Abstract
This communication describes the synthesis and in vitro biological evaluation of novel generation 5 PAMAM dendrimers conjugated with riboflavin as a targeting ligand. Cell-based experiments demonstrated that a dendrimer conjugated with riboflavin is able to undergo cellular binding and uptake in KB cells, and when the dendrimer is also conjugated with methotrexate, the riboflavin dendrimer conjugate can potently inhibit cell growth.
Nanotechnology offers a wide range of unique opportunities for the targeted delivery of various molecules to cancer cells through nanoparticles (NP) coupled with ligands for cancer cell membranemolecules. This approach can deliver anticancer drugs, therapeutic genes, and imaging molecules1-3 and aims to enhance the therapeutic index of a drug or improve the ability of an imaging agent to identify cancer. Recent studies have demonstrated the utility of this approach with anticancer drugs including methotrexate4, doxorubicin5, paclitaxel6, and cisplatin7 as well as imaging agents for detection based on fluorescence2, 8, magnetic resonance imaging (MRI)9, and radioactive isotopes10. The therapeutic or imaging molecules are carried either covalently attached to the particle or encapsulated within a NP. However, the number of ligand molecules that have been demonstrated to actively mediate cell-specific targeting with this approach are limited. Targeting ligands which have been employed for cancer include small molecules like folic acid (vitamin B9), a nanomolar affinity ligand to folic acid receptor (FAR)9, 11, RGD-containing peptides as the class of ligands binding integrin receptors12, 13, Glu-based small peptides and peptido mimetics that bind to prostate specific membrane antigen10, as well as a few protein-based ligands such as antibodies14, transferrin15, and endogenous growth hormone like EGF14, 16. The objective of this study is to evaluate riboflavin (RF) as a new small molecule-based targeting ligand by demonstrating its applicability in targeted uptake of an anticancer drug.
Riboflavin (vitamin B2) belongs to a family of B vitamins and is an essential molecule necessary for the biosynthesis of flavin-based redox cofactors like FMN and FAD. It is taken up with high efficiency by riboflavin binding proteins (RBP; MW = 27.5 ~ 40 kDa, Kd ≈ 1 nM) that are present as both soluble or membrane-bound forms,17, 18 referred to as either RF carrier protein19, 20 or RF transporter21. Prior studies based on RF conjugates composed of RF linked to rhodamin dye22, or albumin protein23 suggest that such RF conjugates are taken up by cells after interacting with a RF receptor.
We have been interested in exploring RF as a ligand for cancer targeting for a number of reasons. As a small molecule, RF has several functional groups that are amenable for covalent modifications. A crystal structure of RF in complex with chicken RBP18 shows that in a bound state the ligand xylene domain of the isoalloxazine unit is stacked between hydrophobic planes comprising a cleft of RF binding protein, while its D-ribose domain is exposed to the aqueous medium and can be utilized as a tethering site.22 This structural knowledge allows the rational design of a linker for ligand attachment. Enhancing the potential utility of RF as a ligand is a recent report that RF carrier proteins are over-expressed in certain human cell lines from breast19, and prostate cancer20, potentially making the proteins a tumor biomarker.
RF-targeted NP conjugates designed for this study are based on a generation 5 (G5) polyamidoamine (PAMAM) dendrimer (diameter 5.4 nm).24 Having a large number of primary amines (theoretically 128) present on its surface provides an approach to a variety of ligand conjugation methods that result in NP conjugates that have non-toxicity, non-immunogenicity, and display extended duration of circulation.25-28 We approached the design of RF-G5 dendrimer conjugates to provide two key capabilities required for targeted delivery: 1) the ligand conjugate maintains the ability to bind to RF receptors on the cell surface and undergo cellular uptake, 2) the conjugate retains biocompatibility and solubility when used for cellular delivery of therapeutic or imaging agents.
