Abstract
DNA replication requires priming of DNA templates by enzymes known as primases. Although DNA primase structures are available from archaea and bacteria, the mechanism of DNA priming in higher eukaryotes remains poorly understood in large part due to the absence of the structure of the unique, highly conserved C-terminal regulatory domain of the large subunit (p58C). Here, we present the structure of this domain determined to 1.7-Å resolution by X-ray crystallography. The p58C structure reveals a novel arrangement of an evolutionarily conserved 4Fe-4S cluster buried deeply within the protein core and is not similar to any known protein structure. Analysis of the binding of DNA to p58C by fluorescence anisotropy measurements revealed a strong preference for ss/dsDNA junction substrates. This approach was combined with site-directed mutagenesis to confirm that the binding of DNA occurs to a distinctively basic surface on p58C. A specific interaction of p58C with the C-terminal domain of the intermediate subunit of replication protein A (RPA32C) was identified and characterized by isothermal titration calorimetry and NMR. Restraints from NMR experiments were used to drive computational docking of the two domains and generate a model of the p58C–RPA32C complex. Together, our results explain functional defects in human DNA primase mutants and provide insights into primosome loading on RPA-coated ssDNA and regulation of primase activity.
Keywords: polymerase, replisome, replication initiation, primed DNA, iron-sulfur cluster
The replisome is a dynamic assembly of proteins that carries out eukaryotic DNA replication. The first step in the initiation of DNA replication on both the leading and lagging strand ssDNA is the synthesis of a short (8–10 nucleotide) RNA primer by a polymerase termed a DNA primase (1–3). DNA primases are unique because they are the sole polymerases capable of de novo synthesis on a ssDNA template. Although integral to the replisome, the activity of DNA primase is not limited to DNA replication; primase is also an essential component of the DNA damage response and plays a role in telomere maintenance (4, 5).
Although all primases serve a common function, their architectures and modes of interaction with other protein factors vary among species (6, 7). The human DNA primase is part of the heterotetrameric polymerase α-primase (pol-prim) complex, which generates short (∼30 nucleotide) stretches of DNA primed with chimeric RNA–DNA hybrids that are handed off to the primary replicative DNA polymerases. The p48 and p58 subunits of human pol-prim form the DNA primase heterodimer. Although the p48 subunit contains the RNA polymerase catalytic active site, both subunits are required for synthesis of short RNA primers (8, 9). The p58 subunit is proposed to play various roles, including stabilizing the p48 subunit, recognizing the DNA template, and promoting initiation and elongation of primers (9, 10).
Considerable knowledge about the molecular mechanism of DNA priming has been obtained from structural studies of archaeal and bacterial primases (11, 12). In contrast, the mechanism of DNA priming in higher eukaryotes remains poorly understood because the p58 subunit of their DNA primases contain a highly conserved C-terminal domain (p58C) that is lacking in the archaeal proteins that have been structurally characterized to date (3, 11, 13). In fact, the function of bacterial primases is entirely distinct. Recently, a 4Fe-4S cofactor was discovered in p58C and disruption of the cluster by mutation of conserved cysteine residues was shown to abolish primase activity (14, 15). Several lines of evidence suggest that iron-sulfur clusters are essential for the function of many types of DNA processing proteins, but the specific roles of these essential cofactors have not been elucidated (16–18). Hence, even though the importance of the cluster has been established, the role of p58C in initiation of DNA replication remains poorly understood, primarily due to a lack of information about its structure and binding partners.
Physical interactions with replication and repair factors are key to the diverse functions of primase. One of the most critical primase interactions is with ssDNA templates coated with replication protein A (RPA). RPA is a modular protein, and its ability to directly interact with many different proteins facilitates assembly and progression of DNA processing machinery (19, 20). For the simian virus 40 (SV40) model system, interactions of pol-prim with RPA and the SV40 large T-antigen (Tag) helicase are required for efficient initiation of replication on the SV40 genome (21, 22). In previous studies, we showed that the physical interaction between the RPA32C protein interaction domain and the Tag origin binding domain (Tag-OBD) allows the pol-prim complex to access the 3′ end of DNA so that primase can initiate replication by synthesizing the RNA primer (19). However, the interactions of RPA with intact pol-prim or the primase subunits have not been investigated in detail.
