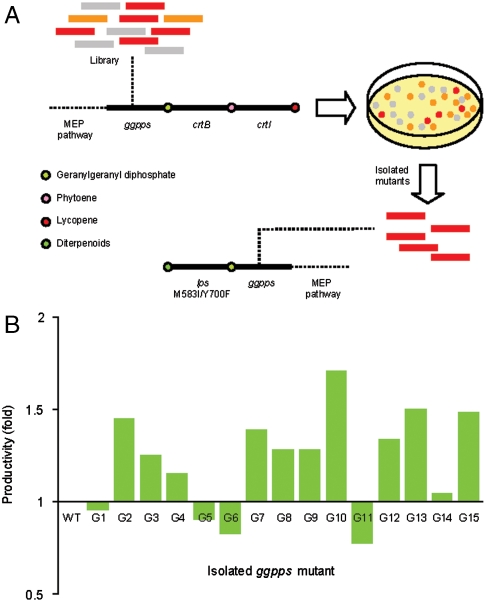
We next opted to mutate GGPPS, with the goal of further increasing pathway productivity. Although the structure of a plant GGPPS of angiosperm origin (from Sinapis alba) is available (41), the crystal structure for a gymnosperm GGPPS has not been solved. Furthermore, the folding similarity of gymnosperm GGPPS enzymes and their angiosperm analogs is not known. Despite catalyzing essentially the same enzymatic reaction, GGPPS enzymes are known to exhibit wide structural diversity among organisms (41). Therefore, based on secondary structure analysis (42), the notable division of gymnosperm from angiosperm GGPPS enzymes may imply significant tertiary fold differences. In the case of T. canadensis GGPPS, its amino acid sequence only exhibited approximately 56% homology with that of S. alba GGPPS, with frequent gaps throughout the entire sequence (Fig. S3). As a result, the lack of a suitable structural guide prompted us to devise a random approach to mutate T. canadensis GGPPS. To enable a facile high-throughput screening method for isolating improved GGPPS variants, we utilized a lycopene biosynthetic pathway consisting of crtB and crtI as a colorimetric reporter (Fig. 4A). In this system, the expression of wild-type GGPPS resulted in colonies with light red coloration. Improved GGPPS variants from the mutagenesis were identified by the improvement of lycopene production in the cell, as determined by red coloration.
Fig. 4.
Generation of GGPPS library based on stochastic mutation. (A) Creation of a facile high-throughput screening assay by fusing a lycopene pathway (crtB and crtI) with ggpps libraries. Mutant ggpps genes that conferred improved lycopene production (red colonies) were isolated. These variants were then coexpressed with lps carrying M593I/Y700F mutations for diterpenoid production assay. (B) Production phenotype of the preengineered E. coli strains coexpressing selected ggpps variants and lps M593I/Y700F. WT represents the strain expressing the wild-type ggpps and lps M593I/Y700F.
We isolated fifteen ggpps variants from colonies exhibiting red coloration. To assess the potential for improving levopimaradiene production in vivo, the fifteen mutant GGPPS isolates were coexpressed with the high-producing LPS M593I/Y700F mutant in the preengineered E. coli strain. Five GGPPS variants did not confer a levopimaradiene increase, indicating false positives obtained from the colorimetric screening. However, the coexpression of ten GGPPS mutants resulted in diterpenoid production improvement (Fig. 4B). The expression of mutant G10 resulted in the highest diterpenoid production increase of approximately 1.7-fold over the pathway harboring the wild-type GGPPS and the LPS M593I/Y700F, an equivalent of an approximately 17.7-fold increase over the pathway harboring wild-type GGPPS and LPS (Fig. 4B and Table S7). Sequence analysis of G10 revealed that two positions were mutated, namely S239C and G295D (Fig. S4). Amino acid alignment with GGPPS sequences from other plants (43) showed that most beneficial mutations are located in the region in between the two highly conserved aspartate-rich DDXXXXD and DDXXD domains (Fig. S4). A structural analysis of a prenyltransferase suggested that the two aspartate-rich regions bound three Mg2+ ions to facilitate the anchoring of the diphosphate groups of the IPP and DMAPP substrates (44). Therefore due to the close proximity to the aspartate motifs and Gly295 replacement with aspartate, the mutations in G10 may affect GGPPS catalysis by improving the binding efficiency of the magnesium ions needed for substrate anchoring. Overall, approximately 10-fold overexpression of the MEP genes and the use of mutant downstream enzymes consisting of GGPPS S239C/G295D and LPS M593I/Y700F increased levopimaradiene production approximately 2,600-fold over the expression of wild-type GGPPS and LPS alone (Table S7).