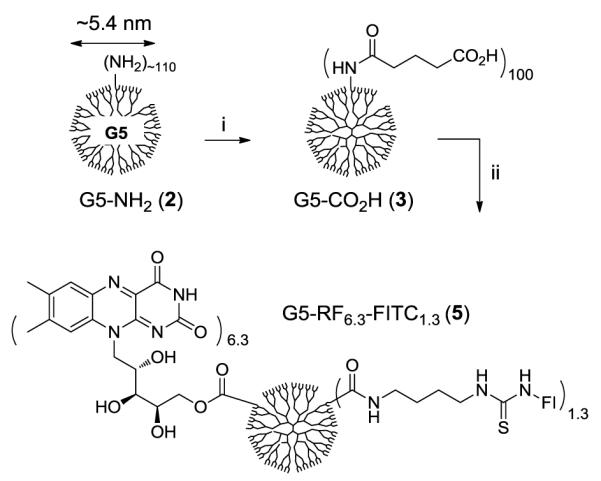
First, we designed a fluorescently labeled PAMAM G5-RF conjugate 5 (Scheme 1) that presents a mean of six copies of RF and approximately one fluorescein molecule on the surface of each dendrimer. This conjugate was prepared by single coupling reaction in which both RF and fluoresceinisothiocyanate (FITC)-diaminobutane 429 were covalently linked by an EDC(1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) to G5 dendrimer where the surface amines have been converted to glutaric acid 3 (Scheme 1: MALDI-TOF mass spectrometry: m/z = 40200 gmol−1; gel permeation chromatography: Mw = 42,729, polydispersity index = 1.046)30. This RF attachment is based on the formation of an ester bond formed between a carboxylic acid present on the dendrimer surface and one of the hydroxyl groups from D-ribityl unit of RF. While the attachment of RF at its ribose unit is not characterized, previous conjugation studies of RF noted that reactions at D-ribose unit22, 23, 31 occur regioselectively at a primary hydroxyl group perhaps because it is more sterically favored than secondary alcohols located adjacent to RF isoalloxazine unit. Analysis of both UV/vis spectral data (FITC) and MALDI mass spectrometric data (5, MW = 43,200 gmol−1) led to an estimate of mean numbers of RF (6.3) and FITC (1.3) molecules present on the dendrimer conjugate 5.32
Scheme 1.
Synthesis of PAMAM G5-RF-FITC nanoconjugate 5. Reagents and conditions: i) glutaric anhydride, Et3N, MeOH, rt, 3 d; ii) riboflavin, FITC-NH(CH2)4NH2 (4), EDC, DMAP, DMF, rt.
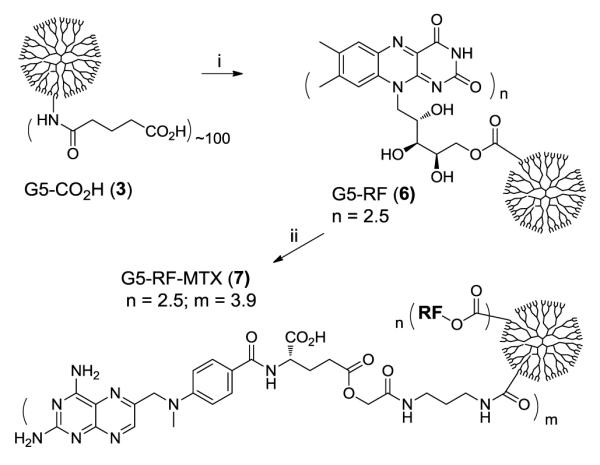
Second, we designed RF-targeted PAMAM G5 conjugates 7 (Scheme 2) that contain methotrexate (MTX), a cytotoxic drug carried as the payload by the conjugate. MTX is a potent inhibitor of dihydrofolate reductase (DHFR; Ki of 4.8 pM) that inhibits cell growth.33 In order to attach MTX to an acid-terminated dendrimer 3, we synthesized a MTX-linker 1 that contains a primary amine located at the terminus of a 7-atom linker tethered to γ-glutamic acid.34 We preferred to use this MTX γ-isomer rather than an α-isomer or a mixture of α-/γ-isomers because MTX derivatives modified at γ-acid maintain cytotoxic activity compared to α derivatives.35, 36 Synthesis of the conjugate 7 from RF and 1 was performed by an EDC-based method involving two coupling reactions performed in a sequence (step i, ii). This sequential approach allows preparation of both conjugates 6 and 7, and allows the sequential characterization of each conjugate by UV/vis and MALDI mass spectral analysis (m/z, gmol−1: 6: 41,100; 7: 43,300).32 However, the coupling reactions could be performed in one step as well that also led to the synthesis of a similar conjugate (G5-RFn-MTXm: n = 4, m = 10).32 In both of the methods by controlling the molar ratio between RF and 1 to G5-CO2H 3, we were able to prepare several different types of RF-conjugated dendrimers with varied numbers of RF and MTX per dendrimer.
