Summary
MicroRNAs (miRNAs) are key regulators of many important biological processes from insulin secretion and fat metabolism to cellular proliferation and differentiation. Given the critical role these small regulatory RNAs play in biology, it is not surprising that the alteration of miRNA expression patterns can have pathogenic consequences. The association between miRNA dysregulation and pathogenesis has been most widely studied in tumorigenesis and a large number of miRNAs have been identified whose expression levels are changed in various tumor types. Although the role that miRNAs play in the development of metastasis is more poorly defined, recent studies have begun to identify miRNAs that can regulate key steps in the metastatic cascade. This review will focus on two emerging stories, the regulation of the epithelial-to-mesenchymal transition (EMT) by members of the miR-200 family and the pleiotropic nature of the metastasis suppressor miR-31.
Introduction
Metastatic development from a primary tumor to a metastatic lesion occurs through a multistep process – local invasion, intravasation, survival in the circulatory system, extravasation and colonization of a secondary anatomical site (1). Each step in this process represents a physiological barrier that must be overcome by the tumor cell for successful metastasis. Malignant cells overcome these barriers through the accumulation of genetic and epigenetic changes, including changes in miRNA expression profiles. MicroRNAs are a class of highly conserved, small RNA molecules that regulate key biological processes, including differentiation and development, cellular proliferation, and metabolism (2). These ~22-nt regulatory RNAs are expressed predominantly by RNA polymerase II (contain a 5′ 7-methyl guanosine cap and a 3′ poly adenosine tail) as elongated hairpin structures which are processed into the mature miRNA through two sequential cleavage steps mediated by RNase III-type endoribonucleases(3). The long primary miRNA (pri-miRNA) transcript is cleaved by Drosha in conjunction with the double stranded RNA binding protein DGCR8 (DiGeorge syndrome critical region 8), yielding an ~70-nt precursor miRNA (pre-miRNA) hairpin that is exported to the cytoplasm by the nuclear transport receptor exportin-5 and the co-factor RanGTP. Once in the cytoplasm, the pre-miRNA is cleaved by the RNase III enzyme Dicer into the transiently double stranded miRNA:miRNA* molecule. The mature single-stranded miRNA is loaded into the Argonaute protein of the RNA-induced silencing complex (RISC) to direct the postranscriptional repression of target mRNAs that bear complementary sites. Although miRNAs can direct mRNA cleavage at sites with extensive complementarity, most miRNAs direct translational repression, mRNA destabilization, or a combination of these two mechanisms (3). The small size of the miRNA and the ability to target mRNAs that have limited complementarity allows a single miRNA to potentially silence the expression of large numbers, even hundreds, of target genes.
MiRNAs are intimately linked with the regulation of development and differentiation. Therefore, it was inevitable that the dysregulation of microRNA expression would be associated with oncogenic transformation and miRNAs that act as tumor suppressors (e. g. miR-15a and miR-16-1) and oncogenes (e. g. miR-155 and miR-21) have been identified in many types of tumors (4). In addition, miRNAs have been found that regulate specific steps in the metastatic pathway by either promoting (for example, miR-10b, -373 and -520c) or suppressing (for example, miR-335, - 206, and -146a/b) metastasis (5). In many cases, these miRNAs are specific for metastatic development and are not involved in tumorigenesis prompting Hurst and colleagues to coin the term “metastamir” to describe this set of miRNAs (5). This review will focus on the role that miRNAs play in the regulation of discrete stages in the metastatic cascade as highlighted by recent studies on the miR-200 family of miRNAs and miR-31.
