Abstract
Glutathione plays a crucial role in free radical scavenging, oxidative injury, and cellular homeostasis. Previously, we identified a non-synonymous polymorphism (P462S) in the gene encoding the catalytic subunit of glutamate cysteine ligase (GCLC), the rate-limiting enzyme in glutathione biosynthesis. This polymorphism is present only in individuals of African descent. Presently, we report that this ethnic-specific polymorphism (462S) encodes an enzyme with significantly decreased in vitro activity when expressed by either a bacterial or mammalian cell expression system. In addition, overexpression of the 462P wild-type GCLC enzyme results in higher intracellular glutathione concentrations than overexpression of the 462S isoform. We also demonstrate that apoptotically stimulated mammalian cells overexpressing the 462S enzyme have increased caspase activation and increased DNA laddering compared to cells overexpressing the wild-type 462P enzyme. Finally, we genotyped several African and African-descent populations and demonstrate that the 462S polymorphism is in Hardy-Weinberg dysequilibrium, with no individuals homozygous for the 462S polymorphism identified. These findings describe a glutathione production pathway polymorphism present in individuals of African descent with significantly decreased in vitro activity.
INTRODUCTION
Glutathione is an important free radical scavenger which plays a pivotal role in protecting against cellular injury induced by oxidative stressors. As a result, glutathione production is intimately related to maintaining overall cellular health and function. Indeed, modulation of intracellular levels of glutathione can affect cellular susceptibility to apoptotic stimuli.[1, 2] Therefore, maintenance of adequate levels of glutathione via de novo synthesis and recycling is critical in maintaining cellular viability.
De novo synthesis of glutathione occurs in a two-step process. The first step is catalyzed by glutamate cysteine ligase (GCL), which combines glutamate and cysteine to produce γ-glutamylcysteine.[3] This rate-limiting enzyme is a 100kDa heterodimeric enzyme composed of a catalytic subunit (GCLC) and a regulatory, or modifier, subunit (GCLM).[4] Glutathione synthesis is completed by glutathione synthetase, which catalyzes the addition of glycine to γ-glutamylcysteine to produce glutathione.
As the catalyst in the rate-limiting step of de novo glutathione synthesis, GCL is an important regulator of glutathione levels. We propose that genetic polymorphisms in the gene encoding the catalytic subunit of the GCL enzyme can cause defects in glutathione synthesis, thereby predisposing an individual to increased cellular injury in the setting of oxidative stress, and consequently leading to increased disease severity as well as worsened clinical outcome.
Our laboratory has been interested in the model of environmentally determined genetic expression (EDGE). Under this model, genetic polymorphisms may only have clinical phenotypic expression during times of environmental stress.[5] Due to the pivotal role of glutathione in cellular health, we screened the GCLC gene for genetic polymorphisms and discovered an ethnic-specific, non-synonymous polymorphism, P462S (cDNA C1384T, rs17883718). Found only in patients of African descent, this polymorphism changes the highly conserved proline residue 462 to a serine residue. By genotyping both African-Americans and Africans from Ghana, we have previously demonstrated that an allele frequency of 5% in these populations for the 462S allele.[6] In this study, we demonstrate that the 462S isoform of the GCLC enzyme has decreased activity in vitro, and overexpression of this enzyme isoform results in lower intracellular glutathione levels and increased cellular susceptibility to apoptosis as compared to the wild-type enzyme. We also demonstrate the 462S polymorphism is not in Hardy-Weinberg equilibrium with a selection bias against individuals homozygous for the 462S polymorphism.
METHODS
Plasmid Construction
In selecting a cloning strategy for the GCLC proteins, we considered and discarded using a tagged system (such as histidine tagging). The position of the polymorphism under study and the nature of the interaction between the GCLC and GCLM subunits made it likely that this would have a significant effect on enzyme function and artificially skew our observations.[7] Using PCR, we cloned the coding regions of human GCLC and GCLM into the expression vector, pcDNA3.1(+) (Invitrogen, Carlsbad, California). These genes were subcloned into the bacterial expression vectors, pET21a and pET24a (Novagen, San Diego, CA), respectively. Inserts were PCR amplified from the existing clones in pcDNA3.1(+) with the primers adding the appropriate restriction site sequences and extra stop codons onto the 3′ end of the insert. The GCLC forward primer sequence was 5′-AGGCCATACATATGGGGCTGCTGTCC-3′; the GCLC reverse primer sequence was 5′-GCGAATTCTCATTACTAGTTGGATGAGTC-3′. The GCLM forward primer sequence was 5′-AGGCCATACATATGGGCACCGACAGC-3′; the GCLM reverse primer sequence was 5′-GCGAATTCTCACTATTAAGAACCCCTTCT-3′. This cloning strategy removes both the N-terminal and C-terminal peptide tags that are intrinsic to the pET21a and pET24a vectors because it has been previously shown that tags placed on either end of the GCLC protein destroy enzymatic activity.[7] The purified inserts and empty vectors were simultaneously digested with NdeI and EcoRI (New England Biolabs, Ipswich, MA) for >3 hours at 37°C and used for ligation.
The 462S (1384T) version of the polymorphism was created through site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Primers were designed according to the guidelines in the kit's manual, although the calculated Tm for the primers was lower than recommended; this guideline could not be achieved due to the sequence of the area around the nucleotide change. The forward primer sequence was 5′-GGATTTTCTCATTTCACTGTCAAAGGTTGATGAG-3′; the reverse primer sequence was 5′-CTCATCAACCTTTGACAGTGAAATGAGAAAATCC-3′. All steps were carried out according to manufacturer's protocol; 12 cycles were chosen for the PCR step as was recommended for a single point mutation. After the cloning and mutagenesis steps were completed, all vectors were sequenced to confirm that no extraneous changes had been introduced.