Scheme 2.
Synthesis of PAMAM G5-RFn-MTXm nanoconjugate 7. Reagents and conditions: i) EDC, 4-dimethylaminopyridine (DMAP), riboflavin, DMF, rt, 24 h; ii) 1, EDC, DMAP, DMF, rt, 24 h.
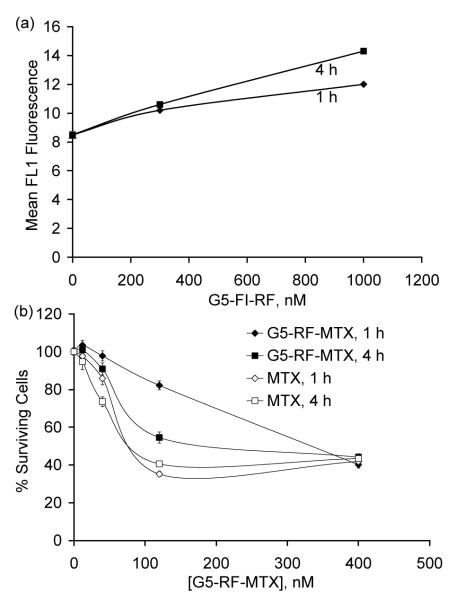
We used the fluorescence-labeled G5-RF6.3-FI1.3 5 (Scheme 1) with flow cytometry to analyze the cellular binding and uptake (taken together, the specific cellular association) in KB cells, a sub-line of the cervical carcinoma HeLa cells, that has shown uptake of RF and other RF conjugates22, 23 (Figure 2a). This analysis showed that the RF conjugate 5 bound to the cells in a dose- and time-dependent manner, and that longer incubation times and higher conjugate concentrations led to greater cellular association. In separate studies (not shown), we compared the cellular uptake of 5 in different human cancer cell lines. The human cancer cell lines KB, IGROV-1 (ovarian), SCC15 (head and neck) and MCF 7 (breast) were incubated in riboflavinfree medium for a week and the cellular association of 5 (1 μM) was tested following a 2 h incubation; the background-subtracted mean fluorescence was 12.3, 21.5, 37.6 and 4.9, respectively in these cell lines. In a following study to determine whether RF-mediated cellular uptake can be applied for targeted drug delivery, we evaluated RF/MTX conjugate 7 (G5-RF2.5-MTX3.9) to determine any effect on KB cell growth by using an XTT assay (Figure 2b).8 The MTX conjugate 7 potently inhibited the cell growth at low nanomolar concentrations in a time- and dose-dependent manner, with a maximal inhibition showing at 100 nM of the conjugate (equivalent to 400 nM MTX). Under similar conditions, equivalent doses of free MTX showed higher cytotoxicity. The apparent IC50 values estimated from the dose response curve (4 h incubation) gave values of 72 nM and 48 nM for the 7 and free MTX, respectively.
Figure 2.
(a) Dose-dependent binding and uptake of G5-RF6.3-FI1.3 5 in KB cells. The cells were incubated with different concentrations (300, and 1000 nM per conjugate basis) of RF dendrimer 5 for 1 or 4 h in HBSS buffer. The cells were then rinsed and resuspended in PBS buffer, and fluorescence intensity was measured in a flow cytometer; (b) Dose-dependent cytotoxicity of G5-RF2.5-MTX3.9 7 and free MTX in KB cells. Cells were treated with 7 for 1 or 4 h at four different concentrations (per MTX basis) in a RF-free HBSS-BSA medium, washed and followed by incubation in a conjugate-free growth medium for 3d. The inhibition of cell growth induced was quantified by an XTT assay, which is based on the conversion of XTT to formazan by the active mitochondria of live cells.