To begin to fill critical gaps in knowledge of human DNA primase function, we have crystallized and determined a high-resolution structure of the p58C domain by X-ray crystallography. In addition, the DNA binding preferences of p58C were measured using a fluorescence anisotropy approach and functional interactions with RPA32C were mapped with NMR. Taken together, our results explain defects in function of primase mutants and provide insight into DNA priming and primosome loading on RPA-coated ssDNA.
Results
Structure of p58C is Unique.
After extensive screening, a p58C construct encoding human DNA primase residues Ser272-Glu464 was crystallized and the structure determined by single wavelength anomalous dispersion (SAD) using the anomalous signal of the four iron atoms present in the 4Fe-4S cluster. The resulting model was refined against native diffraction data extending to 1.7-Å resolution, resulting in a crystallographic residual (Rcryst) of 13.0% and Rfree of 16.9% (Table S1). Density was observed for 169 residues out of 193, with residues Glu330-Lys340, Gly354-Arg359, and Gly458-Glu464 missing presumably because they are disordered.
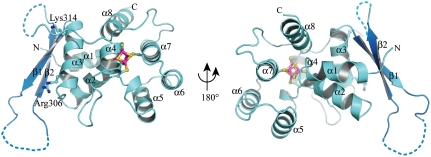
The structure of p58C reveals a single globular domain composed of α-helices flanked by a two-stranded β-sheet, which are separated by a long flexible loop (Fig. 1). The β-sheet region packs against the N-terminal side of the α-helical region (α1–α3). The essential iron-sulfur cluster is located at the core of the protein and is coordinated by four conserved cysteine ligands (Cys286, Cys367, Cys384, and Cys424; Fig. S1). The presence of the 4Fe-4S cluster was confirmed based on the unbiased electron density map (Fig. S2A). We note that this fold is unique for a 4Fe-4S cluster motif in a DNA processing protein, in particular because it is buried well within the hydrophobic core with little direct exposure to the surface (Fig. S2B).
Fig. 1.
The three-dimensional structure of p58C. Helices and β-strands are colored cyan and blue, respectively. The 4Fe-4S cluster and the side chains of key residues Arg306 and Lys314 are shown in ball and stick representation.
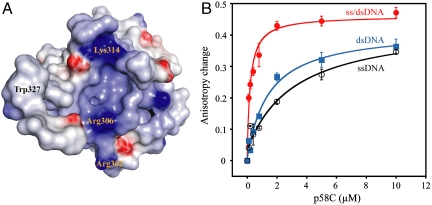
Remarkably, submission of the p58C structure to the Dali server did not return any similarities to structures available in the Protein Data Bank (23). Of note, although previous studies showed that residues Met288-Lys343 in p58C align with a specific pol β-like motif (9), the p58C structure reveals no structural similarity to this motif. However, these residues map to a strikingly basic surface (Fig. 2A), which represents a likely site for DNA binding. The functional relevance of this surface is supported by studies showing these residues are required to synthesize primers of defined unit length, and that single-site mutants in this region cause reduced activity in DNA priming assays (9). Therefore, the reduction in activity associated with the mutations is presumably due to defects in the binding of DNA, although the defects in the binding of other essential priming factors cannot be ruled out at this time.
Fig. 2.
Binding of DNA by p58C. (A) Molecular surface of p58C colored on the basis of electrostatic field potential (red, -7 kBT; blue +7 kBT) with the positions of Arg302, Lys314, and Trp327 highlighted. (B) Plots of increase in fluorescence anisotropy as a function of p58C concentration for labeled 12-mer ssDNA (open black circles), 12-mer dsDNA (closed blue squares), and ss/dsDNA junction with 12 base-pair duplex (closed red circles).
p58C Domain Preferentially Binds ss/dsDNA Junctions.