Bacterial Expression of GCLC and GCLM
The BL21(DE3) strain of E. coli was used for expression of human GCLC and GCLM. This strain of bacteria was co-transformed with GCLC in pET21a and GCLM in pET24a and was submitted to antibiotic selection of 50 μg/ml carbenicillin and kanamycin selection at a concentration of 30 μg/ml. Successful transformation was confirmed by recovery of plasmid from the bacteria with restriction digest confirmation.
100 ml luria broth containing 50 μg/ml carbenicillin and 30 μg/ml kanamycin was inoculated with colonies from the streaked plates. Cultures were incubated at 37°C for 30 minutes. 10 μl of 1 M isopropyl-b-D-thiogalactopyranoside (IPTG) was added to the cultures, and the cultures were incubated overnight at room temperature with shaking.
Cultures were harvested through centrifugation at 2500g. Pellets were washed with cold phosphate-buffered saline and divided into smaller pellets of roughly 200 mg. These pellets were stored dry at −70°C. The pellets were then lysed in 320 mM sucrose, 10 mM Tris-Cl, pH 7.4, 1 mM EDTA containing bacterial protease inhibitors (Sigma-Aldrich, St. Louis, MO) through sonication on ice. Lysates were centrifuged at 14000g for 15 minutes to remove cellular debris. Supernatants were removed, concentrated using Microcon YM-10 columns (Millipore, Billerica, MA), and stored at −80°C.
Western Blotting
10 μg of protein was added to SDS sample buffer (375 mM Tris, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol) for a final volume of 20 μl and was heated at 95°C for 10 minutes. Protein samples were electrophoresed on 5% SDS polyacrylamide gels and transferred to a nitrocellulose membrane using the tank transfer system. The membrane was blocked with 5% non-fat dried milk in Tris-buffered saline with 0.1% Tween-20. Anti-GCLC antibody (LabVision, Fremont, CA) was added to the blocking buffer for a final concentration of 2.5 μg/ml and was incubated at room temperature for 1 hour. The membrane was washed three times in blocking buffer for 10 minutes per wash and incubated in 1:2000 dilution of horseradish peroxidase conjugated anti-rabbit antibody prepared in blocking buffer for 1 hour at room temperature. The membrane was washed three times in blocking buffer for 10 minutes per wash. The membrane was then incubated in a 1:1 mixture of ECL western blotting reagents (Amersham Pharmacia, Piscataway, NJ) and exposed to x-ray film. To quantitate band intensity, the QuantityOne application (Bio-Rad, Hercules, CA) was used to measure the number of pixels present in each GCLC band. These measurements were subsequently used to calculate a ratio of uncleaved:total GCLC in each sample. Significance was calculated using an unpaired t-test.
GCL Activity Assay and γ-Glutamylcysteine Measurement
Protein concentration was estimated in a microplate format using the BCA Protein Assay kit (Pierce). To determine GCL activity, 40 μg of protein lysate in 10 μl 320 mM sucrose, 10 mM Tris-Cl, 1 mM EDTA at pH 7.4 was added to 90 μl assay buffer (0.1 M Tris-Cl, 0.15 M KCl, 20 mM MgCl2, 2 mM EDTA, pH 8.2, 10 mM ATP, 10 mM L-cysteine, 40 mM L-glutamate). This assay is a modification of that described by Gegg et al.[8] Assays were incubated at 37°C for 90 minutes. Assays were then stopped with the addition of 5-sulfosalicylic acid to a final concentration of 50 mM. Samples were incubated on ice for 15 minutes and centrifuged at 14000g for 15 minutes at 4°C. The supernatant was then collected and stored at −80°C. γ-glutamylcysteine (Bachem) concentration standards were prepared in assay buffer at the same time as the actual samples were prepared to control for minor differences in assay conditions. γ-Glutamylcysteine levels were detected using an assay based on NDA derivitization.[9] 90 γl of each GCLC Activity Assay was added to 180 γl of NDA Derivitization solution (50mM Tris, pH=10, 0.5N NaOH, 10m NDA) in a 96-well plate. Assays were incubated at room temperature for 30 minutes, and samples were analyzed using a fluorescent plate reader with excitation at 485λ and detection at 520λ.
Amino Acid Analysis
To validate our NDA-based fluorescent assay and test our mammalian cell transfections, samples were assayed using a Hitachi L-8800 High Performance Amino Acid Analyzer. Absorbance was measured at 570 nm for primary amino groups and 440 nm for secondary amino groups. Prior to analysis by the Amino Acid Analyzer, 50 μl of the sample was mixed with 2γl of 2M hydrochloric acid to acidify the sample. 20 μl of each sample was injected for amino acid analysis. Alanine concentrations were also measured for use as an internal standard. As determined by γ-glutamylcysteine standards, the retention time of γ-glutamylcysteine was determined to be at approximately 8.5 minutes.