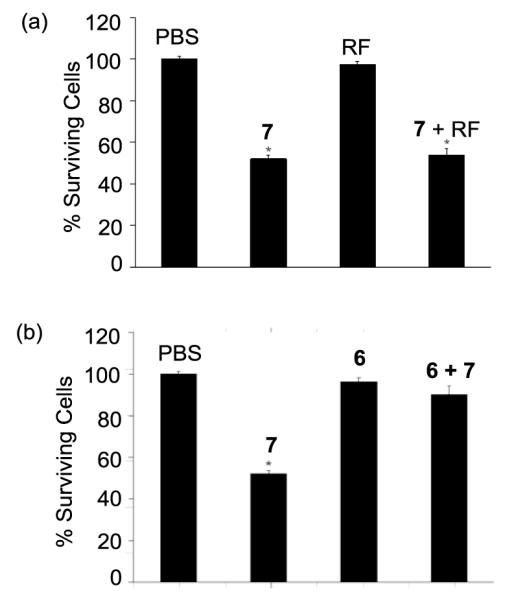
In a study to understand the possible mode(s) of action associated with the cytotoxicity of G5-RF2.5-MTX3.9 7, we performed a series of control experiments in KB cells under a 4 h incubation condition with PBS only, RF (30 μM, Figure 3a), or G5-RF2.5 6 (30 μM, Figure 3b). This showed that either RF or 6 (a RF conjugate containing no MTX) did not show any growth inhibitory effect in KB cells even at a concentration three orders of magnitude higher than 30 nM where ~50% inhibition of cell growth was achieved with the MTX conjugate. 7. We evaluated if this cytotoxicity can be competitively inhibited by co-incubation of 7 (30 nM) and either free RF or RF conjugate 6. Free RF caused virtually no apparent effect on cell growth of 7 (Figure 3a, fourth entry) but 6 reversed the cytotoxicity of 7 almost completely (~50% to 90% cell growth; Figure 3b, fourth entry). This result suggested that only the conjugated RF had adequate avidity to inhibit the binding of 7, and is in a close agreement with a previous observation made with 125I-labeled RF-conjugated albumin in KB cells, where the cellular association of radio-labeled RF-albumin conjugate was not inhibited by free RF but only by unlabeled RF-albumin conjugate.23 Therefore, these results suggest that cellular association mediated by RF dendrimer conjugate 6, is due to a multivalent interaction between the RF conjugate and multiple RF receptors on the cell surface with greater avidity than a monovalent interaction.
Figure 3.
Cytotoxicity of G5-RF2.5-MTX3.9 (7, 30 nM) in KB cells tested under various control conditions. (a) Effect on cell growth of PBS (control), G5-RF2.5-MTX3.9 (7, 30 nM), RF (30 μM), and a mixture of 7 (30 nM) and RF (30 μM), (b) Effect on cell growth of PBS, G5-RF2.5-MTX3.9 (7, 30 nM), G5-RF2.5 (6, 30 μM), and a mixture of 6 (30 μM) and 7 (30 nM). *The p value for each of the data set is <0.005 compared to PBS.
We also examined if the MTX moiety of the conjugate 7 could play a role in cellular uptake and consequently cytotoxicity in this assay condition. Our studies used a G5-MTX7.5 conjugate as a control molecule that had MTX alone on the dendrimers. It failed to show any significant cytotoxicity in KB cells up to 30 nM in FA-free medium, otherwise under identical conditions of the assay (data not shown). This suggests that the cytotoxicity induced by 7 is a consequence of the internalization of the MTX conjugate through the RF receptor. Overall the results obtained from fluorescence binding and cytotoxicity studies suggest that G5-RF2.5-MTX3.9 7 undergoes cellular uptake mediated by RF receptors on the cell surface, and subsequently causes cell growth inhibition by blocking the catalytic activity of DHFR enzyme in cytosol. These are the first studies examining RF-dendrimer conjugates, and therefore this activity is particularly novel.
In summary, we have synthesized a class of RF-targeted NP conjugates based on a generation 5 PAMAM dendrimer using sequential and one-step synthetic methods, and studied the biological activities of these conjugates in vitro in KB cells. Our studies suggest that a monofunctional dendrimer conjugate presenting RF is taken into the cells through a RF receptor mediated mechanism, and can deliver MTX to induce specific and potent cytotoxicity. This opens the potential for RF-conjugated dendrimers to be employed to target malignant cells over-expressing the RF receptor19, 20, to deliver imaging agents and various cancer therapeutics to these cells.
Supplementary Material
Figure 1.
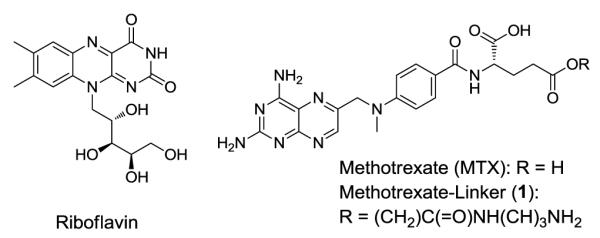
Structure of riboflavin (RF), methotrexate (MTX), and its linker derivative 1.