EMT, E-cadherin and metastasis
Epithelial-to-mesenchymal transitions (EMTs) describe the biological process that facilitates the conversion of epithelial cells into cells that have a more mesenchymal phenotype, including the loss of cell-cell and cell-substratum contacts, loss of cell polarity, and the acquisition of migratory and invasive properties (6). EMTs play a crucial role in development processes (for example, gastrulation and neural crest formation) and wound healing. The dysregulation of this program can have pathogenic consequences. There is increasing evidence that support a role for EMTs in the acquisition of migratory and invasive properties that promote the dissemination of tumor cells and metastasis. A great deal of attention has focused on the role that E-cadherin plays in the maintenance of the epithelial phenotype and EMT. E-cadherin is a member of the calcium-dependent adhesion molecule (Cadherin) family of transmembrane glycoproteins and is critical for the differentiation and maintenance of epithelial characteristics by facilitating the formation of tight junction between cells, promoting interactions with the substratum, and establishing cell polarity (7). E-cadherin expression is regulated by the actions of several transcription factors, including Snail1, Snail2 (Slug), Zeb1, Zeb2 and Twist, that can directly or indirectly repress E-cadherin expression and promote EMT (6, 7). The over-expression of many of these regulators of EMT have been shown to correlate with disease relapse and decreased survival in patients with breast, colorectal and ovarian carcinomas, suggesting that the induction of EMT leads to more aggressive tumors and poorer clinical outcomes.
Recent studies have shown that the members of the miR-200 family are essential regulators of differentiation and the epithelial character of cells in a wide array of cell types and tissues including adipocytes, pancreatic cells, hepatocytes and olfactory neurons (8–11). The highly homologous members of the miR-200 family (miR-141, miR-429, miR-200a, miR-200b and miR-200c) can be divided into two functional groups based on their seed sequences, nucleotides 2 to 7 of the miRNA, which play an important role in target recognition. The 2 groups differ by a single seed nucleotide with miR-200b, miR-429 and miR-200c sharing the 5′-AAUACU-3′ seed sequence, while miR-200a and miR-141 have the 5′-AACACU-3′ seed. In addition, these miRNAs are encoded in two gene clusters found in distinct genetic loci - miR-200c and miR-141 on chromosome 6 in mice and 1 in humans and miR-200b, miR-200a and miR-429 on chromosome 4 of mice and 12 in humans. MiR-200 regulates the epithelial character of cells through the targeted silencing of the zinc finger E box-binding homeobox (Zeb) family of transcriptional factors, Zeb1 and Zeb2. Zeb1 and Zeb2 are master regulators of the mesenchymal phenotype and repress the transcription of genes containing E-box elements in their promoters, including E-cadherin (8–10). In addition to the miR-200 family, miR-205 has been shown to target Zeb1 and Zeb2 through their 3′ UTRs [Gregory, PS yet al. 2008 Nat Cell Biol 10:593–601]. The EMT hypothesis of cancer metastasis would suggest that miR-200 expression would lead to a decrease in the metastatic potential of cells through the targeted silencing of Zeb protein expression resulting in increased E-cadherin expression and the adoption of epithelial characteristics. This has been shown to be the case in several types of cancer cells, whereby the expression of miR-200 is inversely correlated with the invasive and metastatic nature of the tumor. Hanahan and colleagues (2009) found that expression of the miR-200 family was down regulated in metastases and met-like primary tumors in RIP-Tag2 (RT2) transgenic mice that develop neuroendocrine tumors (PNETs) as a consequence of the overexpression of SV40 T antigen in pancreatic b cells (12). Similarly, Gibbons et al (2009) found that the enforced expression of the miR-200b cluster abrogated the metastatic potential of metastasis-prone lung adenocarcinomas in mice that transgenically express constitutively active k-ras and p53 (13). In addition to the miR-200 family, several other miRNAs have been shown to alter E-cadherin expression levels impacting cellular morpohology and malignancy. Similar to the miR-200 family, miR-101 has been shown to be a negative regulator of the polycomb group protein EZH2 (Enhancer of Zeste homolog 2) [Varambally, S et al., 2008. Science 322:1695–9]. EZH2 acts as repressor of E-cadherin expression (among other proteins) by trimethylating histone H3 (lysine 27) at the E-cadherin promoter. Not only is E-cadherin indirectly regulated by miRNA express, but it also serves as a direct target of miR-9. Ma and colleagues have shown that the overexpression of miR-9 inhibits E-cadherin expression promoting cell motility and invasiveness and the formation of pulmonary micrometastases by non-metastatic breast carcinoma cells[Ma, L et al (Weinberg) 2010. Nat Cell Biol. 12:247–56]. The contribution of these different E-cadherin regulatory pathways to tumorogenesis and metastasis remains to be determined. Although these studies and others support the hypothesis that the EMT is an important feature of metastasis and that miRNAs play a key role in regulating this transition, other studies have found the metastatic capacity of a tumor to be governed by processes other than EMT.