To test the ability of p58C to bind DNA, the binding affinities for ssDNA templates, primed DNA, and primed RNA–DNA hybrids were determined from fluorescence anisotropy measurements (Table S2). We found p58C preferentially binds to primed DNA (ss/dsDNA junctions) versus ssDNA and dsDNA (Fig. 2B), with a slight preference for 5′ versus 3′ overhangs. The affinity for ss/dsDNA junctions did not differ significantly with increasing length of the duplex region. Also, the affinity for an RNA–DNA hybrid ss/dsDNA junction was not significantly different than a purely DNA junction.
Having established the preference for ss/dsDNA junctions, we set out to map the DNA binding site on p58C by site-directed mutagenesis, with an emphasis on the highly basic putative DNA binding surface shown in Fig. 2A. The effect of mutations on the DNA binding affinity was determined using our fluorescence anisotropy assay on an eight base pair ss/dsDNA junction with a five nucleotide 5′ overhang (Fig. S3A). Mutation of Lys314, a residue shown to be important for primer initiation (9), and Arg302, another basic residue in the putative DNA binding surface, reduced affinity for the ss/dsDNA junction (Table S2). In both cases, a > 10-fold reduction in affinity was observed for charge reversal to Glu. A third site selected for mutation was the conserved Trp327, which is highly solvent exposed in the structure but resides in close proximity to the basic surface (Fig. 2A). The involvement of this residue in DNA binding was confirmed by intrinsic tryptophan fluorescence (Fig. S3B), NMR (Fig. S3C), and a ∼5-fold reduction in affinity for ss/dsDNA junction for the Trp327Ala mutant. Two double mutations with this Trp and a basic residue, Lys314Glu/Trp327Ala and Arg306Glu/Trp327Ala, further reduced DNA binding about ∼20- and ∼30-fold, respectively, whereas control mutations (Lys328Ala and Lys369Ala) had no effect. Together, the mutational data and extent of solvent exposure of the corresponding residues supports our proposal of DNA binding to the highly basic surface of p58C shown in Fig. 2A.
p58C Domain Binds to the RPA32C Protein Interaction Module.
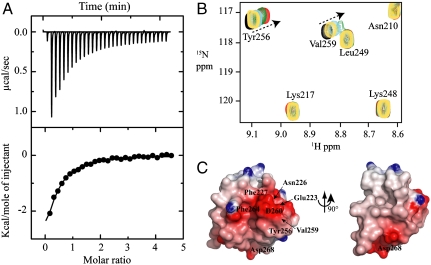
The initiation of replication requires displacement of RPA coated ssDNA so that primase can gain access to the DNA template. Contacts between Tag, pol-prim, and RPA are critical to overcome inhibition of primer initiation on RPA bound ssDNA and efficient primase activity (24). However, the precise region(s) of RPA involved has not been investigated. To determine if RPA interacts with p58C, we performed pull-down experiments in which His-tagged p58C bound to Ni resin was incubated with different protein interaction domains of RPA (RPA70N, RPA70AB, RPA32C). RPA32C exhibited the most significant interaction (Fig. S4A). This domain has previously been shown to be involved in protein–protein interactions with several other DNA processing proteins (Fig. S4B). To quantify the affinity of RPA32C for p58C, we employed isothermal titration calorimetry (ITC). The binding isotherm was fit to simple models, which provided a Kd value of 25 μM (Fig. 3A).
Fig. 3.
Interaction of p58C with RPA32C. (A) Isothermal titration calorimetry binding isotherm for titration of RPA32C into p58C showing the raw heat release (Upper) and the integrated heat release (Lower). (B) Binding of p58C to RPA32C monitored by 15N-1H heteronuclear single quantum coherence NMR spectra of 15N-RPA32C in the absence (black) and presence of 1∶0.25 (yellow), 1∶0.5 (cyan), 1∶1 (green) and 1∶2 (red) molar ratios of p58C. (C) Molecular surface of RPA32C colored according to electrostatic potential (red, -7 kBT; blue +7 kBT) with key residues labeled.
