Abstract
Molecular dynamics simulation of Thermus thermophilus (Tt) RNA polymerase (RNAP) in a catalytic conformation demonstrates that the active site dNMP-NTP base pair must be substantially dehydrated to support full active site closing and optimum conditions for phosphodiester bond synthesis. In silico mutant β R428A RNAP, which was designed based on substitutions at the homologous position (Rpb2 R512) of Saccharomyces cerevisiae (Sc) RNAP II, was used as a reference structure to compare to Tt RNAP in simulations. Long range conformational coupling linking a dynamic segment of the bridge α-helix, the extended fork loop, the active site, and the trigger loop-trigger helix is apparent and adversely affected in β R428A RNAP. Furthermore, bridge helix bending is detected in the catalytic structure, indicating that bridge helix dynamics may regulate phosphodiester bond synthesis as well as translocation. An active site “latch” assembly that includes a key trigger helix residue Tt β’ H1242 and highly conserved active site residues β E445 and R557 appears to help regulate active site hydration/dehydration. The potential relevance of these observations in understanding RNAP and DNAP induced fit and fidelity is discussed.
1. Introduction
Multi-subunit RNA polymerases (RNAPs) synthesize RNA from a DNA template, but many features of the catalytic mechanism and its control remain unknown. For instance, although RNAP functions in an aqueous environment, the importance of active site hydration and dehydration in catalysis has not been fully described. X-ray crystal structures of ternary elongation complexes (TECs) reveal closed and open conformations of the RNAP active site [1–4]. Closing and opening, which are expected to affect, and to be affected by, hydration, rely on the conformation of the trigger helices-trigger loop assembly (Thermus thermophilus (Tt) RNAP β’ 1220 to 1265), which can assume a helical (closed, catalytic) or more looped (open, low activity) conformation. To underscore the importance of active site closing for catalysis, the antibiotic streptolydigin binds to a Tt RNAP TEC with an open trigger loop [2], and the potent Sc RNAP II inhibitor α-amanitin binds to a TEC with an open “wedged” conformation of the trigger loop [1]. In the catalytic structure, the closed trigger helices pack closely with the bridge α-helix, which is a prominent and dynamic feature of RNAP that borders the active site [1, 5, 6]. It has been suggested that bridge helix dynamics, which is expected to be regulated by trigger loop opening and closing, may provide the thermal driving force for translocation of nucleic acids through RNAP [7–9].
Water can regulate the specificity and fidelity of enzymatic reactions [10–15], and many enzymes including RNAP have a buried active site that can close to exclude water [2, 3, 16]. For example, the active site of DNA polymerase (DNAP)-β is substantially dehydrated by active site closing [16], and, in this manner, dehydration could potentially be a determining factor in catalysis and fidelity. Unfortunately, however, biochemical techniques are somewhat limited in their capacity to analyze water-mediated effects. For instance, lower resolution crystal structures may not account for bound waters, and loosely bound water may not be observed crystallographically. To expose RNAP internal sites to hydration, therefore, the TEC structure was immersed in explicit water and subjected to full atomistic molecular dynamics simulations, as has previously been done for Sc RNAP II, T7 RNAP, and DNAPs [17–21]. In this work, simulations strongly indicate that water exclusion from the closed RNAP active site localized to the dNMP-NTP base pair is a determining factor in catalysis. In closing the active site, key trigger helix residues perform a central function in dehydration. In enzymatic mechanisms “induced fit” refers to the cooperative interaction between enzyme and substrates to accurately align reactive groups for chemistry. The current work strongly indicates that dehydration within an enclosed active site provides an essential component of induced fit mechanisms and that, for RNAPs, excessive localized hydration slows elongation by destabilizing the active site base pair, indicating that hydration/dehydration may be important in control of transcriptional fidelity.
A highly conserved residue Tt β R428 (corresponding to Saccharomyces cerevisiae (Sc) Rpb2 R512) is located about 20 Å from Mg2+-I and just C-terminal to the fork loop. The Sc Rpb2 R512C mutation was initially identified in a genetic screen for suppressors of ssu72-2, which encodes a catalytically-impaired form of the RNAP II carboxy-terminal domain (CTD) serine 5 (S5) phosphatase [22–24]. The CTD is a repeating heptapeptide unit (26 repeats in Sc) of consensus 1-YSPTSPS-7. The cyclin-dependent kinase, Kin28, which is a component of the RNAP II transcription initiation factor IIH, phosphorylates S5 of the CTD, and the S5-phosphate is removed by the Ssu72 and Rtr1 phosphatases [25, 26]. The Rpb2 R512C substitution may result in slowing RNA synthesis during early elongation. Presumably, the reduction in elongation rate allows sufficient time for the ssu72-2-encoded phosphatase to remove S5-phosphate, thereby allowing RNAP II to pass through transcription cycle checkpoints. Consistent with this interpretation, rpb2-R512C mutants exhibit defects in transcription in vivo [22] and in vitro [23, 27]. The transcriptional defects of R512C and R512A substitutions are very similar [27].
The β R428A in silico substitution in Tt RNAP was designed based on the Sc RNAP II Rpb2 R512C/A replacements, and Tt R428A provides an essential comparison to wild type (wt) RNAP in molecular dynamics simulations. Although Sc RNAP II (PDB 2E2H), with a closed trigger loop and loaded NTP analogue, might be a reasonable choice for simulations, we selected Tt RNAP (PDB 205J) because its structure was determined at a higher resolution, showing a straighter bridge helix and more planar geometries of active site i and i + 1 base pairs (in the catalytic structure, i indicates the position of the 3’-end of the RNA, and i+1 indicates the position of the dNMP-NTP base pair). In addition, on inspection of the closed Sc and Tt TECs, the Tt RNAP active site appeared to show more favorable atomic contacts than the corresponding Sc RNAP II structure [2, 3]. The closed trigger helix conformation was selected because we reasoned that the catalytic TEC might be more sensitive to mutation than an open, relaxed TEC. Comparing simulations of wt and β R428A Tt RNAP indicates: 1) long range conformational coupling between the bridge helix, the extended fork region, the active site and the trigger helix; 2) the importance of localized active site dehydration in catalysis and fidelity; 3) the importance and potential complexity of bridge α-helix dynamics; and 4) mechanisms for streptolydigin and microcin J25 inhibition of Tt RNAP and α-amanitin inhibition of Sc RNAP II.
2. Materials and Methods
2.1 Purification of Sc RNAP II
Yeast strains YZS84 (RPB2) and YDP19 (rpb2–100), which encodes Rpb2 R512C [22], were used to TAP-tag the C-terminus of the RNAP II Rpb9 subunit, as described [28]. Whole-cell extracts from the resulting strains were prepared as described [29] and used to purify wt RNAP II and the Rpb2 R512C RNAP II complex, according to [28]. Purified RNAP II was dialyzed against buffer consisting of 20 mM HEPES, pH 7.6, 20% (v/v) glycerol, 10 mM EGTA, 10 mM MgSO2, 5 mM dithiothreitol, and 1 mM phenylmethylsulfonylflouride and stored in aliquots at −80°C.
2.2 In vitro TEC assembly
In vitro assembly of Sc RNAP II TECs was done as described [30–33]. The sequence of G9 RNA is 5’-AUCGAGAGG-3’. The DNA non-template strand was 5’-biotinyl-GGTATAGGATACTTACGCCATCGAGAGGGACACGGTGAAAAGAGAACCCAAGCGAC ACC-3’ (G9-sequence is underlined) and the DNA template strand was 5’-GGTGTCGCTTGGGTTCTCTTTTCACCGTGTCCCTCTCGATGGCGTAAGTATCCTATAC C-3’ (G9-complement sequence is underlined). TECs were immobilized on Promega streptavidin-coated Magnesphere beads.
2.3 Rapid chemical quench flow
In vitro transcription experiments with wt and Rpb2 R512C Sc RNAP II were done using rapid chemical quench flow with the Kintek RQF-3 instrument, essentially as described [27, 34, 35]. RNAs were labeled by addition of α32P-GTP (800 Ci/mmole) for 10 min at 25 C in transcription buffer containing 60 mM KCl and 8 mM MgCl2. TECs were then set aside on ice until addition of ATP for the running start to A11. Using this protocol, only active TECs are tracked in the reaction because GTP must be incorporated to visualize RNA on gels. Running starts for 32P-labeled G10 TECs were by addition of 5 µM ATP for 30 s on the bench top. During the 30 s incubation, samples were transferred to the left sample port of the KinTek RQF-3 (KinTek Corporation) rapid chemical quench flow instrument. Reagents for elongation were loaded at twice their working concentration in transcription buffer containing 60 mM KCl and 8 mM MgCl2, in the right sample port. The valves to the sample ports were set to the “fire” position, and the computer-regulated mixing program was initiated. Reactions were stopped with EDTA (500 mM) or HCl (1 M), as indicated. RNA products were analyzed on gels.
2.4 Rationale and strategy for molecular dynamics simulations
These studies demonstrate the requirement for molecular dynamics simulations to adequately interpret mutational structure-function analyses and to gain insight into wt mechanisms. For instance, without dynamics, the analysis of Sc RNAP II Rpb2 R512C/A substitutions would not be complete. Simulations may be particularly informative because the starting Tt RNAP TEC structure (PDB 205J) is in a strained, catalytic conformation that responds very sensitively to the R428A substitution but is stable for wt RNAP. Although not yet done, simulation of R428A in a structure with a relaxed trigger loop conformation (PDB 205I or 2PPB) might be much less informative, because the R428A substitution may not as strongly disrupt an already relaxed TEC. For instance, opening the trigger loop is expected to remove constraints on bridge α-helix dynamics by reducing contacts between the bridge helix and the trigger helices [9]. Based on these results and ideas, we suggest that wt simulations may require a reference structure (in this case R428A) to be most useful and that the choice of starting wt structure (in this case a strained catalytic structure) may be crucial.
Molecular dynamics provides a model based on optimized force field assignments, and, therefore, results of simulations must be considered with some caution. Molecular dynamics provides a picosecond (ps) to nanosecond (ns) time-resolved atomistic model of protein structure and movement with explicit water hydration and counterions. Genetic, biochemical and structural approaches rely on ns scale atomistic events in a hydrated environment but are limited to provide detail comparable to simulations. Using the simulations reported here, comparison of the wt and R428A RNAP gives insight into probable mutant protein defects. More significantly, however, without the R428A RNAP reference, the wt RNAP simulation could not be adequately interpreted. For instance, because excluded waters cannot be observed for wt RNAP, it would not be possible to detect active site dehydration without observing the localized increase of hydration at the i+1 dNMP-NTP base pair in R428A RNAP.