Levopimaradiene Overproduction in Controlled Culture Conditions.
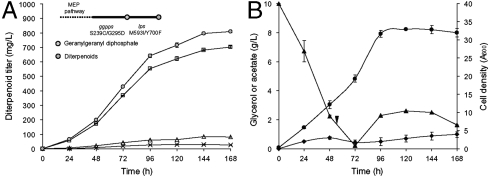
The performance of the preengineered E. coli strain expressing the highest-producing levopimaradiene pathway consisting of GGPPS S239C/G295D and LPS M593I/Y700F was assessed in bioreactors under controlled conditions. The total diterpenoid titer reached a maximum of approximately 800 mg/L in 168 h, and levopimaradiene constituted approximately 700 mg/L (Fig. 5A). Using this engineered strain, 10 g/L glycerol was almost depleted after 56 h. Therefore 3 g/L glycerol was introduced into the culture every 8 h after this time point (Fig. 5B). Despite the relatively rapid consumption of glycerol, acetate only accumulated below 1 g/L throughout the cultivation. Overall, this experiment demonstrated that the production improvement obtained from the new pathway translated well toward larger cultivation.
Fig. 5.
Cultivation of the E. coli strain overexpressing the MEP pathway and the “reprogrammed” plant-derived pathway constituting GGPPS S239C/G295D and LPS M593I/Y700F mutants. (A) Diterpenoid production curves. Total diterpenoid, levopimaradiene, abietadiene, and sandaracopimaradiene are in circles, squares, triangles, and crosses, respectively. (B) Feed, fermentative by-product, and biomass curves. Glycerol, acetate, and cell density are in triangles, diamonds, and circles, respectively. Inverse triangle denotes the time point where 3 g of glycerol was added every 8 h.
Conclusions
Efforts to increase terpenoid production in E. coli previously focused on (i) overexpression of pathway enzymes, and (ii) optimizing the expression of enzymes by codon bias (7, 23, 45, 46). However, these approaches, which both aim to increase enzyme concentration to increase pathway flux, are still limited by the inherent low enzyme activity and specificity of the terpenoid pathways. Thus, in addition to metabolic engineering, the molecular reprogramming of key metabolic nodes such as prenyl transferase (GGPPS) and terpenoid synthase (LPS) though protein engineering is required to achieve substantial overproduction of a desired terpenoid product. Our approach has wide applicability not only in engineering of terpenoid pathways but also in many other secondary metabolic pathways, especially those with promiscuous enzymes that act as regulatory nodes. Our results also dispute the current notion that the MEP pathway is not effective for high-level production of terpenoid molecules (23, 47). In fact, the stoichiometry of the bacterial MEP pathway is 12–14% more efficient in consuming glucose or glycerol without redox imbalance compared to the eukaryotic mevalonate pathway for IPP/DMAPP synthesis. In a broader sense, because terpenoid pathways lead to compounds used in flavors, cosmetics, and potentially biofuels, our engineering approach is directly applicable for the high-level production of many commercially important compounds using microbial biotechnology.
Materials and Methods
Cloning and pathway construction.