To obtain further insight into the binding of p58C to RPA32C, the sequence of p58C was aligned to the RPA32C binding sites of DNA repair factors UNG2 and XPA. A region of p58C (Arg289–Arg302) was found to align well (Fig. S4C). UNG2 and XPA bind RPA32C with affinities in the range of 250 μM using only a single helical element (25). Because p58C binds about 10-fold more tightly, we reasoned that the stronger interaction may require a larger binding surface, suggesting the possibility that p58C utilizes a compound binding surface like that observed for the origin binding domain of the SV40 large T-antigen helicase (Tag-OBD) (19).
NMR-Based Structural Model of the p58C–RPA32C Complex.
To map the p58C-binding site on RPA32C, NMR chemical shift perturbation experiments were performed monitoring 2D 15N-1H heteronuclear single quantum coherence spectra of 15N-enriched RPA32C as increasing amounts of unlabeled p58C were titrated. Significant chemical shift perturbations and reduced peak intensity were observed over the course of titration (Fig. 3B) indicating fast and intermediate exchange rates on the NMR timescale, consistent with the Kd determined by ITC. Analysis of the chemical shift perturbations revealed that acidic and hydrophobic residues (e.g., Glu223, Asn226, Phe227, Thr258, Val259, Asp260, Asp268) contribute to a specific binding surface on RPA32C (Fig. 3C and Fig. S5). Comparison to past studies of RPA32C interactions revealed that the residues perturbed upon binding of p58C are similar to those involved in binding Tag-OBD (Fig. 3C) (19). However, there are some differences, such as the more pronounced perturbation of hydrophobic residues Phe227, Tyr256, and Val259 for the p58C–RPA32C complex. Despite these small differences, the data strongly imply that p58C and Tag-OBD bind to a common surface on RPA32C.
The NMR approach could not be used similarly to map the RPA32C binding site on p58C due to paramagnetic broadening of NMR signals by the 4Fe-4S cluster. We therefore turned to two stages of experimentally guided molecular docking to generate a structural model of the p58C–RPA32C complex. First, because we could not use NMR, we adopted an unbiased computational docking strategy to determine if a unique surface on p58C for binding RPA32C was predicted. These calculations were performed with Hex (26), which uses a scoring function based purely on shape and electrostatics. The approach proved successful because the best scoring complexes all used the same binding surface of p58C. We note that these models contained two clusters with different p58C-interacting surfaces of RPA32C (Fig. S6). However, only one surface was consistent with the NMR data. A second program RosettaDock (27) was then used to generate the final models of the complex.
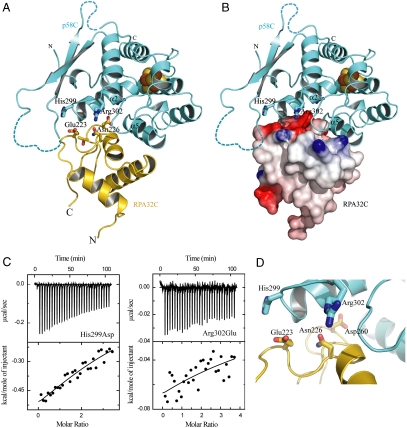
Fig. 4A shows a representative conformer from the ensemble of highest scoring RosettaDock models. The interaction primarily involves residues of α-helices 2 and 5 of p58C and the complementary surface of RPA32C identified by NMR. The complex between the two proteins is stabilized by a combination of electrostatic and hydrophobic interactions. The substantial electrostatic component (Fig. 4B) is consistent with the exothermic heat of binding observed by ITC (Fig. 3A), implying an important role for charged residues in the binding interface.
Fig. 4.
Model of the p58C–RPA32C complex. (A) Ribbon diagram with p58C and RPA32C colored cyan and gold, respectively. Side chains at the interface are shown in ball and stick representation. (B) The same view of A, with RPA32C shown as a molecular surface colored according to electrostatic potential (red, -7 kBT; blue +7 kBT). (C) Isothermal titration calorimetry binding isotherm for titration of RPA32C into p58C mutants His299Asp (Left) and Arg302Glu (Right). Upper and lower panels show the raw heat release and the integrated heat release, respectively. (D) Close-up view of the p58C–RPA32C protein interface.