2.5 Tt RNAP TEC simulations
A model of the Tt RNAP TEC containing a closed, catalytic trigger helix conformation was generated using the structures 205I and 205J [2, 36]. ATP was generated at the active site from AMPcPP (α,β-methylene adenosine triphosphate, a non-incorporatable ATP analogue). The initial structure was placed in an octahedral box with explicit waters and counterions to force charge neutrality and equilibrated while relaxing harmonic restraints placed on the protein over 500 picosecond (ps) time intervals for 1 nanosecond (ns). The temperature was held at 300K. Although T. thermophilus is a hyperthermophile, simulations run at 300 or 340K tend to give similar results [37], and force fields are currently optimized and predominately used around 300K [38], so 300K was selected. Because wt RNAP is stable during simulation, and the R428A RNAP shows instability, 300K appears sufficient for the wt RNAP to mutant comparison.
A model of the β and β’ Tt RNAP TEC (omitting α2 and ω) containing a closed trigger helices conformation, double stranded nucleic acids and ATP was generated using the Molecular Operating Environment MOE 2009.06 program package [39]. Omission of α and ω subunits was done to decrease computation times, and this strategy appeared to be justified because structural integrity was maintained far from the active site during long term simulation, indicating that eliminating subunits did not reduce the overall RNAP stability. To construct the model, two crystallographic structures were necessary: 205J, which has AMPcPP (α,β-methylene adenosine triphosphate, a non-incorporatable ATP analogue) loaded at the active site and which has a closed trigger helices conformation, was made more complete by addition of the cleft loop residues, obtained from 2O5I, β’ 1272–1328, which are disordered in 2O5J [2, 36]. The loop structure was first energy minimized in MOE using the AMBER force field [40] with constraints placed on the rest of the RNAP structure and with implicit waters. The AMPcPP that resides in the active site was used to generate ATP in the same location. Parameters were generated for the complete system in the xLeap module of AMBER8 using the ff03 force field for the protein, RNA, and DNA [41] while the ATP monomer parameters were obtained from the AMBER web site. The long range electrostatics were calculated using the Particle Mesh Ewald method (PME) [42]. Bond lengths involving hydrogens were constrained with SHAKE [43], allowing a 2 femtosecond (fs) time step, and the temperature was controlled using a Berendsen thermostat [44]. This system was energy minimized by first relaxing the water molecules, with a 100 kcal/mol/Å2 harmonic restraint placed on protein and nucleotide atoms, followed by a 500 picosecond (ps) equilibration run, relaxing the restraint from 50 kcal/mol/Å2 to zero at constant pressure, up to the final density of 1 g/ml. A subsequent 500 ps run was performed at constant volume without restraints at 300 K. The total time for system relaxation and equilibration was 1 nanosecond (ns). Production simulations of wt Tt RNAP were performed for a total of 9.5 ns, during which time the wt Tt RNAP TEC varied little, as measured by root mean square deviation, from the original crystal structure (not shown).
The mutant R428A was constructed in MOE with its parameters, and explicit waters generated in xLeap using the ff03 force field. Initially, the relaxation and equilibration runs of R428A RNAP were done as for wt RNAP, leading to a total time of 1 ns. However, it was noted that during equilibration the R428A TEC deviated from its starting structure significantly, altering active site protein and ATP geometries. Subsequently, a second equilibration run was begun from the initial structure with larger restraints (200 kcal/mol/Å2) that were slowly reduced over a longer time frame. After a total of 2 ns, this latter equilibration simulation once again showed similar results to the initial run. The run was continued for another 2 ns, during which time the TEC maintained the new configuration. Utilizing a variety of initial conditions, varying temperature gradients from 0 to 300 K and restraints from 200 to 0 kcal/mol/Å2 through time, we initiated eight separate runs with β R428A varying in their start-up conditions, and durations, and summing to a total of 10 ns. The results from the single simulation reported here were consistent in multiple simulations.
3. Results
3.1 Sc Rpb2 R512C is slow in elongation
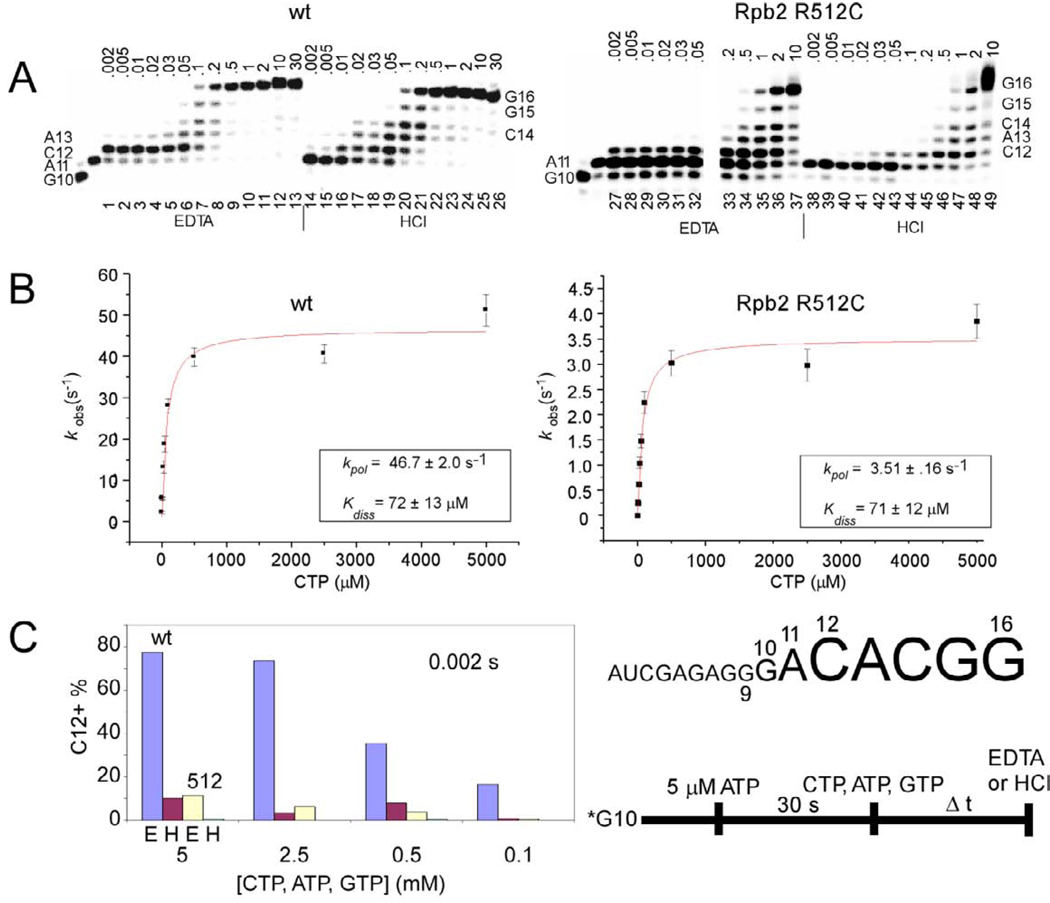
Using a rapid chemical quench flow mixing device, the elongation rate of Sc Rpb2 R512C was compared to wt RNAP II (Fig. 1). TECs were assembled in vitro from G9 RNA (5’-AUCGAGAGG-3’) (G9: a 9 nucleotide RNA ending in 3’-GMP), a DNA template, Sc RNAP II, and the DNA non-template strand. α32P-GTP was added to extend the TEC to G10. After a 30 s running start with 5 µM ATP to advance the TEC to A11, ATP, CTP, and GTP were added to extend transcripts to G16. Reactions were quenched with EDTA or with HCl. EDTA quenching indicates the time of stable NTP-Mg2+ sequestration, and HCl quenching indicates the time of phosphodiester bond synthesis [35, 45].
Fig. 1.
Sc Rpb2 R512C is slow in elongation. The protocol and RNA sequence are shown at the bottom right of the figure. A) Transcriptional defects of Rpb2 R512C. CTP, ATP, and GTP were at 5 mM. Quench times are in seconds. B) Determination of Kdiss and kpol. CTP, ATP, and GTP were varied between 2 µM and 5 mM. Samples were quenched with HCl. Each data point indicates an apparent rate from exponential curve fitting (C12 + longer transcripts (product) versus time). Error bars indicate standard error from exponential fitting. C) Rpb2 R512C may have a defect in CTP-Mg2+ sequestration. At 5, 2.5, 0.5, and 0.1 mM CTP, reactions are quenched at 0.002 s with EDTA (E) or HCl (H).
Wt RNAP II is much faster in elongation than Rpb2 R512C. In Fig. 1A, Fig. 5 mM ATP, CTP, and GTP were added, so the reaction is running close to its maximal rate. For wt, the G16 transcript first appears at 0.1 s (lane 20) and, for Rpb2 R512C, at 0.5 or 1 s (lanes 46–47), indicating a 5–10 fold slower elongation rate for R512C. Because stable CTP-Mg2+ assimilation (EDTA quench) occurs at earlier times than phosphodiester bond synthesis (HCl quench) (compare lanes 1–6 and lanes 14–19), Sc RNAP II stably sequesters CTP-Mg2+ for C12 synthesis prior to formation of the C12 bond [34, 46, 47].
Fig. 5.