All cloning procedures were carried out in E. coli strain DH5alpha (Invitrogen), and pathway engineering was performed in E. coli MG1655 Δ (endA, recA) strains (SI Appendix). Genes were custom-synthesized (DNA 2.0) to incorporate E. coli codon bias, remove restriction sites for cloning purposes, and establish an approximately 50% GC-content. All strains carried the native chromosomal copy of the MEP pathway. Additional copies of pathway genes were inserted as operon dxs-idi-ispD-ispF under a Trc promoter. The single copy MEP strain was constructed by chromosomal localization of the MEP pathway and higher copy numbers were achieved through plasmid-based expression (SI Appendix and Table S8). Relative expression levels were verified through quantitative RT-PCR.
Culture Growth and Library Analysis.
Single transformants of preengineered E. coli strains expressing pTrcGGPPS-LPS, or their mutant variants were cultivated for 18 h at 30 °C in LB. For library characterization, these preinnocula were used to seed fresh 2 mL cultures at a starting A600 of 0.1 in rich medium (SI Appendix). Scale-up experiments (1-L cultures) were done in 3-L bioreactors (SI Appendix). To minimize the loss of diterpenoids due to air-stripping, 20% dodecane was added into the culture. The diterpenoid production was characterized by GC-MS analysis (SI Appendix).
Molecular Modeling.
The homology model of LPS was built based on the structure of 5-epi-aristolochene synthase (48) [EAS (Protein Data Bank ID code 5EAT)]. Sequence alignment (Fig. S5) was performed with the ClustalW (49) method with standard gap penalties. Whereas LPS contains 323 residues in excess of EAS, they aligned almost exclusively at the proximity of the second active side (toward the C-terminus), with a virtually gapless alignment. The CHARMM molecular modeling software (50, 51) with the CHARMM27 parameter set was used to mutate residues. Partial atomic charges needed for the substrate were obtained quantum mechanically with the Gaussian program using the 6-31G* basis set. Fifteen residues within a 10 Å distance from the substrate that contours the binding pocket were determined by using visual molecular dynamics (VMD) (52) (http://www.ks.uiuc.edu/Research/vmd).
Mutant Library Generation and Screening.
The introduction of point mutations and saturation mutagenesis in lps were performed using QuikChange II XL (Stratagene). Nucleotide changes were set by custom designed oligonucleotides (Table S9). Subsequent to sequencing to verify nucleotide changes, the lps variants were used to replace the wild-type lps in pTrcGGPPS-LPS and subjected to expression in the preengineered E. coli for production analysis. The random mutagenesis library of ggpps was created by error-prone (EP) PCR at low mutation rate using GeneMorph II (Stratagene). A pool of plasmid pTrcGGPPS*-CRT was isolated from more than approximately 106 transformants of E. coli DH10B. The plasmid library was then used to transform the E. coli strain overexpressing the MEP pathway for colorimetric screening. Colonies that displayed bright red coloration were isolated after incubation at 25 °C for 3 d (as visualized on LB containing 75 μg/mL ampicillin and 25 μg/mL chloramphenicol). Following plasmid extraction and sequencing, the mutant ggpps genes were used as a pool in the next round of EP PCR. As a control, the integration of wild-type ggpps into the lycopene pathway gave rise to orange colored transformants. The iteration of mutation and screening was stopped after the second round of mutant collection, as no colony that displayed higher red coloration was identified in the third round of EP PCR.
Supplementary Material
Acknowledgments.
We thank S.-H. Yoon (Prather lab) for the construction of E. coli MG1655 Δ (endA, recA). We also thank W. Runguphan, S. O’Connor [Department of Chemistry, Massachusetts Institute of Technology (MIT)], C. Martin, T. L. To (Department of Chemical Engineering, MIT), and D. Groshoff (Harvard University) for constructive discussions, and critical reading of this manuscript. Research in the Prather laboratory is supported by the Camille and Henry Dreyfus Foundation New Faculty Award, and the MIT Energy Initiative (Grant 6917278). Research in the Stephanopoulos laboratory is supported by the Singapore–MIT alliance and the US National Institutes of Health (Grant 1-R01-GM085323-01A1). Research in the Tidor laboratory is supported by the US National Institutes of Health (Grant GM065418).