miR-200 as a promoter and suppressor of metastasis
One of the difficulties in understanding the role that genetic and epigenetic changes play in the metastatic development is the limited number of experimental systems that model the sequential nature of this process. In most cases, paired populations of metastatic and non-metastatic tumor cells are compared without fully understanding the nature of the specific impairment in the non-metastatic cells. Aslakson and Miller (1992) developed a series of isogenic cell lines from a spontaneously arising mammary carcinoma in a BALB/c mouse that have different metastatic potential (14). Although each of these cells are capable of forming primary tumors when injected into the mammary fat pad of BALB/c mice, they differ in their metastatic potential with each cell line unable to complete one or more steps in the metastatic process – invasion and intravasation (67NR), extravasation (168FARN), and colonization of distant sites (4TO7) – except the 4T1 cells which are aggressively metastatic. Based on the miRNA profiles obtained for these cells, the highly metastatic 4T1 cells could be distinguished from the non-metastatic cell lines by the expression of members of the miR-200 family of miRNAs (8). The overexpression of miR-200, either endogenously in 4T1 cells or exogenously (transfection of synthetic miRNA mimics or stable expression from integrated retroviral vectors) in 4TO7 cells, resulted in a suppression of Zeb protein expression (Zeb2), a concomitant increase in E-cadherin expression, and the adoption of an epithelial morphology. However, unlike previous studies, miR-200 expression in these two cell populations enhanced the ability of these mammary carcinoma cells to colonize both the primary (mammary fat pad) and metastatic tissue (lung parenchyma) sites. These results are seemingly contradictory to the EMT hypothesis of cancer metastasis that implies that the induction of epithelial characteristics would inhibit the formation of metastases.
How could the miR-200 family of miRNAs both promote and inhibit metastasis? The answer to this question most likely lies in the cellular context in which miR-200 is expressed. Although the 4TO7 cells are considered non-metastatic by virtue of their inability to form macroscopic metastatic lesions, these cells are able to perform all the steps of the metastatic process up to the formation of micrometastases. Therefore, these cells have already overcome many of the physiological barriers to the development of metastases and the introduction of miR-200 in this context helped the cells to overcome the final hurdle, colonization of the lung parenchyma. On the other hand, the expression of the miR-200 family members in the neuroendocrine tumors of the RP2 mice and the lung adenocarcinomas in the k-ras and p53 transgenic mice enforces the epithelial character of the primary tumors providing a barrier to tumor cell invasion, a necessary step in the initiation of metastasis. The identification of the miR-200 family of miRNAs as promoters of metastasis do less to diminish the role of EMTs in cancer metastases than they do in pointing for the need of re-establishing epithelial characteristics by inducing a mesenchymal-to-epithelial transition (MET) for successful colonization and formation of macroscopic secondary tumors. During developmental processes, EMTs are often reversible and are accompanied by METs permitting migrating cells to reestablish their epithelial character, such as during the development of the kidney epithelium from the metanephric mesenchyme (6). These results may help to reconcile the results from the histological examination of macroscopic metastases where the morphology and E-cadherin expression levels of the metastatic lesion closely resemble those of the primary tumor from which they were derived. This is not the only example of miRNAs playing seemingly contradictory roles in a disease process. One prominent example involves miR-221 and miR-222. In erythroblastic leukaemia, miR-221 and -222 acts as tumor suppressors that inhibit proliferation by silencing KIT expression. On the other hand, in chronic lymphocytic leukaemia (CLL), thyroid carcinomas and hepatocellular carcinomas, miR-221 and -222 induced cellular proliferation and inhibit apoptosis by silencing p27, p57, PTEN and TIMP3 (4). Unlike miR-221 and -222 that mediate their differential effects on tumorigenesis by targeting the silencing of different sets of genes, at first glance, the members of the miR-200 family appear to both inhibit invasion and promote colonization via the silencing of the Zeb1 and 2. However, the full complement of miR-200 target genes is not known so that traits other than morphology may also be playing a role in determining the functional consequences of miR-200 overexpression.