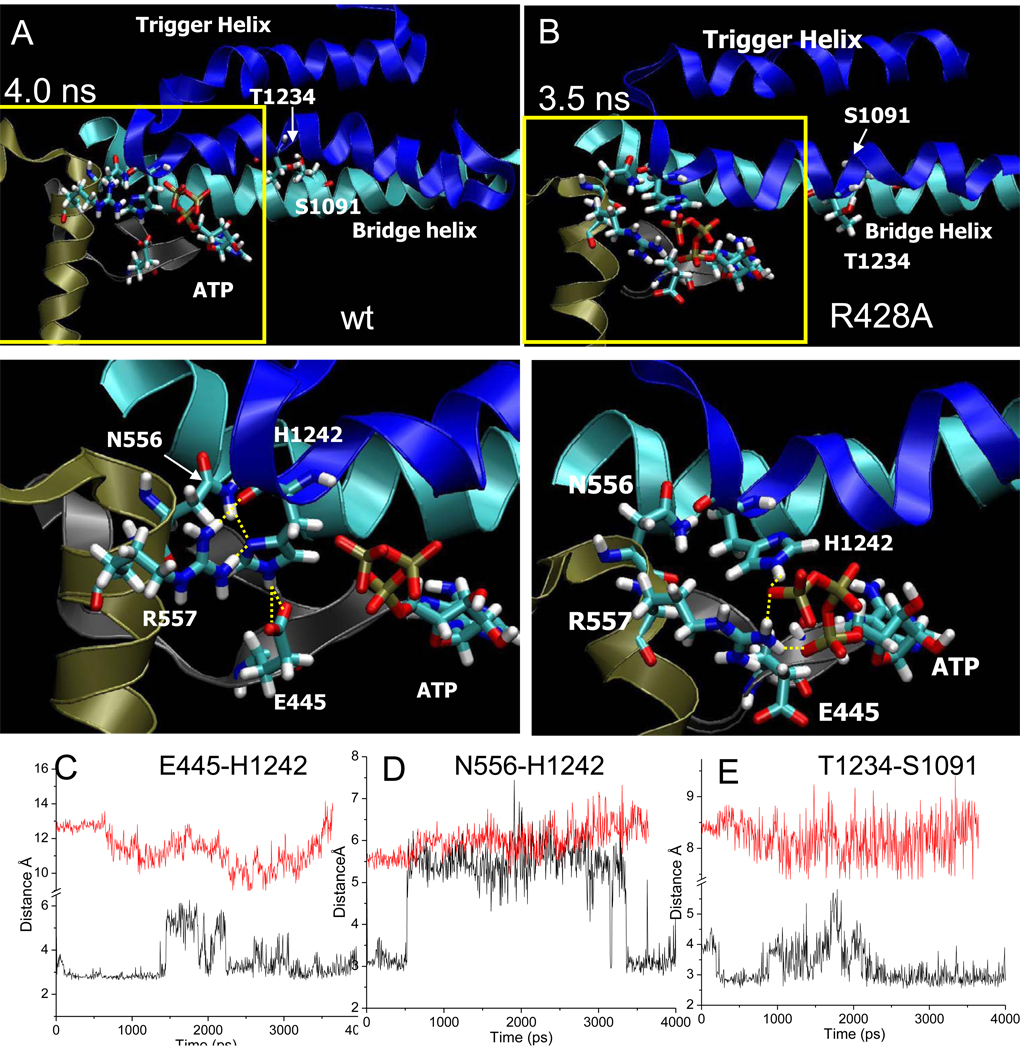
Wt latch and R428A RNAP active site conformations. A) Snapshot of wt RNAP at 4 ns. B) Snapshot of R428A RNAP at 3.5 ns. Trigger helices are dark blue. Bridge helix is cyan. Relevant residues are in stick representation. Lower panels are a magnification of the regions boxed in yellow. Yellow dotted lines indicate hydrogen bonding. C) β E445-P’ H1242 hydrogen bonding for wt (black) and R428A (red) RNAP. D) β N556-β’ H1242 hydrogen bonding for wt (black) and R428A (red) RNAP. For R428A RNAP, latch contacts do not form. E)βT1234-β’ S1091 (trigger helix to bridge helix) hydrogen bonding for wt (black) and R428A (red) RNAP.
In panel B, HCl quench data collected at various CTP concentrations was used to compare apparent elongation rates (kobs) for C12 synthesis to CTP concentration and fit to the hyperbolic equation kobs=(kpol[CTP])/(Kdiss+[CTP]), which is analogous to the Michaelis-Menten equation. Kdiss, a measure of CTP affinity, is 72 +/− 13 µM for wt and 71 +/− 12 µM for Rpb2 R512C, so CTP affinity does not appear to be affected for Rpb2 R512C. kpol, the maximal elongation rate, by contrast, is 46.7 +/− 2.0 s−1 for wt and 3.51 +/− 0.16 s−1 for Rpb2 R512C. Clearly, the R512C substitution significantly reduces elongation rate (about 13-fold for C12 synthesis) despite the 20 Å distance of Rpb2 R512 from the active site.
Relative to wt Sc RNAP II, Rpb2 R512C has only a weak capacity for CTP-Mg2+ preloading for C12 synthesis (Fig. 1C). In this comparison, reactions at various CTP, ATP, and GTP concentrations were quenched with EDTA or with HCl at 0.002 s. Because EDTA quench data gives a higher signal than HCl quench data, wt and Rpb2 R512C RNAP II can stably pre-load CTP-Mg2+ (EDTA quench) before forming the C12 bond (HCl quench). Wt RNAP, however, has a much higher capacity for stable CTP-Mg2+ pre-loading than Rpb2 R512C (compare blue and yellow bars). Because the Kdiss for CTP binding is about 70 µM (Fig. 1B), it is a surprise that NTP concentrations from 100 to 5000 µM so strongly increase the extent of CTP-Mg2+ commitment (Fig. 1C). This interesting result may be attributable to simultaneous templated binding of multiple NTPs [7, 48–52] and/or to allosteric effects of NTPs [53, 54].
3.2 Molecular dynamics simulations
To gain an atomistic understanding of the defects of Rpb2 R512C in elongation, we applied molecular dynamics simulation using a homologous bacterial RNAP system. Unrestrained and fully atomistic simulation provides a model for hydration, functional contacts (such as hydrogen bonding and ionic interactions) and conformation [40, 55, 56]. The starting point for molecular dynamics simulation of the Tt RNAP TEC (PDB 205J) was a structure with a closed trigger helices conformation and bound ATP, replacing the ATP analogue AMPcPP used to block AMP incorporation during crystal structure determination. As simulation of the TEC proceeds, crystal packing forces are relieved, simulated temperatures increase, and explicit waters and ions migrate to available sites. Based on the defects of the Sc Rpb2 R512C/A substitutions (Fig. 1; Table I), simulations were compared for Tt wt and β R428A RNAP TECs. In biochemical experiments, similar defects in transcription are observed with Sc Rpb2 R512C and R512A substitutions (Table I) [27], so the alanine substitution was selected for simulation based on the concept that the greater potential chemical difference of β R428A rather than R428C compared to wt might produce a clearer result. Comparison of wt and R428A RNAP provides insight into mutant protein defects, but much more significantly, R428A RNAP acts as a powerful reference to compare to the wt RNAP simulation, providing insight into active site closing, dehydration and catalysis. Without such a reference, because of its complexity, the wt RNAP simulation would be difficult to interpret.
Table I.
Summary of mutations (in bold). MJ25R (microcin J25 resistance); STLR (streptolydigin resistance).
| Sc RNAP II | Tt RNAP | Ec RNAP | Activity | References |
|---|---|---|---|---|
| Rpb1 G820 | β’ A1077 | β’ A779G/T/P | MJ25R | [70] |
| Rpb1 R821 | β’ R1078 | β’ R780H/C | Rapid elongation rate, altered termination efficiency/ MJ25R |
[71]/[70] |
| Rpb1 G823 | β’ G1080 | β’ G782A | MJ25R | [70] |
|
Rpb1 L1081I/M/T/R/V |
β’ M1238 | β’ M932A | Slow elongation | [58, 59, 76] |
|
Rpb1 F1084Q/A/S/H/I |
β’ F1241 | β’ F935A | Slow elongation | [58, 59, 76] |
|
Rpb1 H1085Y/F/A |
β’ H1242 | β’ H936A/Y | Slow elongation, Sc H1085F/A lethal |
[58, 59, 76] |
| Rpb2 R504A | β R420 | β R540 | Little effect | [27] |
| Rpb2 D505A | β E421 | β E541 | Little effect | [27] |
| Rpb2 K507 | β A423 | β A543P/V/T | STLR | [77] |
| Rpb2 L508 | β G424 | β G544D/R | MJ25R, STLR | [70, 77] |
| Rpb2 A509 | β F425 | β F545S/C/I/L | MJ25R, slow elongation, STLR |
[27, 67, 70] |
| Rpb2 K510A | β D426 | β E546V | Little effect/STLR | [27, 67] |
| Rpb2 R512A/C | β R428 | β R548Q | Slow elongation rate/ MJ25R |
[27], this paper/[70] |
| Rpb2 Q513A | β D429 | β D549H | Little effect/ MJ25R | [27]/[70] |
| Rpb2 L514 | β V430 | β V550E | Mimic ppGpp effect | [78] |
| Rpb2 H515 | β H431 | β H551P/Y | Mimic ppGpp effect, reduced termination |
[72, 78] |
| Rpb2 P524 | β P440 | β P560L/S | Slightly reduced elongation rate, reduced termination |
[72–74] |
| Rpb2 T527 | β T443 | β T563P | Mimic ppGpp effect | [75, 78] |
| Rpb2 P528 | β P444 | β P564S | Suppressor G566D ts |
[74] |
|
Rpb2 E529A/Q/D |
β E445 | β D565 | Sc E529A/D rapid in elongation, E529Q slow in elongation |
[27] |
| Rpb2 G530 | β G446 | β G566D/K | Slow elongation rate, attenuation and termination defects |
[74] |
| Rpb2 Q531 | β A447 | β P567S | Reduced termination | [72] |
| Rpb2 G534 | β G450 | β G570C | STLR | [67] |
| Rpb2 L535 | β L451 | β L571Q/R/P | Mimic ppGpp effect | [75, 78] |
| Rpb2 V536 | β I452 | β I572S | Mimic ppGpp effect | [78] |
| Rpb2 R766Q/A | β R557 | β R678C | Lethal in Sc, Minor defect in Ec |
This paper, [54] |
Wt Tt RNAP was very stable during long term simulation (9.5 ns), but β R428A RNAP showed active site instability during early stages of simulation (~500 ps). To ensure that apparent instability of β R428A RNAP was not a consequence of any particular simulation start-up condition, the results with R428A were reproduced multiple times with different protocols. For presentation, we selected a simulation with a 1 ns start up and a >3.8 ns production run, but results described below were typical of each start up and simulation. Because critical residues and contacts are conserved between Tt RNAP and Sc RNAP II, Tt RNAP R428A appears to be an appropriate model for the Sc RNAP II Rpb2 R512C/A substitutions (Fig. 2), and using a heterologous but homologous system for simulation potentially enhances the generality of conclusions based on mutation of an invariant residue.