Mutations Validate the Model of the p58C–RPA32C Complex.
To test the structural model, mutations of p58 residues Arg286, Lys293, Arg296, His299, and R302 to oppositely charged amino acids were prepared and their affinities for RPA32C measured by ITC. Significant effects on binding of RP32C with p58C mutants His299Asp and Arg302Glu were observed (Fig. 4C), consistent with their role in the structural model as key p58C contacts with Glu223 and Asn226/Asp260 of RPA32C, respectively (Fig. 4D). A third p58C residue well within the binding interface in the model (Arg296) also had a significant effect when mutated, whereas two residues outside the interface (Arg289 and Lys293) did not (Fig. S7). The effect of charge reversal of Arg296, His299, and Arg302 was substantial, resulting in 5-, 10-, and 50-fold reduction in the affinity of RPA32C, respectively (Fig. S7G).
To confirm the conclusion from the affinity measurements, we also performed NMR titration experiments for RPA32C with His299Asp and Arg302Glu. Only weak or no detectable chemical shift perturbations were observed for these mutants even at 1∶2 molar ratio (Fig. S7 E and F), consistent with the weaker binding to RPA32C observed by ITC. To ensure that the structural integrity of the mutants was not compromised, CD spectra were recorded (Fig. S7H); no significant difference between wild-type and mutant proteins were observed. Together, these results serve to confirm the accuracy of the p58C-RPA32C model.
Discussion
p58C is a domain unique to the large subunit of DNA primases in higher eukaryotes. This domain contains a 4Fe-4S cluster that is essential for initiating primer synthesis (15). Although its physiological relevance was established, the biochemical basis for p58C function remains unexplained, in part due to the lack of structural information. The crystal structure of p58C reveals a unique fold for an iron-sulfur cluster motif and suggests that the cluster is integral to the structure of the domain. However, it is unlikely that nature evolved such a complex cofactor purely for maintaining the structural integrity of a small domain. Rather, we believe that the cluster has some yet-to-be-recognized biochemical function. This function might, for example, parallel DNA-mediated electron transport involving the 4Fe-4S cluster, which has been proposed to play a role in the cellular stress response induced by reactive oxygen species (28).
The compound surface of conserved, basic residues of p58C provides an explanation for previous observations that mutation in this region of p58 abolished primase activity. For example, we find that mutation of the conserved residue Lys314, which has a significant defect in synthesizing primers of defined length (9), is also defective in binding DNA. Interestingly, the p58C structure shows the positioning of residues in the basic surface is buttressed by the 4Fe-4S cluster. Thus, like DNA repair proteins containing iron-sulfur clusters, the 4Fe-4S cluster in p58C may play a role in organizing the protein surface to facilitate binding of DNA (9). It is intriguing to consider that conformational changes in various functional states during DNA priming may induce strain in the cluster or alter surface exposure, both of which might alter the redox potential of the 4Fe-4S cluster. Such alterations in the redox potential of the cluster might provide a means to sense aberrant replication amidst DNA damage and to consequently regulate DNA primase activity.
The interaction with RPA32C implies p58C is involved in recruiting pol-prim to the replication fork and/or promoting the release of RPA from template DNA that is required to initiate priming. NMR chemical shift perturbation and isothermal titration calorimetry studies reveal RPA32C binds to p58C with low micromolar affinity. Structural and biochemical data suggest that RPA32C utilizes a common binding surface to interact with p58C and Tag-OBD (19). In SV40 DNA replication, the physical interaction between Tag and RPA was shown to be essential for primosome assembly and initiation of primer synthesis on RPA-coated ssDNA (29, 30). It has been proposed that the interaction of Tag-OBD with RPA32C facilitates displacement of RPA from ssDNA, and in turn enables pol-prim to access ssDNA template (19, 21). The higher affinity of p58C for RPA32C suggests that the p58C domain of primase will out-compete Tag-OBD for binding to RPA-coated ssDNA. Additional studies are required to determine if this switch facilitates loading of the primosome or if the interaction of other pol-prim subunits with RPA is also required.