The question then becomes, why doesn’t the miR-200-mediated enforced expression of E-cadherin inhibit local invasion and intravasation of the 4TO7 cells? This is particularly relevant in light of recent findings that miR-200a expression inhibited the in vitro migration and invasion of nasopharyngeal carcinoma (NPC) cells (15). In addition, miR-200-mediated suppression of WAVE3 (WASF3: Wiskott-Aldrich syndrome protein family member 3) inhibited the invasive capacity of LNCaP prostate adenocarcinomacells and MDA-MB-231 breast cancer cells (16). Two possible explanations could account for the metastatic capacity of the 4T1 cells and the 4TO7 cells that overexpress members of the miR-200 family: 1) a number of cells at the periphery of the tumor undergo a transient EMT that allows them to escape the primary tumor, invade the surrounding stroma and disseminate or 2) the tumor cells can perform all the steps in the metastatic cascade despite their high E-cadherin levels and their epithelial morphology. There is evidence for localized EMTs at the stromal interface of colon, papillary thyroid, cervical and some breast carcinomas associated with a loss of E-cadherin expression and epithelial characteristics. Gibbons et al (2009) have found that miR-200 levels were decreased in cells treated with TGFβ suggesting a mechanism for the downregulation of miR-200 in response to extracellular signals. On the other hand, Wicki and colleagues (2006) showed that tumor cell invasion could occur in an EMT-independent, podoplanin-dependent manner in squamous cell carcinomas (17). In addition, there has been the suggestion that cancer cells can adopt an amoeboid phenotype that would allow them to pass through the rigid extracellular matrix (ECM) during invasion in protease-independent manner [Sanz-Morena et al., 2008. Cell 135:510–523]. However, in vivo evidence supporting this mechanism has not been as readily found and a need for the membrane bound MT1-matrix metaloproteinase (MMP) seems to be necessary for cancer cells to traverse the ECM [Sabeh et al. 2009. JCB 185:11–19].
This suggests that the collective movement and invasion of tumor cells is not always dependent on the down regulation of E-cadherin expression and the alteration of the epithelial characteristics of tumor cells. Although it is possible that the highly metastatic 4T1 cells transiently lose miR-200 expression and induce an EMT, it seems more likely that tumor cells can overcome the physiological barriers to metastatic development without altering E-cadherin expression. This notion is supported by the fact that the 4T1 cells are highly invasive (assessed by transwell migration through matrigel basement membrane components), migratory (assessed by wound healing assays) in the absence of detectably changes in morphology and extracellular signalling molecules. Although morphologically epithelial in nature, gene expression analysis shows that 4T1 cells have a mixture of epithelial (E-cadherin, cytokeratin-18, EGFR) and mesenchymal (Vimentin and Twist) makers (8). This mix of epithelial and mesenchymal traits may permit the invasive and migratory behavior necessary for early events in the metastatic cascade without the loss of E-cadherin expression. Recent studies have found elevated expression of miR-200 family members in ovarian cancers, particularly in serous and endometroid histotypes, compared to normal ovarian tissue (21). Ovarian cancer patients with high miR-200a expression had an ~50% decrease in median survival compared to those lacking miR-200a expression. Bendoraite and colleagues (2010) postulated that the progression from ovarian surface cells (derived from the mesothelial lining of the peritoneal cavity) to ovarian cancer required a miR-200-mediated mesothelial-to-epithelial transition as part of their pathogenesis (18). Although E-cadherin expression may be an important indicator of metastatic potential, it is certainly not a universal marker.