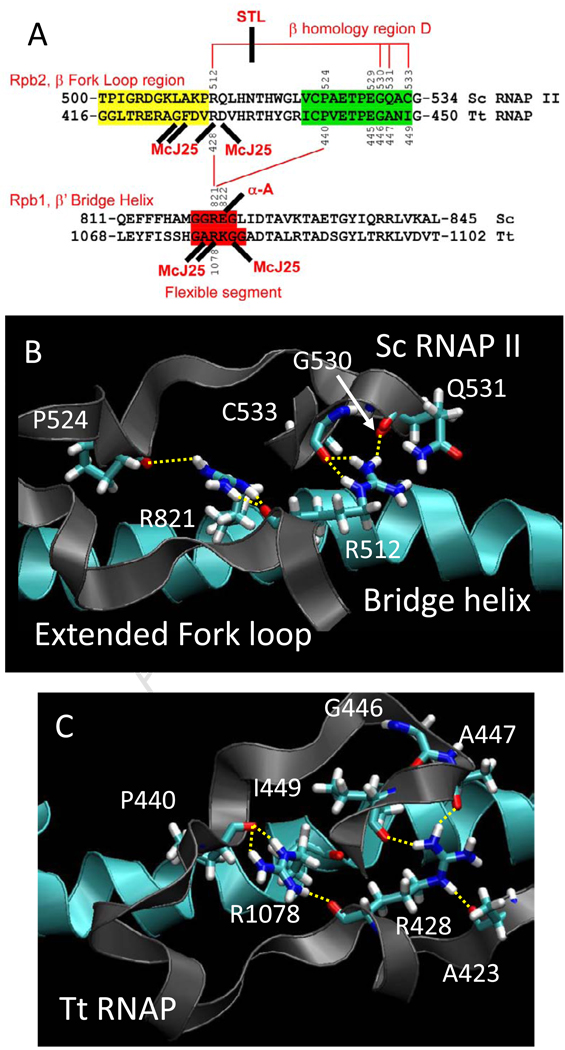
Fig. 2.
Conformational coupling between the bridge α-helix and the extended fork loop region. A) Summary of sequences, proposed interactions and interactions with inhibitors. Fork loop 2 sequence is shaded yellow, a relevant portion of β homology region D is shaded green, and a proposed flexible region of the bridge α-helix (from simulation) is shaded red. Hydrogen bonds are indicated with red lines. Streptolydigin (STL) disrupts a hydrogen bonding network in Tt RNAP. α-amanitin, an RNAP II inhibitor, and Microcin J25 (McJ25), a bacterial RNAP inhibitor, interact at similar positions (sites of some McJ25-resistant mutations are indicated (see Table SI and Fig. 8)). B) A snapshot of Sc RNAP II (PDB 2E2H) at 2 ns. C) A snapshot of wt Tt RNAP at 7 ns. The image shows the bridge helix (blue) and its β’ R1078 residue making backbone hydrogen bonding contact with the extended fork loop region (gray) through terminal NH nitrogens to the backbone oxygens of β P440 and β R428. The terminal NH nitrogen of R428 makes hydrogen bonding contact with the backbone oxygen of β A447 and β I449 while the R428 NE interacts with the backbone oxygen of β A423.
3.3 Bridge α-helix dynamics
Bridge α-helix bending or unwinding against the DNA template within the RNA/DNA hybrid has been proposed to provide the thermal ratchet driving force supporting translocation [5, 8, 57]. Because the trigger helices surround the bridge helix, dynamics of the bridge helix associated with translocation are expected to be restrained in the closed TEC [7, 9]. Interestingly, however, during very early stages of simulation of the catalytic Tt RNAP TEC, the bridge helix bends sharply without disrupting active site geometry. Because bridge helix bending occurs in the catalytic TEC, this observation suggests a role for bridge helix bending and dynamics in catalysis in addition to the proposed role in translocation (Fig. 3). Furthermore, β’ R1078 projects from a dynamic segment of the bridge α-helix to contact β P440 and R428 on the extended fork, linking bridge helix bending and extended fork movement (Fig. 2). By inspection, Tt β’ 1076-GARKGG-1081, which includes β’ R1078 surrounded by 3 glycines, might be expected to be a dynamic feature of the bridge α-helix due to bending and rotation at glycine (Fig. 3). Therefore, defining the β’ 1080-GG-1081 segment as the fulcrum, the bend angle of the bridge helix was measured for wt and β R428A RNAP (Fig. 3B). Upon rapid stabilization (~250 ps simulation), the bend angle changes to about 143 to 157 degrees for wt RNAP (black line) and from about 154 to 170 degrees for β R428A RNAP (red line). During simulation of the catalytic structure, therefore, the β’ 1080-GG-1081 segment is near to the center of a bridge helix pivot that bends more sharply for wt than for β R428A RNAP.
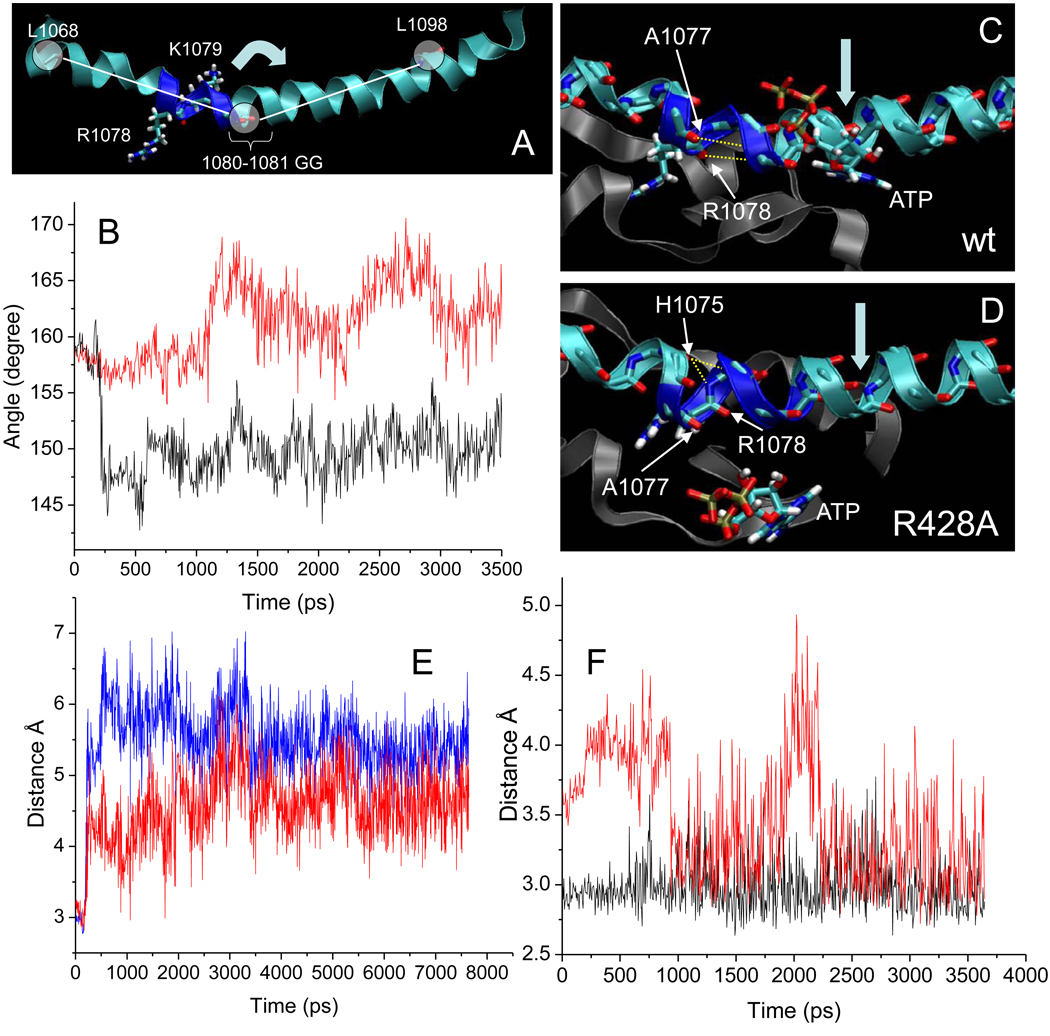
Fig. 3.
Bridge α-helix bending and dynamics at 1076-GARKGG-1081 are altered in β R428A RNAP. A) A bend in the bridge α-helix centered at 1080-GG-1081. β’ 1078-RKGG-1081 residues are dark blue and stick. The center circle represents the ‘pivot’ (see arrow) (center of mass of backbone atoms of 1080-GG-1081) around which the angles between the lines (with center of mass of backbone atoms of L1068 and L1098 shown as circles) were measured. B) The bend angle (as defined in panel A) as a function of simulation time for wt (black) and R428A (red) RNAP. C) In wt RNAP (8 ns), a sharp bend develops in the bridge α-helix. Canonical backbone hydrogen bonds of A1077 to G1081 and R1078 to A1082 break to form the bend. D) In R428A RNAP (3.5 ns), the bridge α-helix bends and bulges. In R428A, hydrogen bonds of A1077 to G1081 and R1078 to A1082 break, and H1075 backbone oxygen makes hydrogen bonding contact with both the G1080 (red) and the K1079 (black) backbone nitrogen. E) In wt RNAP, A1077 to G1081 (red) and R1078 to A1082 (blue) distances are shown as a function of simulation time. F) In R428A RNAP, H1075 to G1080 (red) and H1075 to K1079 (black) distances are shown.
In addition to different bending angles (Fig. 3B), the bends observed in the bridge helix of wt and β R428A RNAP form through somewhat different backbone hydrogen bonding contacts (Fig. 3C–D). In wt RNAP, a bend develops because two hydrogen bonds are broken that are necessary to maintain a canonical α-helical structure. Specifically, as the bend develops early in the simulation, the A1077 O-G1081 N and the R1078 O-1082 N hydrogen bonds, typical of a α-helix, are disrupted (Fig. 3C and E). In the R428A substitution, by contrast to wt RNAP, a bulge forms in the bridge helix within the same region (Fig. 3D and F). As in wt, the A1077 O-G1081 N and the R1078 O-1082 N hydrogen bonds are broken in R428A RNAP. The bulge is stabilized by a bifurcated hydrogen bond between the H1075 O and K1079 N (black line) and the H1075 O and G1080 N (red line) (Fig. 3F). The H1075 O to G1080 N contact is not found in canonical α-helix hydrogen bonding and creates the bulge. It appears therefore that movement in the extended fork region (β 428 to 449) caused by breaking the β R428-A447 and β R428-I449 hydrogen bonds via the β R428➔A substitution alters details of bridge α-helix bending and dynamics, presumably because of strain placed on the hydrogen bond between β’ R1078 and β 428A.