Acknowledgments
This work was supported by the National Cancer Institute, National Institutes of Health under award 1 R01 CA119409 (JRB). We greatly appreciate Dr. Pascale Leroueil for careful proofreading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Information Supplementary data (synthetic methods, and characterization of conjugates 5-7) associated with this article can be found in the online version.
References and notes
- 1.Brigger I, Dubernet C, Couvreur P. Adv. Drug Deli. Rev. 2002;54:631. doi: 10.1016/s0169-409x(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 2.Kukowska-Latallo JF, Candido KA, Cao Z, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR., Jr. Cancer Res. 2005;65:5317. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 3.Cheng H, Zhu J-L, Zeng X, Jing Y, Zhang X-Z, Zhuo R-X. Bioconjugate Chem. 2009;20:481. doi: 10.1021/bc8004057. [DOI] [PubMed] [Google Scholar]
- 4.Majoros IJ, Thomas TP, Mehta CB, Baker JR. J. Med. Chem. 2005;48:5892. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- 5.Luo Y, Bernshaw NJ, Lu Z-R, Kopecek J, Prestwich GD. Pharm. Res. 2002;19:396. doi: 10.1023/a:1015170907274. [DOI] [PubMed] [Google Scholar]
- 6.Majoros IJ, Myc A, Thomas T, Mehta CB, Baker JR. Biomacromolecules. 2006;7:572. doi: 10.1021/bm0506142. [DOI] [PubMed] [Google Scholar]
- 7.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Proc. Natl. Acad. Sci. 2008;105:17356. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas TP, Majoros IJ, Kotlyar A, Kukowska-Latallo JF, Bielinska A, Myc A, Baker JR. J. Med. Chem. 2005;48:3729. doi: 10.1021/jm040187v. [DOI] [PubMed] [Google Scholar]
- 9.Hilgenbrink Andrew R., Low Philip S. J. Pharm. Sci. 2005;94:2135. doi: 10.1002/jps.20457. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Foss CA, Byun Y, Nimmagadda S, Pullambhatla M, Fox JJ, Castanares M, Lupold SE, Babich JW, Mease RC, Pomper MG. J. Med. Chem. 2008;51:7933. doi: 10.1021/jm801055h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RJ, Low PS. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1995;1233:134. doi: 10.1016/0005-2736(94)00235-h. [DOI] [PubMed] [Google Scholar]
- 12.Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, James R, Baker J. Chem. Commun. 2005:5739. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]
- 13.Temming K, Lacombe M, Schaapveld RQJ, Orfi L, Kéri G, Poelstra K, Molema G, Kok Robbert J. ChemMedChem. 2006;1:1200. doi: 10.1002/cmdc.200600201. [DOI] [PubMed] [Google Scholar]
- 14.Shukla R, Thomas TP, Desai AM, Kotlyar A, Park SJ, Baker JR., Jr. Nanotechnol. 2008;19:295102. doi: 10.1088/0957-4484/19/29/295102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian ZM, Li H, Sun H, Ho K. Pharmacol. Rev. 2002;54:561. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 16.Thomas TP, Shukla R, Kotlyar A, Liang B, Ye JY, Norris TB, Baker JR. Biomacromolecules. 2008;9:603. doi: 10.1021/bm701185p. [DOI] [PubMed] [Google Scholar]
- 17.White HB, Merrill AH. Annu. Rev. Nutr. 1988;8:279. doi: 10.1146/annurev.nu.08.070188.001431. [DOI] [PubMed] [Google Scholar]