miR-31 and the multi-step suppression of metastasis
These results suggest that the miR-200 family members can have very different effects on the metastatic potential of tumor cells depending on the cellular context and, potentially, the stage of the metastatic process in which they are expressed. In contrast to the opposing roles that miR-200 plays in metastatic development, Valastyan and coleagues (2009) identified a miRNA, miR-31, which was able to inhibit multiple steps in the metastatic process (19, 20). In a series of elegant experiments, they found that overexpression of miR-31 independently inhibited the invasive capacity of MDA-MB-231 breast cancer cells, the extravasation into or survival in the lung parenchyma, and metastatic colonization. The inhibitory effects of miR-31 on metastatic development could be explained by the inhibition of three mRNA targets, integrin-α5 (ITGA5), radixin (RDX) and RhoA. The siRNA-mediated silencing of each of these genes in MDA-MB-231 cancer cells reduced local invasion, motility and anoikis-mediated cell death in vitro and the development of metastases in am orthotopically implanted xenograft mouse model. When analyzed in more detail, the silencing of ITGA5 or RhoA inhibited early post implantation events as measured by a decrease in the silenced cells in the lung 1 day after intravenous injection of the cells. In contrast, the silencing of RDX had no effect on the persistence of silenced cells in the lungs compared to untreated MDA-MB-231 cells. However, when the size of metastatic lung tumors were measured 3 months after intravenous inoculation, the ITGA5 or RDX silenced cells formed only small micrometastases compared to the RhoA silenced cells which generated macroscopic metastatic lesions. To further characterize the impact of miR31 on the inhibition of metastatic development, miR-31 resistant forms of ITGA5, RDX or RhoA, alone or in combination, were transgenically expressed in miR-31 overexpressing MDA-MB-231 cells. The restoration of RDX and RhoA in the miR-31 expressing MDA-MD-231 cells restored the invasive capacity of the cells while the reintroduction of ITGA5 had no effect on local invasion. In contrast, ITGA5 and RhoA expression was able to rescue the early post-intravasation events (extravasation and maintenance in the lung parenchyma), while RDX expression had no effect on these events. Finally, ITGA5 and RDX but not RhoA re-expression supported metastatic colonization. The re-expression of all three proteins together completely restored the metastatic potential of the miR-31 expressing MDA-MB-231 tumors. These studies indicate that the silencing of different genes by the same miRNA can alter discrete events in the development of metastases.
Conclusions and Perspectives
It is clear that the dysregulation of miRNA expression can have a profound impact on metastatic development. The functional consequence of the miRNA dysregulation lies in the mRNA targets whose expression is altered. This is illustrated by the dysregulation of miR-31 expression in breast cancer cells that influenced different steps of the metastatic cascade through the silencing of distinct targets. The miRNA-mediated regulation of metastatic development is not always the same in every type of tumor and the cellular context can greatly impact the phenotypic consequences resulting from alteration in miRNA expression levels. For example, the over-expression of members of the miR-200 family of miRNAs could act to inhibit metastasis by confining the tumor to the primary location or promote metastasis by facilitating colonization of a second anatomical site. Not surprisingly, miRNAs are being identified that participate at various stages of the metastatic cascade to help tumor cells overcome the physiological barriers to metastases formation. The impact that changes in miRNA expression have on the disease pathology outcome will depend on the cellular context (genetic, epigenetic, environmental etc).