3.4 Conformational coupling between the bridge helix, the extended fork region, the active site and the trigger helix
Because of the removal of the arginine head group, the Tt β R428A substitution disrupts a chain of hydrogen bonds linking β P440-β’ R1078-β R428-β A447 and β R428-β I449 (Fig. 2C). Another hydrogen bond links the β R428 NE to the β A423 main chain oxygen (Fig. 2C). With subtle differences, this network of hydrogen bonds is conserved in Sc RNAP II (compare Fig. 2B–C). β’ R1078 is located on the bridge α-helix within a potentially flexible segment (β’ 1076-GARKGG-1081; Fig. 3). Because β’ R1078 is surrounded on the bridge helix by three glycines, and because flexibility and rotation at glycine disrupts α-helices, β’ R1078 might be expected to be involved in dynamic processes, as is strongly indicated from the simulation (Fig. 3B). During the wt RNAP simulation, the five hydrogen bonds described above and shown in Fig. 2C are stably maintained. Substitution of R428➔A abolishes β R428-β A447, β R428-β I449, and β R428-β A423 hydrogen bonds, but additional consequences result. Specifically, as noted above, the orientation of β’ R1078 on the bridge helix is altered, disruptingβR1078 hydrogen bonding and bridge helix conformation and dynamics (Fig. 3).
3.5 Changes in the β R428A active site
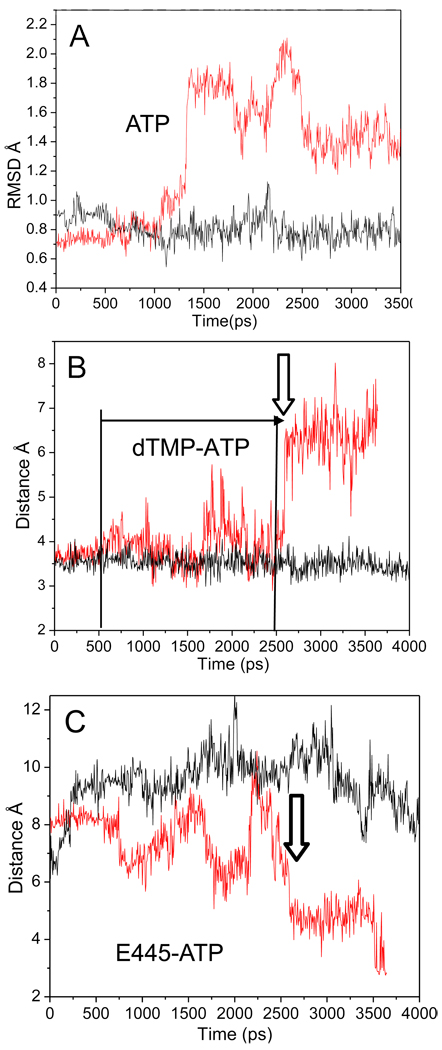
Because Sc Rpb2 R512C is so slow in elongation (Fig. 1), Tt RNAP β R428A might be expected to develop distortions near the active site during simulation (Fig. 4). Fig. 4A shows the root mean square deviation (RMSD) of the i+1 ATP substrate; RMSD is a measure of ATP atom position deviation from the initial crystal structure. As predicted, significant changes in the RMSD are observed in R428A (red line) but not in wt RNAP (black line). The data shown (Fig. 4A–C) for wt RNAP are representative of the entire 9.5 ns simulation, so changes reproducibly observed during the first 0.5 to 3 ns of simulation in the β R428A substitution likely reflect differences of the mutant protein. In Fig. 4B, the R428A substitution is shown to have instability in the i+1 dTMP-ATP base pair, as indicated by fluctuations in the dTMP O4-ATP N6 base pairing distance in R428A (red line) compared to wt (black line) during the 0.5 to 2.6 ns interval. At ~2.6 ns, the distance stretches to more than 6 Å, so the base pair has dissociated and does not re-form during the simulation. In R428A RNAP, the RMSD of ATP at ~ 1.3 ns (Fig. 4A) shows variation from its initial structure before the irreversible loss of dTMP-ATP base pairing, illustrating that alterations in ATP conformation begin prior to dissociation from template (Fig. 4B; vertical arrow).
Fig. 4.
Destabilization of interactions within the R428A RNAP active site. A) RMSD of the ATP molecule of wt (black) and R428A (red). At ~1275 ps, the RMSD of the ATP substrate begins to vary in R428A from its initial structure, while wt RNAP remains stable. B) The distance between the NH2 of ATP to the carboxylate oxygen of TMP (base pair) for wt (black) and R428A (red). dTMP-ATP hydrogen bonding is observed for R428A at the time of the increase in the ATP RMSD of (~1275 ps; panel A), but base pairing becomes erratic (panel B; horizontal arrow; from ~500 ps; distance >4 Å). dTMP-ATP base pairing is lost at ~2600 ps (vertical arrow). C) The β E445 (OE1) to ATP (N6) distance of wt (black) and R428A (red). The vertical arrow indicates the time at which ATP loses hydrogen bonding with dTMP in R428A (panel B) and more closely approaches E445 (panel C).
Another indicator of changes in the active site is the distance of β E445 (proximal to β A447; Fig. 2) to ATP (Fig. 4C). In R428A, the distance between β E445 and ATP decreases to about 3 Å. In wt RNAP, by contrast, the distance does not fall below 6 Å during the entire 9.5 ns simulation. Although many changes in the active site occur in β R428A, the movement of β E445 indicates that at 1.7 ns E445 (carboxylate oxygens) with bound water approaches the dTMP-ATP base pair (Fig. 4C). Simultaneously, there is an increase in fluctuation for the dTMP O4-ATP N6 base pair distance (Fig. 4B). Highly conserved Tt RNAP β E445 corresponds to Sc RNAP II Rpb2 E529 (Fig. 2A; Table I). Interestingly, Rpb2 E529A/D substitutions are faster than wt in elongation. By contrast, the Rpb2 E529Q substitution is slightly slower than wt [27] (Table I). Both functional assays and simulations, therefore, indicate the importance of conserved Tt β E445 (Sc Rpb2 E529), and we propose that this conserved residue assists in regulation of active site hydration as part of a “latch” structure described below.
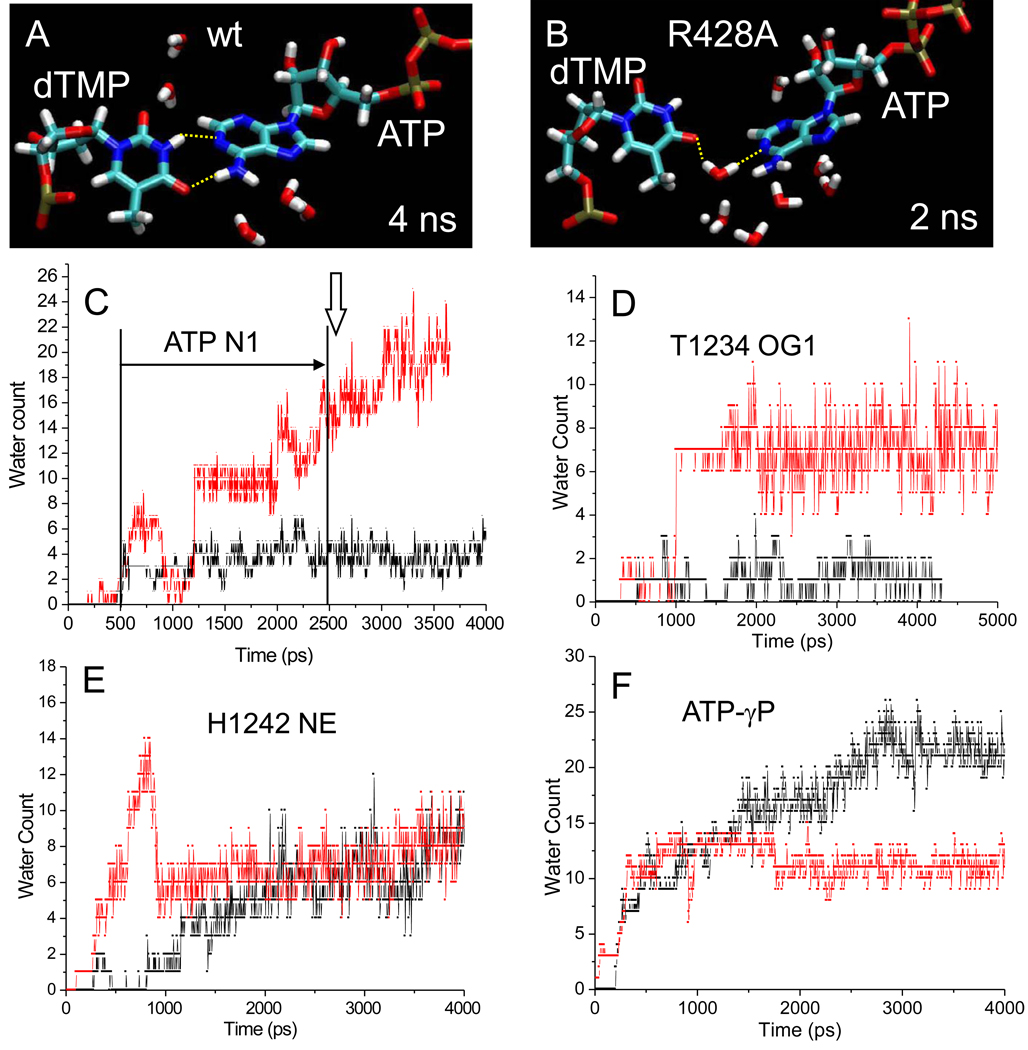
3.6 The “latch” regulates hydration of i+1
In Fig. 5, representative snapshots are shown of the active site for wt and β R428A RNAP TECs. During simulation of wt RNAP, critical trigger helix residueβ’ H1242 [58, 59] assumes consistent contacts with β E445, β N556, and β R557 (Fig. 5). We refer to this network as a “latch” because it appears to form a fully closed and dehydrated catalytic structure for wt RNAP. By contrast, latch contacts are not observed to form for β R428A RNAP. Because mutagenesis of conserved Sc Rpb2 E529 (corresponding to Tt β E445) indicates a regulatory role of this residue (both fast and slow mutants (Table I)), structure-function studies show strong correspondence with simulations. Furthermore, Sc Rpb2 R766Q/A (corresponding to Tt β R557) substitutions are lethal, consistent with an important role for this invariant latch residue.