Mammalian Cell Transfection and Induction of Apoptosis
MRC5-SV2 cells, which are derived from fetal human lung tissue, were cultured in antibiotic-free DMEM containing 10% fetal bovine serum. MRC5-SV2 cells were co-transfected using purified GCLC in pcDNA 3.1 and GCLM in pcDNA 3.1 and the FuGene reagent (Roche). Co-transfection of GCLM was performed to ensure GCLM availability would not limit GCL activity. Transfected cells were incubated at 37°C for 48 hours, and the cell culture media was removed and subjected to amino acid analysis as described above. To induce apoptosis, staurosporine (Sigma-Aldrich) was added directly to the cell media to a final concentration of 2 γM. The cells were incubated at 37°C for 2 hours and harvested using trypsinization. The cells were then pelleted by centrifugation at 2000g for 5 minutes. The cell media was removed, and cells were rinsed in phosphate-buffered saline and re-pelleted. Each cell pellet was resuspended in 200 γl of mammalian cell lysis buffer (50mM Tris pH 7.8, 1mM EDTA, 150mM NaCl, 1% Nonidet-P40) supplemented with a mammalian protease inhibitor mixture (Sigma-Aldrich). Cells were incubated on ice for 60 minutes and centrifuged at 14000g for 5 minutes at 4°C. The supernatant was transferred to a fresh tube and stored at −80°C for Western blot analysis and glutathione measurement.
Glutathione Measurement
Protein content of each MRC5-SV2 cell lysate was quantitated using the BCA protein quantitation assay and these protein levels were used for normalization of measured glutathione levels. Cell lysates were then protein-precipitated by the addition of 5-sulfosalicylic acid to a final concentration of 75mM. Samples were incubated on ice for 30 minutes followed by centrifugation at 14000g for 15 minutes at 4°C. The supernatant was transferred to a fresh tube and diluted 1:200 with working buffer (100mM NaPO4, 1mM EDTA, pH 7.5). Samples were then used in triplicate and assayed by incubating with equal volumes of reagent buffer (67mM NaPO4, 670μM EDTA, 80μg/mL 5,5′ – Dithiobis(2-nitrobenzoic acid), 0.3mg/mL NADPH, 1.5U/mL glutathione reductase) for 6 minutes at room temperature. All reagents were obtained from Sigma-Aldrich. Absorbance was measured at 425λ using a spectrophotometer and concentrations were calculated using a standard calibration curve.
DNA Fragmentation Analysis
Harvested cell pellets were resuspended in 500 γl DNA lysis buffer (20mM Tris pH 7.4, 5mM EDTA, 0.4% Triton X-100). Cells were incubated on ice for 1 hour and then pelleted. The supernatant was transferred to a fresh tube and extracted twice with phenol:chloroform. DNA precipitation was performed by the addition of 5M NaCl and 500 γl isopropanol. Samples were incubated at −80°C overnight and centrifuged at 14000g for 15 minutes at 4°C. Isopropanol was removed, DNA pellets were rinsed with 70% ethanol, and the pellets were resuspended in DNA loading buffer and electrophoresed on 1% agarose.
Genotyping of Kenyan Population
The Kenyans were selected from the Luo, Kikuyu and Masai ethnic groups. The population consisted of 634 randomly selected volunteers from secondary schools and 396 young children diagnosed with malaria as defined by the World Health Organization criteria. Presence of the 462S genetic polymorphism was determined using iPLEX genotyping (Sequenom). This technology is based upon PCR amplification followed by sequence-specific primer extension. Allele detection is then determined by mass difference between products.
Genotyping of Carolina African-Americans
Samples from Carolina African-Americans was provided by the Carolina Breast Cancer Study. This population-based, case-control study was conducted in 24 counties of central and eastern North Carolina, and consisted of 658 African-American women with primary invasive or in situ breast cancer. 602 African-American controls were selected from Division of Motor Vehicles and U.S. Health Care Financing Administration lists. Samples from this population were genotyped using a LightScanner™ instrument (Idaho Technology). This instrument relies on PCR amplification followed by high-melting resolution.
Genotyping of South African Population
Samples from 234 South Africans were genotyped using the Applied Biosystems allelic discrimination assay. These samples were obtained as part of a colon cancer study.
Genotyping of Ghanan Population
Banked DNA from 166 samples from Ghana were genotyped using PCR amplification followed by Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit with [α-33P] dideoxynucleotides (USB Corporation, Cleveland, OH, USA). The three genotyping methods have been cross validated against each other using control samples and sequence confirmation. All research proposals were reviewed and approved by the appropriate institutional review board and informed consent was obtained for each patient sample. All portions of this study were reviewed and approved by the appropriate university and government ethical review boards.
Statistical Methods
GCLC activity measurements were calculated from multiple replications of each experiment. Each series was measured in triplicate, and the average result of each series was used to produce the final mean value. Due to endogenous bacterial and eukaryotic enzymatic activity, values are represented as a percent change from baseline to normalize values. Paired t-test was used to compare the differences between groups. For the genotyping data, all allele frequencies are presented as a proportion of the population. Statistical analysis of Hardy-Weinberg equilibrium was calculated using the Pearson χ2 method with 1 degree of freedom.