Footnotes
A provisional patent application describing elements of this work has been filed.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006138107/-/DCSupplemental.
References
- 1.Chemler JA, Koffas MA. Metabolic engineering for plant natural product biosynthesis in microbes. Curr Opin Biotechnol. 2008;19(6):597–605. doi: 10.1016/j.copbio.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Keasling JD, Chou H. Metabolic engineering delivers next-generation biofuels. Nat Biotechnol. 2008;26(3):298–299. doi: 10.1038/nbt0308-298. [DOI] [PubMed] [Google Scholar]
- 3.Leonard E, Runguphan W, O’Connor S, Prather KJ. Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat Chem Biol. 2009;5(5):292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- 4.Zhou H, Xie X, Tang Y. Engineering natural products using combinatorial biosynthesis and biocatalysis. Curr Opin Biotechnol. 2008;19(6):590–596. doi: 10.1016/j.copbio.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Leonard E, Nielsen D, Solomon K, Prather KJ. Engineering microbes with synthetic biology frameworks. Trends Biotechnol. 2008;26(12):674–681. doi: 10.1016/j.tibtech.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Tyo KE, Alper HS, Stephanopoulos GN. Expanding the metabolic engineering toolbox: more options to engineer cells. Trends Biotechnol. 2007;25(3):132–137. doi: 10.1016/j.tibtech.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Ajikumar PK, et al. Terpenoids: Opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharm. 2008;5(2):167–190. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- 8.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3(7):408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 9.Kirby J, Keasling JD. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 10.Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng. 2009;12(1):70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Atsumi S, Liao JC. Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotech. 2008;19(5):414–419. doi: 10.1016/j.copbio.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohlmann J, Keeling CI. Terpenoid biomaterials. Plant J. 2008;54(4):656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 13.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005;23(5):612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 14.Asadollahi MA, et al. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng. 2009;11(6):328–334. doi: 10.1016/j.ymben.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 16.Tholl D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol. 2006;9(3):297–304. doi: 10.1016/j.pbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Keeling CI, Bohlmann J. Genes, enzymes, and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 2006;170(4):657–675. doi: 10.1111/j.1469-8137.2006.01716.x. [DOI] [PubMed] [Google Scholar]
- 18.Christianson DW. Unearthing the roots of the terpenome. Curr Opin Chem Biol. 2008;12(2):141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallabhaneni R, Wurtzel ET. Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol. 2009;150(2):562–572. doi: 10.1104/pp.109.137042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin DM, Faldt J, Bohlmann J. Functional characterization ofnine Norway Spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 2004;135(4):1908–1927. doi: 10.1104/pp.104.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber DPW, Ralph S, Bohlmann J. Genomic hardwiring and phenotypic plasticity of terpenoid-based defenses in conifers. J Chem Ecol. 2004;30(12):2399–2418. doi: 10.1007/s10886-004-7942-2. [DOI] [PubMed] [Google Scholar]
- 22.Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27(2):157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 23.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 24.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9(2):193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Aponte M, et al. Activation of platelet-activating factor receptor and pleiotropic effects on tyrosine phospho-EGFR/Src/FAK/paxillin in ovarian cancer. Cancer Res. 2008;68(14):5839–5848. doi: 10.1158/0008-5472.CAN-07-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heads JA, Hawthorne RL, Lynagh T, Lynch JW. Structure-activity analysis of ginkgolide binding in the glycine receptor pore. J Neurochem. 2008;105(4):1418–1427. doi: 10.1111/j.1471-4159.2008.05244.x. [DOI] [PubMed] [Google Scholar]