3.7 Regulated and localized dehydration of the active site
An advantage of molecular dynamics over biochemical and crystallographic approaches is that simulation predicts the placement of water. X-ray crystallography fails to identify water molecules that are not resolved or are removed through dehydration during crystallization. In Fig. 6, snapshots of wt and R428A RNAP are compared. At 4 ns, as expected, wt has a planar dTMP-ATP base pair with two canonical hydrogen bonding contacts (Fig. 6A). By contrast, at 2 ns simulation of R428A RNAP, a water molecule has inserted into the dTMP-ATP base pair, breaking both hydrogen bonds. This snapshot of a disrupted base pair with inserted water in R428A RNAP (Fig. 6B) is characteristic of the reversible instability of the dTMP-ATP base pair observed between 0.5 and 2.6 ns of simulation (Fig. 4B; distance >4 Å; horizontal arrow).
Fig. 6.
Localized flooding of the i+1 dNMP-NTP base pair in R428A RNAP. A) wt RNAP snapshot at 4 ns. B) R428A RNAP snapshot at 2 ns. The dTMP-ATP base pair is shown. In panel B a water molecule has inserted between dTMP and ATP, breaking both canonical T A base-pair hydrogen bonds. C-F) Hydration of 6 Å radius spheres surrounding various active site atoms in wt (black) and R428A (red) RNAP. C) Hydration surrounding ATP N1, participating in dTMP-ATP base pairing. In R428A RNAP water-mediated interruption of base pairing occurs off and on from ~500 ps to ~2500 ps, as indicated by the horizontal arrow (distance >4 Å). The vertical arrow indicates the time of apparently irreversible base pair breaking in R428A. D) Hydration surrounding T1234 OG1, involved in hydrogen bonding to S1091 in wt RNAP. E) Hydration surrounding H1242 NE1, involved in the latch. F) Hydration surrounding the ATP-γ phosphorous.
Because water insertion into the active site dTMP-ATP base pair is not observed during the 9.5 ns simulation of wt RNAP, the hypothesis was considered that R428A RNAP was defective because of excessive active site hydration. Therefore, hydration was compared for wt and R428A RNAP (Fig. 6C–F). Spheres of radius 6 Å were defined around 4 atoms: 1) ATP N1 (dTMP-ATP base pairing); 2)β’ T1234 OG1 (hydrogen bonding to β’ S1091 in wt RNAP) (Fig. 5A); 3)β’ H1242 NE1 (latch); and 4) ATP-γ phosphorous. The number of water molecules contacting or within each sphere was counted as a function of simulation time.
Most significantly, the dTMP-ATP base pair becomes locally flooded with water during simulation of R428A RNAP but not wt (Fig. 6C). Note that an increase in hydration in R428A RNAP occurs prior to dTMP-ATP base pair instability (0.5 to 2.6 ns) and continues to increase after breaking of the base pair (~2.6 ns). Similarly, in β R428A RNAP there is a significant increase in hydration surrounding β’ T1234 compared to wt RNAP. In wt RNAP, β’ T1234 on the trigger helix forms a hydrogen bond to β’ S1091 on the bridge helix (Fig. 5E). The contact breaks in R428A RNAP, explaining the increased hydration at this site in R428A (Fig. 6D and Fig. 5E).
Increases in hydration in R428A RNAP, however, are local and regulated by the latch. Critical trigger helix residue β’ H1242 assumes different contacts in wt and R428A RNAP, but has very similar surrounding hydration (Fig. 6E), indicating that the R428A RNAP active site may not be globally flooded and that the R428A structure remains closed and substantially dehydrated (Fig. 5–Fig. 6). 5–11 water molecules surrounding β’ H1242 NE1 represents a dehydrated active site, because surface exposed β H102 NE1 is surrounded by an average of 22 waters. Interestingly, because of formation of the latch, the ATP γ-phosphorous resides in a more hydrated environment in wt than in R428A RNAP (Fig. 6F). The difference in hydration around the γ-phosphate is likely due to specific contacts in R428A RNAP between a γ-phosphate oxygen and β E445 and a γ-phosphate oxygen and β R557, residues that lose these contacts from formation of the latch in wt RNAP (Fig. 5). Because β E445 and β R557 can make alternate contacts either proximal or more distal from ATP, these latch residues may assist in regulating active site hydration and dehydration. In wt RNAP, association with β’ H1242 in the latch dehydrates β E445 and β R557.
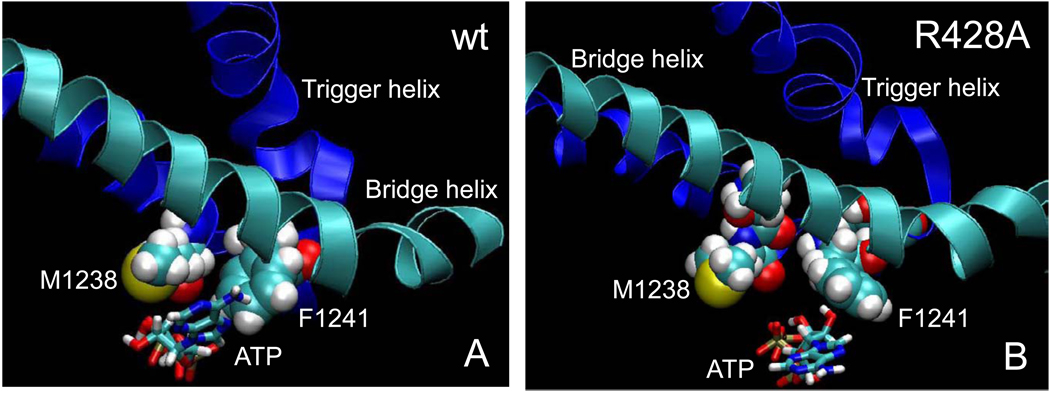
Molecular dynamics simulations also indicate active site dehydration functions of the trigger helices residues β’ M1238 and β’ F1241 (Fig. 7), which have hydrophobic character and which closely approach the i+1 dTMP-ATP base pair [2, 3, 58, 59]. In the β R428A substitution in which the i+1 base pair floods with water, contacts are very different from wt (Fig. 3, Fig. 5 and Fig. 6). Simulation data suggest that excessive hydration localized to the i+1 dNMP-NTP base pair in β R428A may inhibit formation of the fully closed and dehydrated trigger helices conformation by inhibition of formation of the latch, slowing elongation. In summary, formation of the latch structure appears to dehydrate the i+1 dTMP-ATP base pair in wt RNAP. Disruption of the latch in β R428A RNAP appears to result in localized flooding of the i+1 dTMP-ATP base pair with water causing base pair instability. Disruption of the latch and the failure to fully dehydrate the active site in Tt β R428A RNAP appear to explain the transcriptional defect of Sc Rpb2 R512C/A RNAP II (Fig. 1) [27].
Fig. 7.
Hydrophobic trigger helix residues β’ M1238 and F1241 in dehydration of the dNMP-NTP base pair. A) In wt RNAP, M1238 and F1241 closely approach ATP (4 ns). B) In R428A RNAP, M1238 and F1241 are further away, the position and conformation of ATP is altered, the bridge helix is less bent, and the conformation of the trigger helices is changed (3.5 ns).
4. Discussion
4.1 Molecular dynamics simulation and RNAP structure-function
Major current issues in understanding the catalytic mechanism of multi-subunit RNAPs include trigger loop dynamics and function, bridge α-helix dynamics, and long distance conformational coupling. Atomistic molecular dynamics simulation of Tt wt compared to β R428A RNAP provides significant insight into these features of RNAP function. Notably, simulation indicates that trigger loop closing has a fundamental role in dehydration of the RNAP active site. In addition to a previously proposed role in translocation [8, 57], bridge α-helix dynamics and bending in the catalytic TEC appear to contribute to phosphodiester bond formation. Through a chain of hydrogen bonds, movement of the bridge α-helix is coupled to dynamics of the extended fork loop region, ultimately affecting catalytic function, at least partly through regulation of active site hydration.
4.2 Active site closing and dehydration
The major insight that arises from atomistic molecular dynamics simulation of Tt RNAP is that dehydration is likely to be a key feature of induced fit within the buried RNAP active site. We posit moreover that dehydration can drive, and be driven by, active site closing and catalysis. These hypotheses appear generally applicable to multi-subunit RNAPs, single subunit RNAPs (i.e. bacteriophage T7 RNAP) [60], high fidelity DNAPs [61], and many other enzymes with sequestered active sites (e.g. adenylate kinase [62–64], 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase [65]). We posit that induced fit mechanisms for these essential enzymes include a very strong but previously unrecognized dehydration component. Essentially, dehydration appears to shape and align the enclosed RNAP or DNAP active site to support accurate catalysis and to recognize and reject inaccurate substrates.
For Sc RNAP II and Tt RNAP, closing of the trigger helices appears to: 1) dehydrate the active site; 2) form the catalytic structure; and 3) drive chemistry by alignment of reactive groups. Disruption of active site closing and delay of chemistry, therefore, might be accomplished by any change that causes an increase in active site hydration, specifically localized to the i+1 dNMP-NTP base pair, where water can compete for substrate hydrogen bonding. Such an increase in local hydration might occur during pausing, attenuation, termination, and fidelity decisions.
We predict from these studies that binding a dNTP or a non-cognate NTP will, at least in part, be recognized by inhibiting latch formation, increasing hydration of the i+1 position, blocking formation of the catalytic complex, and facilitating replacement of the inaccurately loaded substrate. In this way, hydration of the active site is proposed to be a primary determinant of RNAP fidelity. Substitution of ATP with incorporated AMP and pyrophosphate (the pretranslocated product TEC) may cause an increase in local hydration to signal that the trigger helices should begin to open after completion of the phosphodiester bond.