RESULTS
Expression Studies of P462S Polymorphism
Prokaryotic Studies
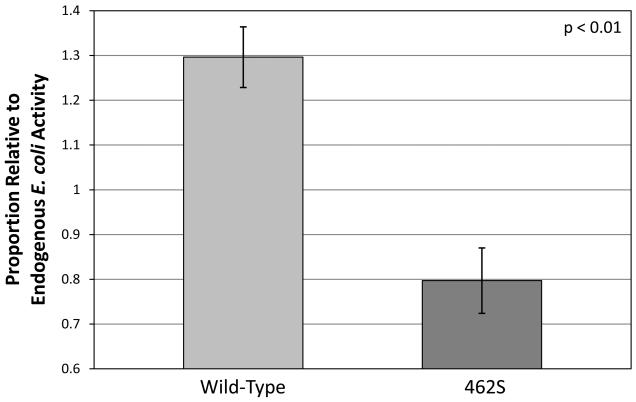
To compare the rates of glutathione biosynthesis in vitro, bacterial plasmids were created to produce recombinant human versions of either the 462P wild-type or the 462S variant of the GCLC enzyme. These plasmids were transformed into E. coli, and protein expression was induced. The bacterial lysates were collected and used to measure GCL activity by quantifying the amount of γ-glutamylcysteine produced in an in vitro fluorescence-based assay. γ-Glutamylcysteine levels were then measured in relation to endogenous production using a fluorescence-based assay. We determined that overexpression of the wild-type enzyme resulted in 1.30 ± 0.07 more γ-glutamylcysteine production as compared to endogenous E. coli lysate. In contrast, overexpression of the 462S-encoded enzyme produced 0.80 ± 0.07 less γ-GC as compared to endogenous E. coli lysate. These results demonstrate that, in a bacterial expression system, the 462S GCLC enzyme has significantly decreased in vitro activity as compared to the 462P wild-type (p<0.01) (Figure 1).
Figure 1. Bacterially-expressed GCLC 462S enzyme has significantly decreased enzymatic activity in vitro.
Plasmids encoding either the 462P wild-type or 462S GCLC polymorphism were used in an E. coli expression system. Bacterial lysates were used to produce γ-glutamylcysteine, which was measured using a fluorescent-based assay. Bar graphs indicate proportion of activity relative to an equivalent amount of endogenous E. coli lysate. Graph values represent the mean ± SEM over several experiments. P-values are based on the difference between the two polymorphisms and were calculated using a paired t-test.
Eukaryotic Studies
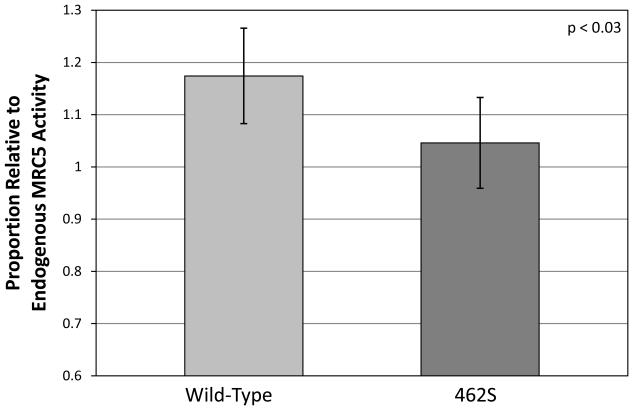
To further study the enzymatic activity of the 462S variant in vitro, we expressed either the recombinant human wild-type (462P) GCLC enzyme or the 462S variant in mammalian cells by transiently transfecting MRC5-SV2 cells (which have two normal copies of the gene). After allowing protein expression for 48 hours, the cell media was collected, and levels of γ-glutamylcysteine were measured using an ion-exchange chromatography-based amino acid analyzer. Mammalian cells expressing the wild-type enzyme produced 1.17 ± 0.09 more γ-glutamylcysteine as compared to empty cassette-transfected negative control samples, whereas cells expressing the 462S variant only produced 1.05 ± 0.09 more γ-glutamylcysteine than control samples. These results demonstrate that the 462S GCLC enzyme produces significantly less glutathione precursors than a wild-type enzyme in a eukaryotic overexpression system (p<0.03) (Figure 2).
Figure 2. Mammalian cell-expressed GCLC 462S enzyme has significantly decreased enzymatic activity in vitro.
Plasmids encoding either the 462P wild-type or 462S GCLC polymorphism were transiently transfected into MRC5-SV2 cells and incubated for 48 hours. γ-Glutamylcysteine levels in the cell media were measured using an amino acid analyzer. Bar graphs indicate proportion of γ-GC production relative to control MRC5 lysate. Graph values represent the mean ± SEM over several experiments. P-values are based on the difference between the two polymorphisms and were calculated using a paired t-test.
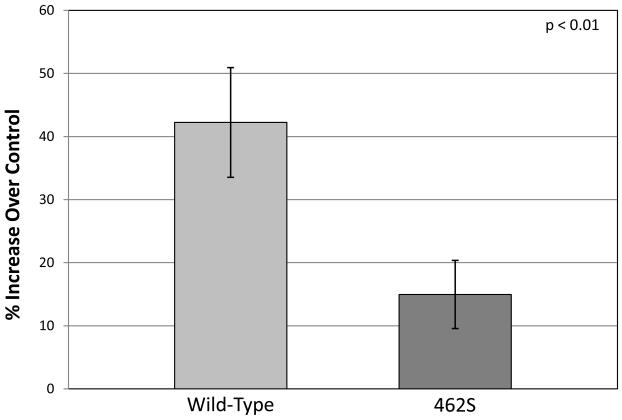
Since the 462S GCLC enzyme has less in vitro activity and produces significantly less glutathione precursors as compared to the 462P wild-type enzyme, we hypothesized that mammalian cells that overexpress the 462S isoform would have lower intracellular glutathione levels than cells overexpressing the wild-type enzyme. We transiently transfected MRC5-SV2 cells with either the 462S or wild-type GCLC enzyme and lysed these cells after allowing 48 hours of protein expression. We measured total intracellular glutathione levels in these cell lysates and determined that cells overexpressing the wild-type 462P isoform have a 1.42 ± 0.09 higher intracellular concentration of glutathione as compared to mock-transfected cells. In contrast, cells overexpressing the 462S isoform only have a 1.15 ±d 0.05 increase in intracellular glutathione (Figure 3). This significant difference (p < 0.01) indicates that overexpression of the wild-type 462P GCLC enzyme results in significantly higher intracellular glutathione levels as compared to overexpression of the 462S isoform.