Our results are also consistent with p58C having a regulatory role in DNA priming. It has been suggested that p58C negatively regulates primase activity by recognizing defined lengths of RNA primer (9). Here, we report that p58C has a preference for primed DNA (ss/dsDNA junctions), which may support this model. It is intriguing to speculate that, as primase reaches its threshold of 8–10 base pairs, the p48/58 complex may undergo a conformational change associated with the high affinity of p58C for the ss/ds junction, which could limit further primer synthesis and promote the switch from RNA priming by the p48 catalytic subunit to DNA primer extension by the p180 subunit of pol-prim. However, it is premature to draw conclusions about priming mechanisms from experiments on isolated p58C and considerably greater experimental evidence would be required before such a model could be confirmed.
While this manuscript was under revision, a crystal structure of yeast PriL-CTD was reported (31). Although the human and yeast iron-sulfur cluster domains share a similar fold, the structure of the DNA binding region in human p58C differs significantly from the corresponding region of yeast PriL-CTD (Fig. S8). In the human protein, the basic putative DNA binding region is composed of a helix and a short antiparallel β-sheet (α3, β1, β2). In contrast, the corresponding region in yeast forms a three helix bundle (α3, α4, α5). Moreover, human p58C binds to DNA with much higher affinity than yeast PriL-CTD. For instance, the fluorescence anisotropy approach showed that yeast PriL-CTD binds a 20-mer ssDNA with a Kd of 70 ± 0.2 μM and a 20-mer dsDNA with a Kd of 178 ± 1.3 μM. Using the same method and same oligonucleotides, p58C binds with ∼70-fold and ∼125-fold higher affinity, respectively. In addition, the conserved Trp327 in human p58C is solvent exposed and its direct involvement in DNA binding is implied by intrinsic tryptophan fluorescence, NMR, and mutational analysis. In contrast, the corresponding residue (Trp376) in yeast PriL-CTD is completely buried in the structure (Fig. S8). Thus, the structural dissimilarity between DNA binding regions is consistent with substantial differences in the DNA binding function of these proteins. We conclude that the yeast protein does not provide an adequate model for DNA priming in humans or other higher eukaryotes.
In summary, our structure and biochemical studies of p58C provide an essential missing link in understanding the molecular basis for DNA primase function in higher eukaryotes. These results provide critical information to guide further studies of intact human DNA primase to test and refine hypotheses about the mechanism of action of the primosome.
Methods
The details of protein expression, site-directed mutagenesis, purification, crystallization, structure determination, DNA interaction analysis by fluorescence anisotropy, RPA32C interaction analysis by isothermal titration calorimetry and NMR, and the computational modeling are described in the SI Text. In short, p58C was expressed in Escherichia coli and purified by several steps of chromatography. The protein was crystallized by sitting drop vapor diffusion and the final diffraction data were collected from an in-house source and at Life Sciences Collaborative Access Team beamline at the Advanced Photon Source. The structure was determined by SAD using the anomalous signals of the Fe atoms. DNA binding affinities were determined by measuring the change in fluorescence anisotropy of labeled oligonucleotides as protein is added to the solution. The interaction of p58C with RPA32C was analyzed by ITC and heteronuclear NMR. The model of the p58C–RPA32C complex was generated by an initial phase of free docking calculations using Hex (26), which predicted a specific RPA32C binding site on p58C, followed by a second round of docking calculations using RosettaDock (27) with the Hex-defined binding surface for p58C and NMR-defined binding surface for RPA32C.
Supplementary Material
Acknowledgments.