- 27.Ivic L, et al. Terpene trilactones from Ginkgo biloba are antagonists of cortical glycine and GABA(A) receptors. J Biol Chem. 2003;278(49):49279–49285. doi: 10.1074/jbc.M304034200. [DOI] [PubMed] [Google Scholar]
- 28.Jensen AA, et al. Probing the pharmacophore of ginkgolides as glycine receptor antagonists. J Med Chem. 2007;50(7):1610–1617. doi: 10.1021/jm070003n. [DOI] [PubMed] [Google Scholar]
- 29.Peters RJ, et al. Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry. 2000;39(50):15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- 30.Ravn MM, Coates RM, Flory JE, Peters RJ, Croteau R. Stereochemistry of the cyclization-rearrangement of (+)-copalyl diphosphate to (-)-abietadiene catalyzed by recombinant abietadiene synthase from Abies grandis. Org Lett. 2000;2(5):573–576. doi: 10.1021/ol991230p. [DOI] [PubMed] [Google Scholar]
- 31.Aharoni A, et al. The “evolvability” of promiscuous protein functions. Nat Genet. 2005;37(1):73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 32.Copley SD. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: The patchwork approach. Trends Biochem Sci. 2000;25(6):261–265. doi: 10.1016/s0968-0004(00)01562-0. [DOI] [PubMed] [Google Scholar]
- 33.Fischbach MA, Clardy J. One pathway, many products. Nat Chem Biol. 2007;3(7):353–355. doi: 10.1038/nchembio0707-353. [DOI] [PubMed] [Google Scholar]
- 34.Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem. 2006;281(12):8024–8033. doi: 10.1074/jbc.M510016200. [DOI] [PubMed] [Google Scholar]
- 35.Greenhagen BT, O’Maille PE, Noel JP, Chappell J. Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc Natl Acad Sci USA. 2006;103(26):9826–9831. doi: 10.1073/pnas.0601605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Maille PE, et al. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat Chem Biol. 2008;4(10):617–623. doi: 10.1038/nchembio.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters RJ, Croteau RB. Abietadiene synthase catalysis: Mutational analysis of a prenyl diphosphate ionization-initiated cyclization and rearrangement. Proc Natl Acad Sci USA. 2002;99(2):580–584. doi: 10.1073/pnas.022627099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keeling CI, Weisshaar S, Lin RP, Bohlmann J. Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc Natl Acad Sci USA. 2008;105(3):1085–1090. doi: 10.1073/pnas.0709466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bloom JD, Romero PA, Lu Z, Arnold FH. Neutral genetic drift can alter promiscuous protein functions, potentially aiding functional evolution. Biol Direct. 2007;2:17. doi: 10.1186/1745-6150-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440(7087):1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 41.Kloer DP, Welsch R, Beyer P, Schulz GE. Structure and reaction geometry of geranylgeranyl diphosphate synthase from Sinapis alba. Biochemistry. 2006;45(51):15197–15204. doi: 10.1021/bi061572k. [DOI] [PubMed] [Google Scholar]
- 42.Liao Z, et al. A new geranylgeranyl diphosphate synthase gene from Ginkgo biloba, which intermediates the biosynthesis of the key precursor for ginkgolides. DNA Sequence. 2004;15(2):153–158. doi: 10.1080/10425170410001667348. [DOI] [PubMed] [Google Scholar]
- 43.Hefner J, Ketchum REB, Croteau R. Cloning and functional expression of a cDNA encoding geranylgeranyl diphosphate synthase from Taxus canadensis and assessment of the role of this prenyltransferase in cells induced for Taxol production. Arch Biochem Biophys. 1998;360(1):62–74. doi: 10.1006/abbi.1998.0926. [DOI] [PubMed] [Google Scholar]
- 44.Hosfield DJ, et al. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J Biol Chem. 2004;279(10):8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 45.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27(8):753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 46.Tyo KE, Ajikumar PK, Stephanopoulos G. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol. 2009;27(8):760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 47.Morrone D, et al. Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol. 2010;85(6):1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starks CM, Back K, Chappell J, Noel JP. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277(5333):1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 49.Larkin MA, et al. Clustal W and Clustal X version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14(1):33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.