Figure 3. Mammalian cells overexpressing the 462S GCLC enzyme have significantly lower intracellular glutathione levels compared to cells overexpressing the wild-type enzyme.
Plasmids encoding either the wild-type or 462S GCLC polymorphism were transiently transfected into MRC5-SV2 cells and incubated for 48 hours. Total glutathione levels were measured in the cell lysate. Bar graphs indicate percent increase in glutathione levels relative to mock-transfected MRC5 lysate. Graph values represent the mean ± SEM over several experiments. P-values were calculated using a paired t-test.
Eukaryotic Apoptosis Studies
Recent data has suggested that a reduction in glutathione levels is an early and important event in apoptosis.[2, 10, 11] Mitochondrial dysfunction, resulting in the release of free radicals into the cytoplasm, occurs during apoptosis, and free radical scavengers are subsequently consumed. Indeed, depletion of intracellular glutathione followed by the addition of low levels of oxidants are sufficient to induce apoptosis in several types of mammalian cells. In addition, increasing levels of intracellular glutathione can rescue cells from apoptosis. Similarly, studies have demonstrated that overexpression of GCLC is sufficient to rescue cells from apoptosis. Furthermore, the GCLC protein is a known target of caspase-mediated cleavage during apoptosis, suggesting that inhibition of GCLC activity may be an integral part of apoptosis.[12]
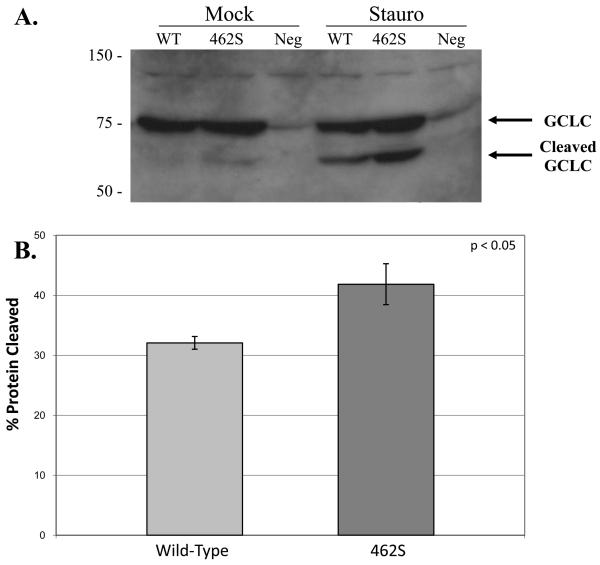
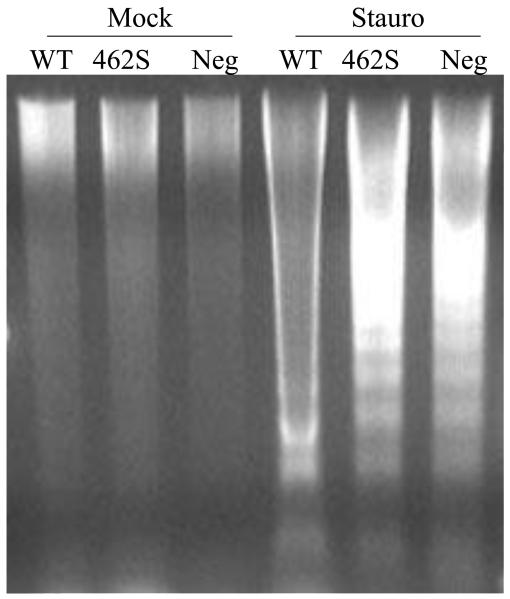
To study whether overexpression of the 462S polymorphism affects eukaryotic cell physiology differently than overexpression of the wild-type 462P isoform, we transfected mammalian cells with expression vectors encoding either the 462P wild-type or 462S variant of the GCLC enzyme. After allowing protein expression for 48 hours, we induced apoptosis by adding staurosporine to the cell media. The cells were then harvested, and cell lysates were collected. Using Western blot analysis, we confirmed equal overexpression of the GCLC enzyme in the transfected cells. In addition, we determined that caspase-mediated cleavage of GCLC was increased in the cells overexpressing the 462S GCLC enzyme as compared to cells overexpressing the wild-type enzyme (Figure 4a). By performing band quantitation, we estimated that 32.1% ± 1.1 of the total GCLC is cleaved when cells overexpressing the wild-type GCLC enzyme are exposed to apoptotic stimuli. In contrast, approximately 41.8% ± 3.4 of GCLC is cleaved in cells that overexpress the 462S GCLC version (Figure 4b). Finally, we subjected the cell lysates to DNA fragmentation analysis and demonstrated that DNA fragmentation occurs more robustly in cells overexpressing the 462S variant as compared to the wild-type (Figure 5). These results suggest that, in the setting of staurosporine-induced apoptosis, cells overexpressing the 462S GCLC variant have increased caspase activity and susceptibility to apoptosis as compared to cells overexpressing the 462P wild-type GCLC enzyme.
Figure 4. Apoptotic MRC5-SV2 cells overexpressing 462S demonstrate increased caspase-mediated cleavage as compared to 462P wild-type.