4.3 Long range conformational coupling
Molecular dynamics simulation indicates tight conformational coupling between a dynamic segment of the bridge α-helix (Tt β’ 1076-GARKGG-1081), the extended fork loop, and the active site, resulting in base-pair dehydration for catalysis, in which the closed trigger helix and the latch participate. In the catalytic structure, bending of the bridge α-helix at β’ 1080-GG-1081 (Fig. 3) re-orients a chain of hydrogen bonds that links β P440-β’ R1078-β R428-β A447 and β R428-β I449 (Fig. 2), resulting in an alteration in the position of nearby β E445 in the active site. β E445 participates with β N556 and R557 and key trigger helix residue β’ H1242 to form the latch that appears to regulate hydration and dehydration of the active site to control fidelity and catalysis. β’ M1238 and F1241 on the trigger helix appear to exclude water from the i+1 dTMP-ATP base pair. The β R428A substitution breaks the hydrogen bonding network, explaining the defects of Tt R428A RNAP in simulation and homologous Sc Rpb2 R512C/A substitutions that have been studied genetically and biochemically (Fig. 1) [27] (Table I). In simulation, the β R428A mutation disables conformational coupling (Fig. 2), changes bridge helix dynamics and conformation (Fig. 3 and Fig. 5), alters active site and trigger helices contacts (Fig. 5), and increases local hydration to destabilize the active site base pair (Fig. 6).
Because of differences in bridge helix sequence, detailed dynamics in the catalytic TEC may be different for Tt RNAP and Sc RNAP II. In this work, we identify a dynamic segment of the bridge helix within Tt RNAP β’ 1076-GARKGG-1081, in which 1080-GG-1081 forms the center of a bend (Fig. 3). In Sc RNAP II, the homologous segment is Rpb1 819-GGREGL-824. Because the glycines surrounding the critical arginine (Sc Rpb1 R821; Tt β’ R1078) are differently distributed, the details of bridge helix bending and dynamics may vary somewhat between these organisms. In both Tt and Sc, however, three of four bridge helix glycines are located within these segments, indicating that this is a dynamic segment of the bridge helix, and the functional connections of Sc Rpb1 R821 and Tt β’ R1078 appear to be very similar.
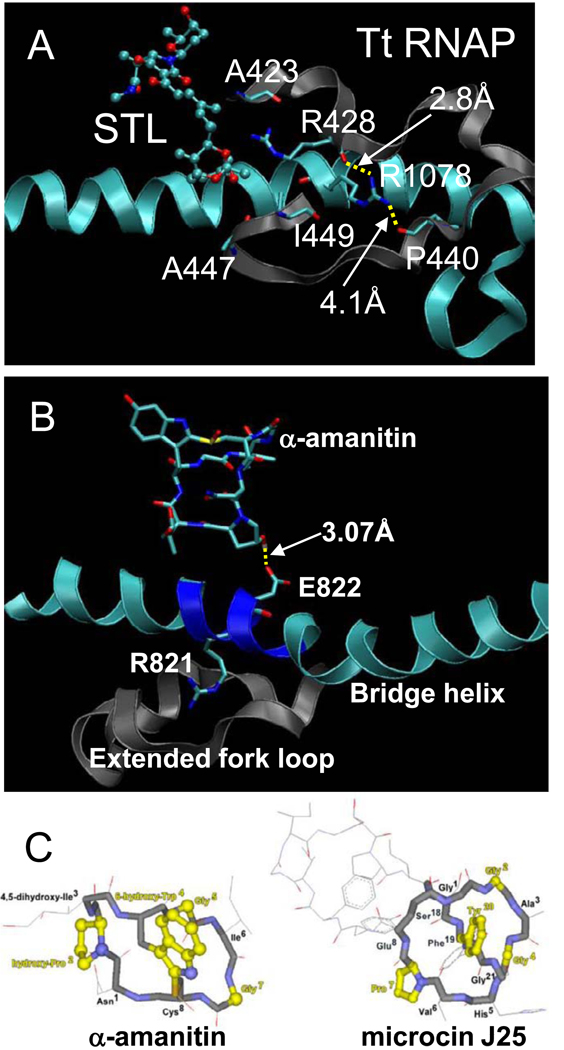
4.5 Inhibitors
Tt RNAP inhibitors streptolydigin and microcin J25 and Sc RNAP II inhibitor α-amanitin interact in the vicinity of the hydrogen bonding network proposed in this work (Fig. 8). Most notably, streptolydigin binding disrupts Tt β R428 hydrogen bonding (Fig. 8A). Specifically, similarly to the R428➔A substitution, streptolydigin disrupts R428-A447 and R428-I449 hydrogen bonds [2, 66, 67]. The mushroom-derived toxin a-amanitin interacts with Rpb1 E822 (Fig. 8B), within 819-GGREG-823, which we propose is a flexible and dynamic segment of the bridge α-helix [1, 68] (Fig. 3). α-amanitin, therefore, is likely to inhibit transcription in part through restraint of bridge helix dynamics. Because the structure of bacterial antibiotic microcin J25 resembles that of a-amanitin (Fig. 8C) and because these inhibitors bind to similar regions of RNAP (Fig. 2), microcin J25 is expected to inhibit transcription in a similar manner to α-amanitin [48, 69]. Microcin J25 and a-amanitin structures include octapeptide rings with similar crossbridges that project hydroxylated, aromatic amino acids (Fig. 8C). Because α-amanitin appears to limit bridge helix dynamics (Fig. 8B), it is likely that microcin J25 may share this mechanism of inhibition [70]. Molecular dynamics simulation, therefore, provides enhanced insight into the mechanisms of potent RNAP inhibitors and drugs.
Fig. 8.
Mechanisms of action of drugs and inhibitors. A) Streptolydigin disrupts R428 hydrogen bonding to A447 and I449. B) The mushroom toxin α-amanitin forms a hydrogen bond to Rpb1 E822 within a flexible segment of the bridge α-helix (819-GGREG-823) (PDB 2VUM). C) Microcin J25, a bacterial antibiotic, is similar in structure to α-amanitin. Residues shown in yellow are similar or identical amino acids at similar positions [69].
4.6 Mutational analyses
Because the bridge helix, extended fork loop, active site and trigger helix are connected and their dynamics are coupled, broader mutational analyses of these regions are summarized in Table I. Simulation indicates the importance of bridge α-helix dynamics in RNAP catalysis (Fig. 3). Consistent with this hypothesis, Ec β’ R780H (Tt β’ R1078; Sc Rpb1 R821) on the bridge α-helix, a residue that hydrogen bonds to Ec β R548 (Tt β R428; Sc Rpb2 R512), is rapid in elongation with altered termination activities [71]. An attempt to reduce the bending and flexibility of Sc Rpb1 819-GGREG-823 by substitution to 819-AAREA-823 resulted in a lethal mutation, indicating the importance of dynamics within this glycine-rich segment.
Consistent with molecular dynamics simulations, residues that are within the conserved Rpb2 R512 (Tt β R428) hydrogen bonding network (Fig. 2) and that function in conformational coupling to the active site appear from mutational analysis to be the most important for transcriptional functions. Residues that are not involved in the network have more minor effects. As examples, Sc RNAP II Rpb2 R504A, D505A, K510A, and Q513A, surrounding R512 but not interacting in the network, have very slight defects in elongation. Escherichia coli (Ec) β P560L/S (Tt β P440; Sc Rpb2 P524) and Ec G566D/K (Tt β G446; Sc Rpb2 G530), which are part of the network, have defects in elongation, attenuation and termination [72–75]. As noted above, latch residues Sc Rpb2 E529 (Tt β E445), Sc Rpb2 R766 (Tt β R557) and Sc Rpb1 H1085 (Tt β’ H1242) and neighboring trigger helix residues β’ M1238 and β’ F1241 are shown to be important for transcription. Structure-function studies, therefore, are in clear correspondence with the molecular dynamics simulations that we report here.
5. Conclusions
Genetics and biochemistry were used to identify Sc Rpb2 R512 as an important invariant amino acid for RNAP II function. Molecular dynamics simulations of Tt wt and β R428A RNAP (the homologous residue) were done to gain an atomistic understanding of the mutation and to gain insight into wt RNAP. This comparison indicates that RNAPs and probably DNAPs with enclosed active sites appear to utilize opening and closing mechanisms in part to dehydrate the i+1 base pair as a fidelity check and to align reactive groups for phosphodiester bond synthesis. Induced fit within a sequestered active site, therefore, appears to have a strong requirement for dehydration. In simulation, increasing local hydration at i+1 is shown to destabilize the active site base pair because nearby water competes with hydrogen bonding. Simulation indicates that hydration/dehydration is sensed and regulated by a latch structure comprised of β E445, N556, R557, and β’ H1242. Within the dynamic segment 1076-GARKGG-1081, simulation strongly indicates bridge helix bending associated with formation of a dehydrated catalytic intermediate. Through this segment, bridge helix dynamics is coupled to movement of the extended fork loop through a system of hydrogen bonds. The extended fork loop reaches to the active site through the β E445 residue, which is part of the latch and which interacts with the key trigger helix residue β’ H1242. Extensive conformational coupling, therefore, links the bridge helix, the extended fork region, the active site and the trigger helix in the catalytic RNAP structure, resulting in regulated and localized active site dehydration.