We thank Marie-Eve Chagot for the assistance with generating constructs, the Life Sciences Collaborative Access Team staff at the Advanced Photon Source for help with data collection, and Ellen Fanning for many stimulating discussions. This research was supported by operating grants from the National Institutes of Health (NIH) (R01 GM65484 to W.J.C.; R01 GM080570 to B.F.E.). Additional NIH support was provided by the Structural Biology of DNA Repair Machines Program (P01 CA92584), the Vanderbilt Center in Molecular Toxicology (P50 ES00267), and the Vanderbilt-Ingram Cancer Center (P30 CA68485).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3L9Q).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002009107/-/DCSupplemental.
References
- 1.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2009;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 3.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 4.Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 5.Michael WM, Ott R, Fanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 6.Keck JL, Berger JM. Primus inter pares (first among equals) Nat Struct Biol. 2001;8:2–4. doi: 10.1038/82996. [DOI] [PubMed] [Google Scholar]
- 7.Arezi B, Kuchta RD. Eukaryotic DNA primase. Trends Biochem Sci. 2000;25:572–576. doi: 10.1016/s0968-0004(00)01680-7. [DOI] [PubMed] [Google Scholar]
- 8.Foiani M, Lucchini G, Plevani P. The DNA polymerase alpha-primase complex couples DNA replication, cell-cycle progression and DNA-damage response. Trends Biochem Sci. 1997;22:424–427. doi: 10.1016/s0968-0004(97)01109-2. [DOI] [PubMed] [Google Scholar]
- 9.Zerbe LK, Kuchta RD. The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry. 2002;41:4891–4900. doi: 10.1021/bi016030b. [DOI] [PubMed] [Google Scholar]
- 10.Arezi B, Kirk BW, Copeland WC, Kuchta RD. Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: Implications for the role of the p58 subunit. Biochemistry. 1999;38:12899–12907. doi: 10.1021/bi9908991. [DOI] [PubMed] [Google Scholar]
- 11.Lao-Sirieix SH, et al. Structure of the heterodimeric core primase. Nat Struct Mol Biol. 2005;12:1137–1144. doi: 10.1038/nsmb1013. [DOI] [PubMed] [Google Scholar]
- 12.Corn JE, Pelton JG, Berger JM. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat Struct Mol Biol. 2008;15:163–169. doi: 10.1038/nsmb.1373. [DOI] [PubMed] [Google Scholar]
- 13.Lipps G, et al. Structure of a bifunctional DNA primase-polymerase. Nat Struct Mol Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 14.Klinge S, et al. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol. 2007;14:875–877. doi: 10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner BE, et al. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J Biol Chem. 2007;282:33444–33451. doi: 10.1074/jbc.M705826200. [DOI] [PubMed] [Google Scholar]
- 16.Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Curr Opin Chem Biol. 2005;9:145–151. doi: 10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Fan L, et al. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pugh RA, et al. The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the single-stranded DNA-double-stranded DNA junction. J Biol Chem. 2008;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- 19.Arunkumar AI, et al. Insights into hRPA32 C-terminal domain—mediated assembly of the simian virus 40 replisome. Nat Struct Mol Biol. 2005;12:332–339. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisshart K, Taneja P, Fanning E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J Virol. 1998;72:9771–9781. doi: 10.1128/jvi.72.12.9771-9781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dornreiter I, et al. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 24.Weisshart K, et al. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J Biol Chem. 2000;275:17328–17337. doi: 10.1074/jbc.M000717200. [DOI] [PubMed] [Google Scholar]
- 25.Mer G, et al. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie DW. Evaluation of protein docking predictions using Hex 3.1 in CAPRI rounds 1 and 2. Proteins. 2003;52:98–106. doi: 10.1002/prot.10379. [DOI] [PubMed] [Google Scholar]
- 27.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genereux JC, Boal AK, Barton JK. DNA-mediated charge transport in redox sensing and signaling. J Am Chem Soc. 2010;132:891–905. doi: 10.1021/ja907669c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins KL, Kelly TJ. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 31.Sauguet L, et al. Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS ONE. 2010;5(4):e10083. doi: 10.1371/journal.pone.0010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.