A) Transfected MRC5-SV2 cells were incubated with staurosporine to induce apoptosis. α-GCLC western blot analysis was performed on lysates. Molecular weight in kD is indicated on left. B) Band signals were quantitated and cleaved:total protein ratios were calculated. Graph values indicate mean cleaved:total protein ratios ± SEM over several experiments. P-value was calculated using an unpaired t-test.
Figure 5. Apoptotic MRC5-SV2 cells overexpressing 462S undergo increased DNA fragmentation as compared to wild-type.
Transfected MRC5-SV2 cells were incubated with staurosporine to induce apoptosis. Cell lysates were collected, and DNA was separated on a 1% agarose gel and visualized by UV light following ethidium bromide staining. Similar results were obtained using Hœchst nuclear staining (data not shown).
Population Studies and Allele Frequencies
Since the 462S polymorphism encodes an enzyme which has little or no activity in vitro resulting in decreased glutathione production and significant physiologic differences when overexpressed in eukaryotic cells, we hypothesized that the 462S allele may also have detrimental effects at the level of the organism. Our initial genetic screen of the GCLC gene revealed a heterozygosity rate ranging from 4-10% in 2 populations of individuals of African descent.[6] To better understand the allelic distribution of the 462S polymorphism and study whether the polymorphism exists in Hardy-Weinberg equilibrium, we broadened our screening to include other populations of African descent.
As demonstrated in Table 1, the 462S allele was detected among various populations of African descent. The rate of heterozygosity for the 462S allele was lowest among African-Americans in the Middle Tennessee population at 4.3%, whereas 10.2% of the Ghanan population was heterozygous for the 462S allele. Of note, after genotyping 2805 individuals, we have identified no homozygotes for the 462S polymorphism, and χ2 analysis reveals that the 462S polymorphism is in Hardy-Weinberg disequilibrium (p<0.05). These results indicate a selection bias against homozygosity for the 462S allele and suggest that this genotype may be lethal.
Table 1. Comparative frequency of the 462S polymorphism among different populations of African descent.
The prevalence of the 1384T polymorphism was determined among various populations of African descent. The table demonstrates the number of individuals from each group with a specific genotype. Percent of individuals are indicated in parentheses.
| Population | GCLC Genotype | ||
|---|---|---|---|
| CC | CT | TT | |
| Endemic Kenya | 958 (93.0%) | 72 (7.0%) | 0 (0%) |
| Carolina | 1167 (92.6%) | 93 (7.4%) | 0 (0%) |
| Middle Tennessee† | 110 (95.7%) | 5 (4.3%) | 0 (0%) |
| Ghana† | 149 (89.8%) | 17 (10.2%) | 0 (0%) |
| South Africa | 212 (90.6%) | 22 (9.4%) | 0 (0%) |
| Total | 2596 (92.5%) | 209 (7.5%) | 0 (0%) |
indicates data previously published. [6]
DISCUSSION
Genetic polymorphisms can lead to drastically different phenotypes between individuals despite similar clinical environments. Understanding the role of genetic polymorphisms in key biochemical pathways can lead to both diagnostic and therapeutic clinical utilities. We have previously identified the non-synonymous P462S polymorphism in GCLC, the rate-limiting enzyme in glutathione biosynthesis. We have previously characterized the prevalence of this polymorphism and found that it occurs exclusively in patients of African descent and is found in up to 10% of this population, changing a highly conserved proline residue to a serine residue.[6]
Here, we have established that the P462S polymorphism encodes an enzyme that has markedly decreased activity in vitro when expressed in both a bacterial and mammalian expression system. In addition, we have shown that the decreased production of γ-glutamylcysteine, in turn, results in significantly decreased intracellular glutathione levels. From our preliminary studies, the mechanism of inactivation of the GCLC enzyme by this polymorphism is unknown. Currently, the exact structure of mammalian GCLC has not been elucidated, though previously published computer models indicate that the 462P residue lies remotely from the ligand binding site.[13, 14] However, mutation of a proline to a serine residue would likely result in steric effects that can affect protein conformation. Likewise, the 462P residue may be involved in allosteric regulation of the GCLC enzyme.
We have also demonstrated that mammalian cells that overexpress the 462S GCLC enzyme are physiologically distinct from cells that overexpress the 462P wild-type enzyme, as they are more susceptible to apoptotic stimuli. We acknowledge that our estimates of enzymatic activity are limited by the overexpression systems we have chosen. We plan to further these experiments by identifying fibroblast cell lines that contain the 462S polymorphism. Studies using these cell lines will allow us to study the effect of this polymorphism under more native conditions.
Finally, we have demonstrated that the 462S allele is not in Hardy-Weinberg equilibrium with an apparent selection bias against the homozygous 462S genotype. As our in vitro data has shown that the 462S isoform of the GCLC enzyme has markedly decreased activity, we would expect an individual homozygous for this polymorphism to have a significant phenotype. Indeed, human GCLC deficiency has only been identified in at least 8 patients.[15-19] Hemolytic anemia has been common to each of these patients, and GCLC deficiency may also be associated with aminoaciduria and neurodegeneration.[18] In addition, targeted knockout of the GCLC gene in mice has resulted in an embryonically lethal phenotype, reinforcing the importance of an intact glutathione biosynthesis pathway.[20, 21]
While the homozygous 462S genotype may have a lethal phenotype, the heterozygous phenotype is unknown. Given a heterozygosity rate of up to 10% among some African populations, the heterozygous genotype may not have a readily identifiable phenotype. However, the 462S polymorphism may predispose individuals to clinical disease related to oxidative stress. Indeed, several polymorphisms in the GCLC gene have been associated with increased risk of several diseases, including myocardial infarction, schizophrenia, and diabetes.[22-24]
The high heterozygosity rate of the 462S genotype among Africans as compared to African-Americans is also interesting. In areas where malaria is endemic, the heterozygous genotype could theoretically provide some clinical benefit. Indeed, in the case of sickle-cell trait and glucose 6-phosphate deficiency, increased red cell sensitivity to oxidant stress is thought to play a major role in preventing malarial parasite replication.[25, 26] We speculate that individuals who are heterozygous for the 1384T polymorphism may also have impaired red blood cell response to oxidative stress and be more resistant to malarial infection through enhanced red cell lysis. This could potentially account for the increased presence of this polymorphism in individuals from Ghana which is centered in the regions where malaria is prevalent. Further studies will be needed to determine whether the P462S polymorphism is associated with malaria and other clinical diseases.