Acknowledgements
This work was in part supported by the National Institutes of Health Grant GM57461 (to Z.F.B.) and GM39484 (to M.H.) and the National Science Foundation MCB 0447799 (to M.F.). Z.F.B. receives support from Michigan State University, the Michigan State University Agricultural Experiment Station, and the Michigan State University College of Osteopathic Medicine. A.M.N. was in part supported by the National Science Foundation Grant sponsored, Michigan State University, Undergraduate Program in Biological Modeling. S.A.S. was supported by the Gene Expression in Development and Disease Initiative, Michigan State University. This project utilized the Michigan State University High Performance Computing Center. The contents of this publication do not necessarily reveal the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Abbreviations
- TEC
ternary elongation complex
- RNAP
RNA polymerase
- DNAP
DNA polymerase
- wt
wild type
- Tt
Thermus thermophilus
- Sc
Saccharomyces cerevisiae
- Ec
Escherichia coli
- AMPcPP
α,β-methylene adenosine triphosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nature structural & molecular biology. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 2.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 3.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brueckner F, Ortiz J, Cramer P. A movie of the RNA polymerase nucleotide addition cycle. Curr Opin Struct Biol. 2009;19:294–299. doi: 10.1016/j.sbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan CD, Kornberg RD. A bridge to transcription by RNA polymerase. J Biol. 2008;7:39. doi: 10.1186/jbiol99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochimica et biophysica acta. 2010 doi: 10.1016/j.bbagrm.2010.01.007. (in press) [DOI] [PubMed] [Google Scholar]
- 8.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 9.Feig M, Burton ZF. RNA polymerase II flexibility during translocation from normal mode analysis. Proteins. 2010;78:434–446. doi: 10.1002/prot.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helms V, Wade RC. Hydration energy landscape of the active site cavity in cytochrome P450cam. Proteins. 1998;32:381–396. [PubMed] [Google Scholar]
- 11.Okimoto N, Nakamura T, Suenaga A, Futatsugi N, Hirano Y, Yamaguchi I, Ebisuzaki T. Cooperative motions of protein and hydration water molecules: molecular dynamics study of scytalone dehydratase. J Am Chem Soc. 2004;126:13132–13139. doi: 10.1021/ja048053u. [DOI] [PubMed] [Google Scholar]
- 12.Milac AL, Buchete NV, Fritz TA, Hummer G, Tabak LA. Substrate-induced conformational changes and dynamics of UDP-Nacetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase-2. Journal of molecular biology. 2007;373:439–451. doi: 10.1016/j.jmb.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chopra S, Dooling RM, Horner CG, Howell EE. A balancing act between net uptake of water during dihydrofolate binding and net release of water upon NADPH binding in R67 dihydrofolate reductase. The Journal of biological chemistry. 2008;283:4690–4698. doi: 10.1074/jbc.M709443200. [DOI] [PubMed] [Google Scholar]
- 14.Keshwani MM, Harris TK. Kinetic mechanism of fully activated S6K1 protein kinase. The Journal of biological chemistry. 2008;283:11972–11980. doi: 10.1074/jbc.M800114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Schlick T. Quantum mechanics/molecular mechanics investigation of the chemical reaction in Dpo4 reveals water-dependent pathways and requirements for active site reorganization. J Am Chem Soc. 2008;130:13240–13250. doi: 10.1021/ja802215c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, Beard WA, Wilson SH, Broyde S, Schlick T. Polymerase beta simulations suggest that Arg258 rotation is a slow step rather than large subdomain motions per se. Journal of molecular biology. 2002;317:651–671. doi: 10.1006/jmbi.2002.5450. [DOI] [PubMed] [Google Scholar]
- 17.Arora K, Schlick T. In silico evidence for DNA polymerase-beta's substrate-induced conformational change. Biophys J. 2004;87:3088–3099. doi: 10.1529/biophysj.104.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radhakrishnan R, Arora K, Wang Y, Beard WA, Wilson SH, Schlick T. Regulation of DNA repair fidelity by molecular checkpoints: "gates" in DNA polymerase beta's substrate selection. Biochemistry. 2006;45:15142–15156. doi: 10.1021/bi061353z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suenaga A, Okimoto N, Futatsugi N, Hirano Y, Narumi T, Ohno Y, Yanai R, Hirokawa T, Ebisuzaki T, Konagaya A, Taiji M. Structure and dynamics of RNA polymerase II elongation complex. Biochem Biophys Res Commun. 2006;343:90–98. doi: 10.1016/j.bbrc.2006.02.124. [DOI] [PubMed] [Google Scholar]
- 20.Woo HJ, Liu Y, Sousa R. Molecular dynamics studies of the energetics of translocation in model T7 RNA polymerase elongation complexes. Proteins. 2008 doi: 10.1002/prot.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatramani R, Radhakrishnan R. Computational delineation of the catalytic step of a high fidelity DNA polymerase. Protein Sci. 2010 doi: 10.1002/pro.361. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappas DL, Jr, Hampsey M. Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in Saccharomyces cerevisiae. Molecular and cellular biology. 2000;20:8343–8351. doi: 10.1128/mcb.20.22.8343-8351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes-Reyes M, Hampsey M. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Molecular and cellular biology. 2007;27:926–936. doi: 10.1128/MCB.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell. 2002;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 25.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 26.Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell. 2009;34:168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domecq C, Kireeva M, Archambault J, Kashlev M, Coulombe B, Burton ZF. Site-directed mutagenesis, purification and assay of Saccharomyces cerevisiae RNA polymerase II. Protein Expr Purif. 2010;69:83–90. doi: 10.1016/j.pep.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 29.Woontner M, Jaehning JA. Accurate initiation by RNA polymerase II in a whole cell extract from Saccharomyces cerevisiae. The Journal of biological chemistry. 1990;265:8979–8982. [PubMed] [Google Scholar]
- 30.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. The Journal of biological chemistry. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 31.Kireeva ML, Lubkowska L, Komissarova N, Kashlev M. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 2003;370:138–155. doi: 10.1016/S0076-6879(03)70012-3. [DOI] [PubMed] [Google Scholar]
- 32.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient Reversal of RNA Polymerase II Active Site Closing Controls Fidelity of Transcription Elongation. Mol Cell. 2008 doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kireeva M, Nedialkov YA, Gong XQ, Zhang C, Xiong Y, Moon W, Burton ZF, Kashlev M. Millisecond phase kinetic analysis of elongation catalyzed by human, yeast, and Escherichia coli RNA polymerase. Methods. 2009 doi: 10.1016/j.ymeth.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kireeva M, Nedialkov YA, Gong XQ, Zhang C, Xiong Y, Moon W, Burton ZF, Kashlev M. Millisecond phase kinetic analysis of elongation catalyzed by human, yeast, and Escherichia coli RNA polymerase. Methods. 2009;48:333–345. doi: 10.1016/j.ymeth.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 37.Tang L, Liu H. A comparative molecular dynamics study of thermophilic and mesophilic ribonuclease HI enzymes. J Biomol Struct Dyn. 2007;24:379–392. doi: 10.1080/07391102.2007.10507127. [DOI] [PubMed] [Google Scholar]
- 38.Best RB, Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J Phys Chem B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C.C. Group. Molecular Operating Environment. http://www.chemcomp.com/
- 40.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meagher KL, Redman LT, Carlson HA. Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem. 2003;24:1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 42.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics. 1993;98:10089–10092. [Google Scholar]
- 43.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 44.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-Dynamics with Coupling to an External Bath. Journal of Chemical Physics. 1984;81:3684–3690. [Google Scholar]
- 45.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold JJ, Gohara DW, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochemistry. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burton ZF, Feig M, Gong XQ, Zhang C, Nedialkov YA, Xiong Y. NTPdriven translocation and regulation of downstream template opening by multi-subunit RNA polymerases. Biochem Cell Biol. 2005;83:486–496. doi: 10.1139/o05-059. [DOI] [PubMed] [Google Scholar]
- 49.Gong XQ, Zhang C, Feig M, Burton ZF. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol Cell. 2005;18:461–470. doi: 10.1016/j.molcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 50.Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. Journal of molecular biology. 2008;381:1106–1113. doi: 10.1016/j.jmb.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 51.Xiong Y, Burton ZF. A Tunable Ratchet Driving Human RNA Polymerase II Translocation Adjusted by Accurately Templated Nucleoside Triphosphates Loaded at Downstream Sites and by Elongation Factors. The Journal of biological chemistry. 2007;282:36582–36592. doi: 10.1074/jbc.M707014200. [DOI] [PubMed] [Google Scholar]
- 52.Zhang C, Burton ZF. Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. Journal of molecular biology. 2004;342:1085–1099. doi: 10.1016/j.jmb.2004.07.070. [DOI] [PubMed] [Google Scholar]
- 53.Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 54.Holmes SF, Santangelo TJ, Cunningham CK, Roberts JW, Erie DA. Kinetic investigation of Escherichia coli RNA polymerase mutants that influence nucleotide discrimination and transcription fidelity. The Journal of biological chemistry. 2006;281:18677–18683. doi: 10.1074/jbc.M600543200. [DOI] [PubMed] [Google Scholar]
- 55.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L. K. Schulten, Scalable molecular dynamics with NAMD, Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 58.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 60.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 61.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci U S A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cukier RI. Apo adenylate kinase encodes its holo form: a principal component and varimax analysis. J Phys Chem B. 2009;113:1662–1672. doi: 10.1021/jp8053795. [DOI] [PubMed] [Google Scholar]
- 63.Krishnamurthy H, Lou H, Kimple A, Vieille C, Cukier RI. Associative mechanism for phosphoryl transfer: a molecular dynamics simulation of Escherichia coli adenylate kinase complexed with its substrates. Proteins. 2005;58:88–100. doi: 10.1002/prot.20301. [DOI] [PubMed] [Google Scholar]
- 64.Lou H, Cukier RI. Molecular dynamics of apo-adenylate kinase: a distance replica exchange method for the free energy of conformational fluctuations. J Phys Chem B. 2006;110:24121–24137. doi: 10.1021/jp064303c. [DOI] [PubMed] [Google Scholar]
- 65.Su L, Cukier RI. An Enhanced Molecular Dynamics Study of HPPK-ATP Conformation Space Exploration and ATP Binding to HPPK (dagger) J Phys Chem A. 2009 doi: 10.1021/jp808664k. [DOI] [PubMed] [Google Scholar]
- 66.Temiakov D, Zenkin N, Vassylyeva MN, Perederina A, Tahirov TH, Kashkina E, Savkina M, Zorov S, Nikiforov V, Igarashi N, Matsugaki N, Wakatsuki S, Severinov K, Vassylyev DG. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 67.Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD, Jr, Dharia C, Napoli A, Laptenko O, Lee J, Borukhov S, Ebright RH, Arnold E. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci U S A. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gong XQ, Nedialkov YA, Burton ZF. Alpha-amanitin blocks translocation by human RNA polymerase II. The Journal of biological chemistry. 2004;279:27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- 70.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the beta' subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 72.Landick R, Stewart J, Lee DN. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 73.Tavormina PL, Landick R, Gross CA. Isolation, purification, and in vitro characterization of recessive-lethal-mutant RNA polymerases from Escherichia coli. J Bacteriol. 1996;178:5263–5271. doi: 10.1128/jb.178.17.5263-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tavormina PL, Reznikoff WS, Gross CA. Identifying interacting regions in the beta subunit of Escherichia coli RNA polymerase. Journal of molecular biology. 1996;258:213–223. doi: 10.1006/jmbi.1996.0244. [DOI] [PubMed] [Google Scholar]
- 75.Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like "stringent" RNA polymerases in Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nature structural & molecular biology. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heisler LM, Suzuki H, Landick R, Gross CA. Four contiguous amino acids define the target for streptolydigin resistance in the beta subunit of Escherichia coli RNA polymerase. The Journal of biological chemistry. 1993;268:25369–25375. [PubMed] [Google Scholar]
- 78.Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. The EMBO journal. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]