- 18.Monaco HL. The EMBO Journal. 1997;16:1475. doi: 10.1093/emboj/16.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karande AA, Sridhar L, Gopinath KS, Adiga PR. Int. J. Cancer (Pred. Oncol.) 2001;95:277. doi: 10.1002/1097-0215(20010920)95:5<277::aid-ijc1047>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 20.Johnson T, Ouhtit A, Gaur R, Fernando A, Schwarzenberger P, Su J, Mohamed, Ismail F, El-Sayyad HI, Karande A, Elmageed ZA, Rao P, Raj M. Frontiers in Bioscience. 2009;14:3534. doi: 10.2741/3477. [DOI] [PubMed] [Google Scholar]
- 21.Duurkens RH, Tol MB, Geertsma ER, Permentier HP, Slotboom DJ. J. Biol. Chem. 2007;282:10380. doi: 10.1074/jbc.M608583200. [DOI] [PubMed] [Google Scholar]
- 22.Huang S-N, Phelps MA, Swaan PW. J. Pharmacol. Exp. Ther. 2003;306:681. doi: 10.1124/jpet.103.051581. [DOI] [PubMed] [Google Scholar]
- 23.Holladay SR, Yang Z.-f., Kennedy MD, Leamon CP, Lee RJ, Jayamani M, Mason T, Low PS. Biochimica et Biophysica Acta (BBA) - General Subjects. 1999;1426:195. doi: 10.1016/s0304-4165(98)00147-0. [DOI] [PubMed] [Google Scholar]
- 24.Tomalia DA, Naylor AM, William A, Goddard I. Angew. Chem. Int. Ed. 1990;29:138. [Google Scholar]
- 25.Cloninger MJ. Curr. Opin. Chem. Biol. 2002;6:742. doi: 10.1016/s1367-5931(02)00400-3. [DOI] [PubMed] [Google Scholar]
- 26.Esfand R, Tomalia DA. Drug Discovery Today. 2001;6:427. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 27.Majoros I, Baker J. James, editors. Dendrimer-Based Nanomedicine. Pan Stanford; Hackensack, NJ: 2008. [Google Scholar]
- 28.Medina SH, El-Sayed MEH. Chem. Rev. 2009;109:3141. doi: 10.1021/cr900174j. [DOI] [PubMed] [Google Scholar]
- 29.Gapski GR, Whiteley JM, Rader JI, Cramer PL, Hendersen GB, Neef V, Huennekens FM. J. Med. Chem. 2002;18:526. doi: 10.1021/jm00239a020. [DOI] [PubMed] [Google Scholar]
- 30.Choi SK, Thomas T, Li M, Kotlyar A, Desai A, James R, Baker J. Chem. Commun. 2010:46. doi: 10.1039/b927215c. DOI:10.1039/b927215c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riva S, Chopineau J, Kieboom APG, Klibanov AM. J. Am. Chem. Soc. 2002;110:584. [Google Scholar]
- 32.See supplementary information for full details.
- 33.Rosowsky A, Forsch RA, Wright JE. J. Med. Chem. 2004;47:6958. doi: 10.1021/jm040122s. [DOI] [PubMed] [Google Scholar]
- 34.The MTX-linker 1 was synthesized in a two step process, O-alkylation of MTX with 3-(bromoacetyl)amino-1-N-Boc-propane, and subsequent deprotection of the N-Boc group with TFA. 1H NMR (400 MHz, CD3OD): δ 8.60 (s, 1H), 7.74-7.72 (d, 2H, J = 7.2 Hz), 6.84-6.81 (d, 2H, J = 7.2 Hz), 4.89 (s, 2H), 4.66-4.62 (d, 1H, J = 15.2 Hz), 4.55-4.51 (d, 1H, J = 15.2 Hz), 4.53-4.49 (t, 1H, J = 5.6 Hz), 3.34-3.31 (t, 2H, J = 5.6 Hz), 3.24 (s, 3H), 2.93-2.89 (t, 2H, J = 5.6 Hz), 2.49-2.46 (t, 2H, J = 5.6 Hz), 2.26-2.23 (m, 1H), 2.16-2.12 (m, 1H), 1.88-1.81 (quin, 2H, J = 5.6 Hz). 13C NMR (100 MHz, CD3OD): δ 175.20, 171.76, 169.33, 163.51, 156.37, 152.25, 151.75, 148.93, 145.51, 128.99, 121.98, 120.72, 116.86, 115.76, 113.50, 111.34, 62.62, 55.19, 52.94, 36.76, 35.36, 29.85, 29.80, 27.17, 25.49, 15.19 ppm. MS (ESI): m/z (relative intensity, %) = 569.3 (100) [M+H]+, 1137.5 (3) [2M+H]+. HRMS (ESI) calcd for C25H33N10O6 569.2585, found 569.2573.
- 35.Rosowsky A, Forsch RA, Freisheim JH, Galivan J, Wick M. J. Med. Chem. 1984;27:888. doi: 10.1021/jm00373a014. [DOI] [PubMed] [Google Scholar]
- 36.Nagy A, Szoke B, Schally AV. Proc. Natl. Acad. Sci. 1993;90:6373. doi: 10.1073/pnas.90.13.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.