In summary, we have demonstrated that an ethnic-specific, non-synonymous polymorphism in GCLC not only affects the production of glutathione intermediates in vitro, but also affects cellular susceptibility to apoptotic stimuli. We propose that individuals who possess this polymorphism may have deficiencies in glutathione production, thereby predisposing such individuals to increased cellular injury in the setting of oxidative stress and leading to increased disease severity as well as worse clinical outcome. This polymorphism and its deleterious effects might lead to the predisposition to or the earlier onset of a number of diseases associated with oxidant injury and antioxidant depletion, including diabetes, neurologic and cardiovascular degenerative diseases.
ACKNOWLEDGMENTS
*This work was supported in part by grants from the National Institutes of Health Grant 5T32HL007256, Clinical Nutrition Research Unit (National Institutes of Diabetes and Digestive and Kidney Diseases), the National Institutes of Health Fogarty International Center Grant R24 TW007988, the National Cancer Institute, and divisional funding from the Division of Pediatric Critical Care Medicine, Vanderbilt Children's Hospital.
Non-Standard Abbreviations
- GCL
Glutamate Cysteine Ligase
- GCLC
Catalytic Subunit of Glutamate Cysteine Ligase
- GCLM
Modifier Subunit of Glutamate Cysteine Ligase
- γ -GC
γ-Glutamylcysteine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The 462S SNP in the GCLC gene reported in this paper can be found in the NCBI dbSNP Entrez database at accession number RS17883718.
REFERENCES
- 1.Chiba T, Takahashi S, Sato N, Ishii S, Kikuchi K. Fas-mediated apoptosis is modulated by intracellular glutathione in human T cells. Eur J Immunol. 1996;26:1164–1169. doi: 10.1002/eji.1830260530. [DOI] [PubMed] [Google Scholar]
- 2.Ghibelli L, Fanelli C, Rotilio G, Lafavia E, Coppola S, Colussi C, Civitareale P, Ciriolo MR. Rescue of cells from apoptosis by inhibition of active GSH extrusion. Faseb J. 1998;12:479–486. doi: 10.1096/fasebj.12.6.479. [DOI] [PubMed] [Google Scholar]
- 3.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 4.Gipp JJ, Bailey HH, Mulcahy RT. Cloning and sequencing of the cDNA for the light subunit of human liver gamma-glutamylcysteine synthetase and relative mRNA levels for heavy and light subunits in human normal tissues. Biochem Biophys Res Commun. 1995;206:584–589. doi: 10.1006/bbrc.1995.1083. [DOI] [PubMed] [Google Scholar]
- 5.Summar ML, Hall L, Christman B, Barr F, Smith H, Kallianpur A, Brown N, Yadav M, Willis A, Eeds A, Cermak E, Summar S, Wilson A, Arvin M, Putnam A, Wills M, Cunningham G. Environmentally determined genetic expression: clinical correlates with molecular variants of carbamyl phosphate synthetase I. Mol Genet Metab. 2004;81(Suppl 1):S12–19. doi: 10.1016/j.ymgme.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Willis AS, Freeman ML, Summar SR, Barr FE, Williams SM, Dawson E, Summar ML. Ethnic diversity in a critical gene responsible for glutathione synthesis. Free Radic Biol Med. 2003;34:72–76. doi: 10.1016/s0891-5849(02)01178-4. [DOI] [PubMed] [Google Scholar]
- 7.Tu Z, Anders MW. Expression and characterization of human glutamate-cysteine ligase. Arch Biochem Biophys. 1998;354:247–254. doi: 10.1006/abbi.1998.0676. [DOI] [PubMed] [Google Scholar]
- 8.Gegg ME, Clark JB, Heales SJ. Determination of glutamate-cysteine ligase (gamma-glutamylcysteine synthetase) activity by high-performance liquid chromatography and electrochemical detection. Anal Biochem. 2002;304:26–32. doi: 10.1006/abio.2001.5607. [DOI] [PubMed] [Google Scholar]
- 9.White CC, Viernes H, Krejsa CM, Botta D, Kavanagh TJ. Fluorescence-based microtiter plate assay for glutamate-cysteine ligase activity. Anal Biochem. 2003;318:175–180. doi: 10.1016/s0003-2697(03)00143-x. [DOI] [PubMed] [Google Scholar]
- 10.Borutaite V, Brown GC. Nitric oxide induces apoptosis via hydrogen peroxide, but necrosis via energy and thiol depletion. Free Radic Biol Med. 2003;35:1457–1468. doi: 10.1016/j.freeradbiomed.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Kwon YW, Masutani H, Nakamura H, Ishii Y, Yodoi J. Redox regulation of cell growth and cell death. Biol Chem. 2003;384:991–996. doi: 10.1515/BC.2003.111. [DOI] [PubMed] [Google Scholar]