The identification of disease-associated changesin miRNA expression has suggested that the manipulation of miRNA expression levels could serve as potential therapeutic agents for the inhibition of metastatic development. The biochemical similarity between miRNAs and siRNAs means that the same delivery reagents developed for use with siRNAs could be applied for the delivery of miRNAs or miRNA inhibitors. In fact, the first phase 1 clinic trial has begun using a miRNA antagonizer (a non-cleavable RNA substrate complementary to the miRNA) to miR-122 resulting in the inhibition of HCV replication. Unlike siRNAs that are designed to silence expression of a single transcript (although off-target effects may increase the number of genes), miRNAs could potentially target a large number of transcripts potentially complicating their therapeutic application. Although the therapeutic alteration of miRNA expression levels represents a tantalizing prospect for the treatment of tumorigenesis and metastatic development, the ability of miRNAs to silence the expression of a multitude of genes, sometimes with opposing functional consequences, suggests that caution should be exercised in the application of miRNA-based therapeutics. It is essential to understand the role that any miRNA is playing in a pathogenic process to be able to determine their feasibility as therapeutic targets.
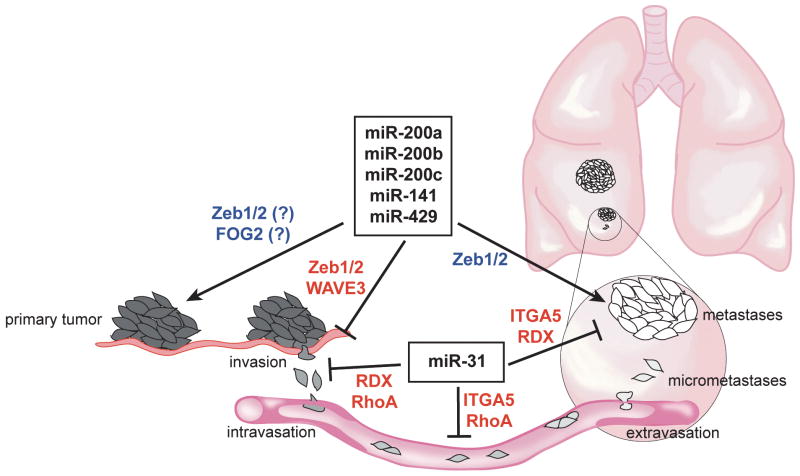
Figure 1.
miRNA-mediated regulation of metastasis. The dysregulation of miRNA expression levels can influence metastatic development at various stages of the metastatic cascade. Members of the miR-200 famly of miRNAs have been shown to promote primary tumor formation and either inhibiting the development of metastases by impairing the invasive or migratory properties of tumor cells or promote metastasis by facilitating the colonization of secondary tissues and organs. On the other hand, miR-31 can inhibit distinct steps in the development of metastasis through the silencing of different sets of genes. The target genes that promote tumorigenesis or metastatic formation are listed in blue and those inhibiting these processes are listed in red.
Acknowledgments
Unfortunately due to space constraints it is impossible to reference every paper in this review, and we apologize for any oversights.
References
- 1.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–8. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Wells A, Yates C, Shepard CR. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin Exp Metastasis. 2008;25:621–8. doi: 10.1007/s10585-008-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dykxhoorn DM, Wu Y, Xie H, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurteau GJ, Carlson JA, Roos E, Brock GJ. Stable expression of miR-200c alone is sufficient to regulate TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle. 2009;8:2064–9. doi: 10.4161/cc.8.13.8883. [DOI] [PubMed] [Google Scholar]
- 10.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 11.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson P, Lu J, Zhang H, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–405. [PubMed] [Google Scholar]
- 15.Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 391:535–41. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 16.Sossey-Alaoui K, Bialkowska K, Plow EF. The miR200 family of microRNAs regulates WAVE3-dependent cancer cell invasion. J Biol Chem. 2009;284:33019–29. doi: 10.1074/jbc.M109.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9:261–72. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Bendoraite A, Knouf EC, Garg KS, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 116:117–25. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev. 2009;23:2592–7. doi: 10.1101/gad.1832709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]