- 12.Franklin CC, Krejsa CM, Pierce RH, White CC, Fausto N, Kavanagh TJ. Caspase-3-Dependent Cleavage of the Glutamate-L-Cysteine Ligase Catalytic Subunit during Apoptotic Cell Death. Am J Pathol. 2002;160:1887–1894. doi: 10.1016/S0002-9440(10)61135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton D, Wu JH, Alaoui-Jamali M, Batist G. A novel missense mutation in the gamma-glutamylcysteine synthetase catalytic subunit gene causes both decreased enzymatic activity and glutathione production. Blood. 2003;102:725–730. doi: 10.1182/blood-2002-11-3622. [DOI] [PubMed] [Google Scholar]
- 14.Manu Pereira M, Gelbart T, Ristoff E, Crain KC, Bergua JM, Lopez Lafuente A, Kalko SG, Garcia Mateos E, Beutler E, Vives Corrons JL. Chronic non-spherocytic hemolytic anemia associated with severe neurological disease due to gamma-glutamylcysteine synthetase deficiency in a patient of Moroccan origin. Haematologica. 2007;92:e102–105. doi: 10.3324/haematol.11238. [DOI] [PubMed] [Google Scholar]
- 15.Hirono A, Iyori H, Sekine I, Ueyama J, Chiba H, Kanno H, Fujii H, Miwa S. Three cases of hereditary nonspherocytic hemolytic anemia associated with red blood cell glutathione deficiency. Blood. 1996;87:2071–2074. [PubMed] [Google Scholar]
- 16.Konrad PN, Richards F, 2nd, Valentine WN, Paglia DE. -Glutamyl-cysteine synthetase deficiency. A cause of hereditary hemolytic anemia. N Engl J Med. 1972;286:557–561. doi: 10.1056/NEJM197203162861101. [DOI] [PubMed] [Google Scholar]
- 17.Richards F, 2nd, Cooper MR, Pearce LA, Cowan RJ, Spurr CL. Familial spinocerebellar degeneration, hemolytic anemia, and glutathione deficiency. Arch Intern Med. 1974;134:534–537. [PubMed] [Google Scholar]
- 18.Ristoff E, Augustson C, Geissler J, de Rijk T, Carlsson K, Luo JL, Andersson K, Weening RS, van Zwieten R, Larsson A, Roos D. A missense mutation in the heavy subunit of gamma-glutamylcysteine synthetase gene causes hemolytic anemia. Blood. 2000;95:2193–2196. [PubMed] [Google Scholar]
- 19.Ristoff E, Larsson A. Patients with genetic defects in the gamma-glutamyl cycle. Chem Biol Interact. 1998;111-112:113–121. doi: 10.1016/s0009-2797(97)00155-5. [DOI] [PubMed] [Google Scholar]
- 20.Dalton TP, Dieter MZ, Yang Y, Shertzer HG, Nebert DW. Knockout of the mouse glutamate cysteine ligase catalytic subunit (Gclc) gene: embryonic lethal when homozygous, and proposed model for moderate glutathione deficiency when heterozygous. Biochem Biophys Res Commun. 2000;279:324–329. doi: 10.1006/bbrc.2000.3930. [DOI] [PubMed] [Google Scholar]
- 21.Shi ZZ, Osei-Frimpong J, Kala G, Kala SV, Barrios RJ, Habib GM, Lukin DJ, Danney CM, Matzuk MM, Lieberman MW. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc Natl Acad Sci U S A. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bekris LM, Shephard C, Janer M, Graham J, McNeney B, Shin J, Zarghami M, Griffith W, Farin F, Kavanagh TJ, Lernmark A. Glutamate cysteine ligase catalytic subunit promoter polymorphisms and associations with type 1 diabetes age-at-onset and GAD65 autoantibody levels. Exp Clin Endocrinol Diabetes. 2007;115:221–228. doi: 10.1055/s-2007-970574. [DOI] [PubMed] [Google Scholar]
- 23.Gysin R, Kraftsik R, Sandell J, Bovet P, Chappuis C, Conus P, Deppen P, Preisig M, Ruiz V, Steullet P, Tosic M, Werge T, Cuenod M, Do KQ. Impaired glutathione synthesis in schizophrenia: convergent genetic and functional evidence. Proc Natl Acad Sci U S A. 2007;104:16621–16626. doi: 10.1073/pnas.0706778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura S, Kugiyama K, Sugiyama S, Miyamoto S, Koide S, Fukushima H, Honda O, Yoshimura M, Ogawa H. Polymorphism in the 5′-flanking region of human glutamatecysteine ligase modifier subunit gene is associated with myocardial infarction. Circulation. 2002;105:2968–2973. doi: 10.1161/01.cir.0000019739.66514.1e. [DOI] [PubMed] [Google Scholar]
- 25.Anastasi J. Hemoglobin S-mediated membrane oxidant injury: protection from malaria and pathology in sickle cell disease. Med Hypotheses. 1984;14:311–320. doi: 10.1016/0306-9877(87)90135-6. [DOI] [PubMed] [Google Scholar]
- 26.Ruwende C, Hill A. Glucose-6-phosphate dehydrogenase deficiency and malaria. J Mol Med. 1998;76:581–588. doi: 10.1007/s001090050253. [DOI] [PubMed] [Google Scholar]